Abstract
The c-myb gene encodes a transcription factor required for the normal development of T cells in the thymus, and for subsequent peripheral T-cell activation and survival. However, the profile of genes known to be transcriptionally regulated by c-Myb in T cells does not adequately explain the pleiotrophic nature of the effects of c-Myb. We present here a detailed molecular characterization of the regulation of a novel target gene, the histone variant H2A.Z. We show that c-Myb is able to bind to and activate the H2A.Z promoter in T cells both in vitro and in vivo, and present evidence that perturbation of Myb activity during T-cell development results in reduced H2A.Z expression. As H2A.Z is absolutely required for the early stages of mammalian development, and plays essential roles in the regulation of chromatin structure in gene promoters in yeast, its regulation by c-Myb is likely to be of some importance during T-cell development.
Keywords: Myb, H2A.Z, T cells, transcription
Introduction
c-Myb is an essential transcription factor required for normal haematopoiesis. The function of c-Myb is well characterized in T-cell development where it is required at four distinct points during thymocyte maturation.1,2 Immature thymocytes can be broadly classified as double negative (DN) thymocytes, which lack expression of the CD4 and CD8 coreceptors, and double positive (DP) thymocytes, which express both coreceptors.3 c-Myb is required for two distinct transitions in DN thymocytes: for the earliest committed thymocytes to progress beyond the DN1 stage (CD44+ CD25–),1 then for selection and expansion of thymocytes with successful rearrangement of the T-cell receptor β chain (TCR-β) at the DN3 stage (CD44– CD25+).2,4–6 Later, at the double positive (DP) stage, c-Myb is required for cell survival and may contribute to TCR-α gene rearrangement.2,6 c-Myb is further required for differentiation from the late DP precursor to the final CD4 single positive stage, but not for the corresponding DP to CD8 single positive transition.2,6,7 In mature T cells, specific deletion of c-myb or inhibition of c-Myb by expression of a dominant negative transgene reduces the activation-induced proliferative response.4,6
c-Myb activates transcription through the hexameric consensus Myb-binding site (MBS), defined as YAACT/GG. However, molecular studies into the transcriptional targets of c-Myb have yet to explain its profound effects on haematopoietic development. c-Myb has been proposed to regulate transcription of the CD4 and recombination activating gene (RAG-2) genes.8–10 However, while specific deletion of c-Myb in thymocytes in vivo resulted in impaired rearrangement of both chains of the TCR and a block in differentiation of CD4 T cells, no decrease in RAG-2 or CD4 expression was detected.2,7 Apart from the Bcl2 family members whose products mediate the antiapoptotic function of c-Myb,11–13 few other promising target genes exist that could be responsible for the essential functions of c-Myb in T cells. To address this issue, we have recently published a subtraction cloning-based screen identifying 29 novel targets in a model system of haematopoiesis.14 To extend this screen to a T-cell system, we immobilized cDNA molecules isolated from the initial screen onto nylon filters and probed filters with labelled total cDNA produced from mouse splenic T cells activated ex vivo by interleukin (IL)-2 treatment. The screen identified 81 putative Myb-responsive genes, including the histone variant H2A.Z, which was isolated a total of three times.
H2A.Z is highly conserved throughout eukaryotic evolution and recent data implicate H2A.Z in a wide range of chromatin-related processes including transcriptional regulation, positioning of heterochromatin boundaries, chromosomal stability and cell cycle progression.15–29 The H2A.Z gene differs markedly from the major H2A and other core histone genes in that it is not associated with the replication-dependent gene clusters and possesses introns, a conserved TATA box and a polyadenylation signal sequence.30–32 In addition, the structure of the H2A.Z promoter appears to be complex, with basal expression linked to Sp1 association with the proximal promoter33 and upstream regions mediating increased transcription in a cell type- and differentiation-dependent manner.34
The identification of H2A.Z as a potential c-Myb target was of great interest, as c-Myb has not previously been implicated as a regulator of genes involved in epigenetic regulation. Therefore, we chose to investigate the relationship between c-Myb and H2A.Z in detail. Here we show that Myb proteins bind directly to MBSs upstream of the H2A.Z gene in vitro and in chromatin immunoprecipitation (ChIP) assays. Expression of v-Myb, the activated form of c-Myb, induces H2A.Z in CD4 T cells in vivo. Conversely, blocking or knocking out Myb activity reduces H2A.Z expression in T-cell lines and in specific thymocyte subsets in vivo.
Materials and methods
Mouse lines
The T-cell specific vMyb4 transgenic mouse line, the floxed c-myb mouse line (MybF/F) and RAG-2–/– mice have been described previously.4,35–37 T-cell specific deletion of c-Myb was induced by breeding MybF/F mice to CD2Cre or CD4Cre transgenic mice.38,39
Cell culture
HD3 macrophages were routinely cultured in Dulbecco's modified Eagle's minimal essential medium (DMEM) supplemented with 8% fetal calf serum (FCS) and 2% chicken serum (Invitrogen, Paisley, UK). EL4 cells stably transfected with the MERT dominant negative transgene have been described.12MERT was activated by addition of 1–2 µm 4-hydroxytamoxifen (Sigma-Aldrich, Cambridge, UK).
Electrophoretic mobility shift assays (EMSAs)
EMSA was performed as described previously40 using as probe 0·1 pmol of MBSwt, an IRD700-labelled double-stranded oligonucleotide of sequence 5′- CTAGGACATTATAACGGTTTTTAGTCTAG-3′, and, as a source of Myb DNA-binding activity, 2 µl of rabbit reticulocyte lysate (Promega, Southampton, UK) either unprogrammed or programmed with plasmid pT7βMT.4 Where appropriate, 1 pmol (10×), 10 pmol (100×) or 100 pmol (1000×) of unlabelled competitors was added. Unlabelled oligonucleotide competitors were: MBSwt as above; H2AZ1, 5′-CAGAAGCCTCAGTTGTTTTCGGCAT-3′; H2AZ2, 5′-GTACATCTACTAACTGCACGTCTAA-3′; H2AZ3, 5′-CCAGCATTTGTAACTGATTACATGA-3′; H2AZ4, 5′-GAACCATCTGTAACTGCTTGTGAAT-3′; H2AZ5, 5′-GAATCATGTTACAGTTACCCACTG-3′. All mutant MBSs contained a single base pair change in the YAAC core to YCAC. Bands were visualized on a Licor Odyssey scanner (LI-COR Biosciences UK Ltd, Cambridge, UK).
Chloramphenicol acetyl transferase (CAT) reporter assays
For CAT assays, 5 × 105 HD-3 cells were transfected using DMRIE-C transfection reagent (Invitrogen) in 24-well plates according to the manufacturer's instructions. Cells were harvested after 48 hr, washed in cold phosphate-buffered saline (PBS), resuspended in 200 µl of 0·25 m Tris-HCl (pH 7·8) and lysed in three freeze–thaw cycles. Protein concentrations were determined (BCA protein assay; Pierce, Cramlington, UK) and CAT activity was assayed as described previously.41 Chloramphenicol products were separated by thin layer chromatography in 5% methanol and 95% chloroform and autoradiographed. The percentage of acetylated chloramphenicol products was calculated and corrected for transfection efficiency using a β-galactosidase internal control.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as described previously7 using extracts from primary thymocytes. Primers were: H2AZMBS1, 5′-GTAGAGTTGACTAGCATTCTGC-3′ and 5′-CTTATTGGAACTAAGGAATTG-3′; H2AMBS2, 5′-ATGCGAAATTCGCAAGACTCAG-3′ and 5′-ATAGACTTGTACACACGGTAC-3′; H2AZ MBS33-5, 5′-GGACAGTACCGTGTGTACAAG-3′ and 5′-TAAAGCTATTATGTGTCAGC-3′.
Flow cytometry and cell sorting
Single cell suspensions were prepared from whole thymus and spleen. In some experiments, CD69lo and CD69hi thymocyte populations were separated by positively selecting CD69hi cells by magnetic antibody cell sorting (MACS) using LS columns (Miltenyi Biotech, Surrey, UK), followed by flow sorting of individual populations. Cells in all experiments were stained at 4° in PBS containing 2% FCS, 5 mm ethylenediaminetetraacetic acid (EDTA) and 0·1% sodium azide. Antibodies were: CD69-biotin, CD4-PE, Thy1·2-PE, CD8a-FITC, CD4-APC, CD25-PE, CD44-FITC and streptavidin-PE-Cy7 (BD Biosciences, Oxford, UK). Fluorescence-activated cell sorting (FACS) was performed on a FACSVantage electronic cell sorter (BD Biosciences) or a MoFlo (DakoCytomation, Glostrup, Denmark). DN thymocytes were sorted by FACS after depletion of CD4- and CD8-expressing thymocytes by MACS. Data were analysed using flowjo (Tree Star, Ashland, WA).
Northern blotting
RNA was processed using standard protocols. The mouse H2A.Z probe was a purified, 262-base polymerase chain reaction (PCR) product labelled with [32P]-dCTP (PCR primer sequences: H2AZF, ATCTAGGACAACCAGCCACG; H2AZR, CTGTTGTCCTTTCTTCCCGATC). The H2A.Z probe was hybridized to nylon membranes in QuikHyb hybridization solution (Stratagene, La Jolla, CA) in accordance with the manufacturer's instructions, and visualized by autoradiography.
Quantitative reverse transcription–polymerase chain reaction (qRT-PCR)
Total RNA was extracted from between 5 × 104 and 5 × 105 sorted T cells using TriZol reagent (Invitrogen). RNA was reverse-transcribed using the Omniscript kit (Qiagen, Crawley, UK). qRT-PCR reactions were performed on a Taqman 7900HT analyser (Applied Biosystems, Foster City, CA). All qRT-PCR assays were obtained from Applied Biosystems. Where indicated, qRT-PCR data were subjected to the paired Student's t-test to determine significance of differences in H2A.Z mRNA expression between the transgenic and littermate control groups.
Results
Consensus MBSs upstream of the H2A.Z gene
We identified five conserved, putative MBSs (defined as YAACT/GG) in the 1·5 kb upstream of the H2A.Z proximal promoter, and a further 11 sequence elements containing the conserved MBS core (YAAC) that diverge from the conserved hexamer at only one of the last two positions. Four of the completely conserved hexamers cluster to a short, 300-bp region approximately 1 kb upstream of the first exon (Fig. 1a). This 300-bp region is highly conserved between mouse and human with conservation of three of the four MBSs. At least one MBS is also present upstream of the chicken H2A.Z gene (h2af), although there is little sequence homology overall.
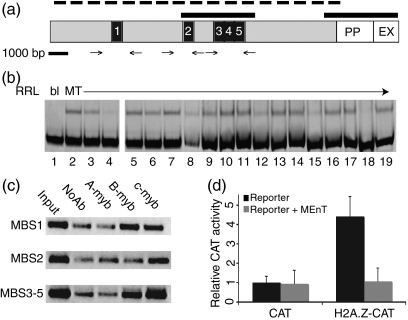
Figure 1.
Myb proteins interact directly with Myb-binding sites (MBSs) immediately upstream of the histone variant H2A.Z proximal promoter. (a) Schematic of the genomic DNA sequence up to 2000 bases upstream of the murine H2A.Z cDNA. Regions of homology with the human gene are indicated by solid black bars. MBS1–5 are indicated by black boxes. Approximate positions of chromatin immunoprecipitation (ChIP) polymerase chain reaction (PCR) primers are represented by arrows and the broken line indicates the region cloned upstream of the chloramphenicol acetyl transferase (CAT) gene in CAT reporter assays. (b) Electrophoretic mobility shift assays (EMSAs) showing that in vitro translated Myb DNA-binding domain (DBD) fragment MT recognizes H2A.Z Myb-binding sites. Binding reactions all contained 0·1 pmol labelled Mybswt probe: lane 1, unprogrammed reticulocyte lysate; lanes 2–19, MT-programmed lysate. Lanes 4–19 contained added unlabelled oligonucleotide competitors as described in the text. (c) hIP assays showing B- and c-Myb proteins binding the H2A.Z promoter in primary thymocytes. (d) Quantification of CAT assays showing activation of parental CAT plasmid (CAT) and an H2A.Z promoter-linked CAT gene (H2AZ-CAT) in HD3 cells, in the absence (black bars) or presence (grey bars) of the MEnT dominant negative protein. CAT assays were performed three times. Bars represent mean values with the standard deviations indicated as error bars. bl, blank; EX, Exon; NoAb, no antibody; PP, proximal promoter; RRL, red reticulocyte lysate.
Myb interacts with H2A.Z MBSs in vitro and in vivo and is functionally important for H2A.Z transcription
We performed EMSAs to determine whether a 26-kDa Myb DNA-binding domain (DBD) fragment, MT, translated in rabbit reticulocyte lysate, could be competitively prevented from binding to a strong canonical MBS probe (MBSwt) by an excess of unlabelled H2A.Z MBSs. Figure 1(b), lane 2 shows the band-shift obtained with the complex between MBSwt probe and MT, and lanes 3 and 4 demonstrate competition with a 10-fold and 100-fold molar excess of unlabelled MBSwt. None of the H2A.Z MBSs was able to compete when added at either a 10-fold (data not shown) or 100-fold molar excess (Fig. 1b, lanes 5, 8, 11, 14 and 17). However, when added in a 1000-fold molar excess, H2A.Z MBSs 3, 4 and 5 were able to compete well (lanes 12, 15 and 18), and MBS1 and MBS2 competed extremely weakly (lanes 6 and 9). As controls, no competition was observed with a 1000-fold excess of any of the H2A.Z MBSs when they were mutated to destroy the core AAC of their Myb-binding sites (Fig. 1b, lanes 7, 10, 13, 16 and 19), indicating the specificity of the interaction, and no retarded band was observed in unprogrammed reticulocyte lysate (Fig. 1b, lane 1), demonstrating that the interaction was with the Myb DNA-binding domain fragment.
We used ChIP to determine whether the weak interactions observed in vitro were relevant to c-Myb activity in T cells. Cross-linked DNA–protein complexes from primary murine thymocytes were immunoprecipitated with antibodies against A-, B- and c-Myb and purified DNA was tested for enrichment of the H2A.Z upstream sequence by PCR with specific primers (Fig. 1a, arrows). The c-Myb antibody immunoprecipitated three fragments containing MBS1, MBS2 and MBS3–5, respectively (Fig. 1c), indicating that c-Myb occupies this region of the H2A.Z upstream sequence in primary T cells. The closely related Myb family member B-Myb was also able to bind to the regions encompassing MBS1 and MBS3–5 but appeared absent from the central MBS2 fragment (Fig. 1c). As control, an antibody against A-Myb, which is not expressed in T cells, did not immunoprecipitate any fragments. Therefore, B- and c-Myb are bound to the H2A.Z promoter region in primary thymocytes.
Transient transfection assays were used to test whether the interactions between c-Myb and the H2A.Z upstream region had functional importance for H2A.Z transcription. Transfection of a reporter gene comprising approximately 2 kb of the H2A.Z upstream region (Fig. 1a, broken line) driving expression of the chloramphenicol acetyl transferase (H2AZ-CAT) gene into HD3 macrophages, a cell line expressing high levels of endogenous c-Myb protein, resulted in greater than 4-fold activation of CAT activity compared with a control reporter gene lacking the H2A.Z upstream region. This activation was blocked by cotransfection of the MEnT protein (Fig. 1d), indicating that activation of the H2A.Z promoter was Myb dependent. MEnT is an active repressor of Myb transcriptional activity consisting of the c-Myb DBD fused to the Drosophila Engrailed repressor domain which has been used previously for interrogation of c-Myb function in thymocytes.4
H2A.Z transcripts are elevated in specific T-cell subsets in a transgenic mouse model of Myb function in vivo
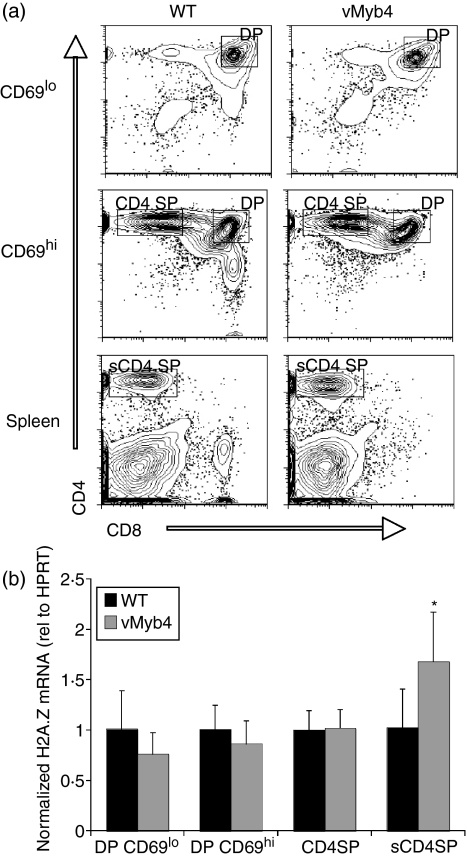
We used the previously developed vMyb4 transgenic mouse line35 to test whether increasing Myb transcriptional activity in thymocytes and splenic T cells in vivo would lead to increases in H2A.Z mRNA levels. Thymocyte subsets and splenic T cells were isolated by FACS from vMyb4 transgenic mice and wild-type littermate controls; CD8 lineage T cells were not compared as this lineage is absent in vMyb4 mice (Fig. 2a). We used the activation marker CD69 to distinguish DP thymocytes that had yet to rearrange their TCR-α chains (CD69lo) from post-rearrangement DPs (CD69hi). We observed no significant difference in H2A.Z transcript levels between vMyb4 transgenic mice and non-transgenic controls in CD69lo DP, CD69hi DP or CD4SP thymocytes (Fig. 2b; P = 0·273, 0·418 and 0·424, respectively) or DN thymocytes (data not shown). However, in mature, splenic CD4 T cells, H2A.Z transcript levels were elevated by 68% compared with wild-type littermate controls (Fig. 2b). This difference was significant (P = 0·032), indicating that increased Myb transcriptional activity results in elevated H2A.Z mRNA in CD4 T cells in vivo.
Figure 2.
Elevated histone variant H2A.Z mRNA in vMyb4 T-cell populations in vivo. (a) Flow cytometry of thymocytes (top and centre panels) and splenocytes (bottom panels) from vMyb4 transgenic mice and wild-type littermate controls (WT), stained with anti-CD4 and anti-CD8 antibodies. (b) Quantitative reverse transcription–polymerase chain reaction (qRT-PCR) analysis of H2A.Z mRNA expression using RNA from sorted T lymphocytes. Data are presented as means from triplicate qRT-PCR reactions performed from each of three independent sorts. Error bars represent standard error. DP, double positive; HPRT, hypoxanthine-guanine phosphoribosyltransferase.
Active repressors of Myb transcription reduce H2A.Z transcripts
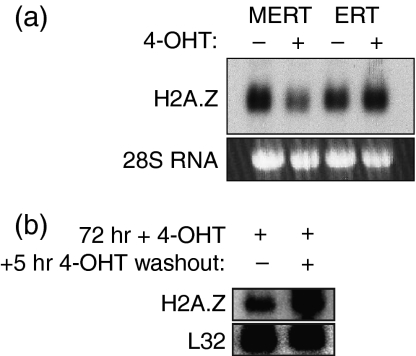
EL4 thymoma cells stably transfected with the MERT inducible active repressor of Myb transcription, termed EL4-MERT cells, have been used previously to investigate regulation of the c-Myb target gene Bcl2.12 MERT comprises the c-Myb DBD fused to the Drosophila Engrailed repressor domain and a modified version of the oestrogen-receptor hormone-binding domain that is activated only by 4-hydroxytamoxifen (4-OHT). Activation of the MERT protein by 4-OHT for 24 hr efficiently reduced steady-state H2A.Z transcripts (Fig. 3a). This reduction was dependent on the Myb DBD, as a control protein, ERT, which lacks the DBD, had no effect on H2A.Z transcripts when induced by 4-OHT. Next, to determine whether the loss of H2A.Z mRNA was likely to be a result of MERT directly repressing the H2A.Z gene, MERT protein was activated by 4-OHT for 72 hr, 4-OHT was washed out of the cell culture for 5 hr, and the level of H2A.Z mRNA was assessed by northern blot. H2A.Z mRNA was efficiently re-induced at the 5-hr time-point (see Fig. 5b below; ‘+’ lane), suggesting that a direct interaction might be occurring.
Figure 3.
Northern blot analysis of histone variant H2A.Z RNA in EL4 cells stably transfected with the MERT active repressor. (a) MERT was induced with 4-hydroxytamoxifen (4-OHT) (+) or mock induced with vehicle (–) for 24 hr. EL4-ERT cells (an inactive version of MERT lacking the Myb DNA-binding domain) were included as a control. Data are representative of five independent experiments. (b) Rapid re-induction of H2A.Z following MERT switch-off. MERT was induced for 72 hr, and then 4-OHT was either washed out of the cultures (+) or not (–). RNA was prepared 5 hr after washout.
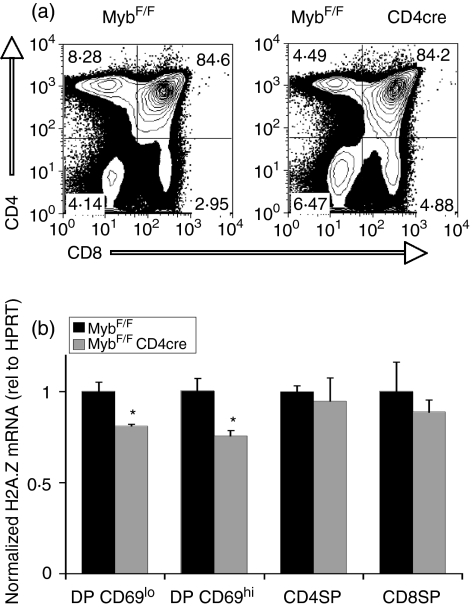
Figure 5.
Deletion of c-myb at the double negative 4 (DN4) stage reduces histone variant H2A.Z expression in thymic double positive (DP) subsets. (a) Fluorescence-activated cell sorting (FACS) profiles of thymocytes stained with antibodies against CD4 (y-axis) and CD8 (x-axis) from MybF/F mice in the absence or presence of the CD4Cre transgene. (b) Quantitative reverse transcription–polymerase chain reaction (qRT-PCR) analysis of H2A.Z mRNA expression using RNA isolated from sorted T lymphocytes of MybF/F CD4Cre mice. Data are presented as means from triplicate qRT-PCR reactions performed from each of three independent sorts. Error bars represent standard error. HPRT, hypoxanthine guanine phosphoribosyltransferase.
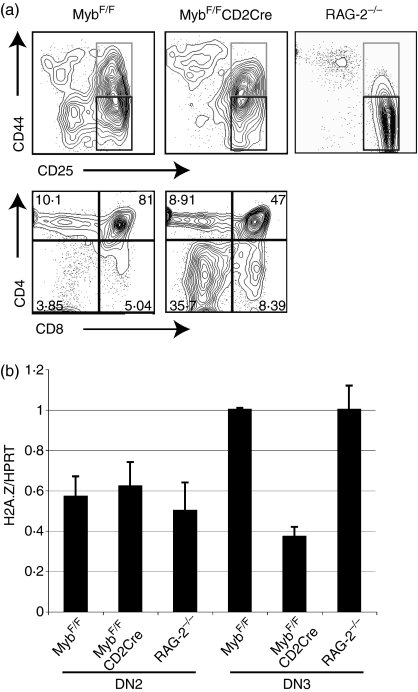
Specific deletion of c-Myb leads to a reduction in H2A.Z mRNA
To determine whether loss of c-Myb in primary T cells could affect H2A.Z transcription, mice with floxed c-myb alleles (MybF/F)36 were crossed to mice carrying either a T-cell specific CD2Cre or a CD4Cre transgene.38,39 In MybF/F CD2Cre crosses, c-myb is deleted during the DN2 stage of T-cell development, and in MybF/F CD4Cre mice, c-myb is lost at the DN4 stage. These two crosses can therefore be used to test for the effects of loss of c-myb at two crucial control points for which it has previously been shown to be important:5,7 the DN3 (CD25+ CD44–) stage, at which thymocytes with a productively rearranged TCR-β chain are signalled to proliferate and differentiate to become DP, and the DP stage itself, where the process of positive selection occurs.
First, to examine the DN3 control point, DN2 and DN3 thymocyte subsets were purified by flow sorting from MybF/F CD2cre mice and MybF/F littermate controls. In the former, thymuses are very small, and the percentage of DP thymocytes is roughly halved (Fig. 4a, lower right panel), as thymocytes are blocked at the DN3 stage (Fig. 4a, upper panel; compare left and centre plots). As a control for any non-specific effects of a DN3 block, DN2 and DN3 thymocytes from RAG-2–/– mice, which have an absolute block at the DN3 stage because of their inability to rearrange the TCR-β locus, were also purified (Fig. 4a, upper right panel). mRNA from the sorted populations was then extracted and H2A.Z transcript levels assayed by qRT-PCR. Figure 4(b) shows that, relative to both the RAG-2 and the control MybF/F subsets, H2A.Z mRNA was substantially decreased in the MybF/F CD2Cre DN3 postdeletion population, but not in DN2 cells, which still contain c-Myb.
Figure 4.
Deletion of c-myb at the double negative 2 (DN2) stage reduces histone variant H2A.Z expression in DN3 thymocytes. (a) Fluorescence-activated cell sorting (FACS) profiles of thymocytes. Upper panel: DN thymocytes stained with antibodies against CD44 (y-axis) and CD25 (x-axis) from MybF/F mice in the absence or presence of the CD2Cre transgene. A recombination activating gene (RAG)-2–/– profile is also shown. DN2 (grey boxes) and DN3 (black boxes) populations used for mRNA preparation are indicated. Lower panel: FACS profiles of total thymocytes stained with antibodies against CD4 (y-axis) and CD8 (x-axis) from MybF/F mice in the absence or presence of the CD2Cre transgene. Percentages of cells in each quadrant are shown. (b) Quantitative reverse transcription–polymerase chain reaction (qRT-PCR) analysis of H2A.Z mRNA expression using RNA isolated from sorted DN2 and DN3 cells of the indicated genotypes. Data are presented as means from triplicate qRT-PCR reactions. Error bars represent standard deviation.
Next, we purified DP thymocytes by flow sorting from MybF/F CD4Cre animals or control MybF/F littermates. These mice contain similar numbers and percentages of DP cells, but have a defect in progression to the CD4 SP lineage7(Fig. 5a). Similar to the situation in DN3 cells, we observed small but consistent reductions in H2A.Z expression, evident in a 19% reduction in CD69lo DPs and a 25% reduction in CD69hi DPs (Fig. 5b). Both of these differences were significant (P = 0·017 and P = 0·012, for CD69lo and CD69hi DPs, respectively). No significant differences were observed in SP T cells, either in the thymus (Fig. 5b) or in the spleen (data not shown), consistent with low levels of endogenous c-Myb expressed in these cells.2,7 Taken together, these data suggest that c-Myb contributes to H2A.Z expression at both the DN3 and DP stages of thymocyte development in vivo, such that loss of c-Myb results in a significant drop in levels of H2A.Z mRNA.
Discussion
Despite the essential role of c-Myb at multiple points in T-cell development, transcriptional targets that mediate this role have proved difficult to identify. Here we describe the regulation by c-Myb of the histone variant H2A.Z in T-cell lines and in thymocytes in vivo. c-Myb and its close relative B-Myb interact with conserved MBSs upstream of the H2A.Z promoter, and endogenous c-Myb activates the upstream region in reporter assays. Repression of Myb activity using the MERT protein also led to repression of endogenous H2A.Z in EL4 T cells, and the gene is rapidly re-induced following inactivation of MERT. Mouse models of Myb overexpression and deletion were used to characterize the regulation of H2A.Z by c-Myb in progressive stages of thymocyte differentiation. Expression of v-Myb in transgenic mice led to elevated expression of H2A.Z transcripts in mature splenic CD4 T cells, whereas deletion of c-myb in the thymocytes of conditional knockout mice reduced H2A.Z expression specifically in DN3 and DP thymocytes.
Transcriptional regulation of H2A.Z is clearly multifaceted. Ubiquitous transcription factors such as Sp1 may be important in the basal expression of H2A.Z.33 However, evidence exists that different regions of the H2A.Z upstream sequence are required for maximal expression depending on differentiation status.34 H2A.Z may have a basal cellular function that requires a certain level of ubiquitous expression in addition to specialized functions that require higher levels of H2A.Z during differentiation. As c-Myb expression is highly restricted to immature and activated cells of the haematopoietic system and some immature epithelial cell types, it is likely to contribute to H2A.Z transcription in a highly restricted context, and thus may stimulate a specialized function of H2A.Z in differentiating cells, such as the thymocytes reported here.
The developmental stage-specific regulation of H2A.Z expression we observed here may reflect differences in the endogenous c-Myb activity between thymocyte subsets. For example, increasing Myb activity (by expressing v-Myb) has no effect in DP thymocytes where endogenous c-Myb activity is high, whereas strong effects are observed in resting splenic CD4SP T cells, which have low c-Myb activity. The opposite situation is observed when c-myb is deleted: H2A.Z expression is reduced in DN3 and DP thymocytes where c-Myb activity is high, but there is no detectable difference in mature SP T cells, where c-Myb activity is low. The residual H2A.Z expression observed even after deletion of c-Myb suggests that c-Myb is one member of a group of transcriptional activators that contribute to H2A.Z expression, and furthermore that there is probably some functional redundancy between c-Myb and B-Myb, both of which bind the H2A.Z promoter in thymocytes. The recent development of a conditional B-Myb knockout mouse model42 will make it possible to test this latter idea directly.
What c-Myb-specific functions might H2A.Z mediate during T-cell development? H2A.Z is an interesting target gene for a number of reasons. Deposition of H2A.Z into nucleosomes decreases the stability of inter-nucleosomal interactions,43,44 leading to a state apparently poised for activation dependent on the activity of other regulatory proteins.22,23 As H2A.Z nucleosomes are distributed widely throughout the genome, the consequences of activation of H2A.Z by c-Myb may be the indirect transcriptional regulation of a large number of genes. This can be interpreted in the context of a recent report that c-Myb regulates the acetylation of histone H3,45 perhaps establishing a new general function of c-Myb as a regulator of histones, by both transcriptional and post-translational mechanisms. It would also be interesting to determine whether H2A.Z has a role in mediating TCR rearrangement, as deletion of c-myb has been reported to affect this process.2 There exists some evidence that histone modification events regulate accessibility and selection of TCR gene segments in V(D)J recombination,46–49 and it is tempting to speculate that histone variants such as H2A.Z could also be important in regulating chromatin structure during this process. However, as yet, the function of H2A.Z in mammals is largely unknown. As the physiological role of c-Myb in T cells is well characterized, the cell- and developmental stage-specific regulation of H2A.Z expression by c-Myb may provide important clues as to the function of H2A.Z itself.
Acknowledgments
We thank Demelza Bird for technical assistance, Fredrik Wallberg, Ayad Eddaoudi and Derek Davies for help with flow sorting, and Georgina Lang for helpful comments on the manuscript. This work was funded by Cancer Research UK.
References
- 1.Allen RD, Bender TP, Siu G. c-Myb is essential for early T cell development. Genes Dev. 1999;13:1073–8. doi: 10.1101/gad.13.9.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol. 2004;5:721–9. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 3.Anderson G, Jenkinson EJ. Lymphostromal interactions in thymic development and function. Nat Rev Immunol. 2001;1:31. doi: 10.1038/35095500. [DOI] [PubMed] [Google Scholar]
- 4.Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T cell development. Genes Dev. 1994;8:770–82. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- 5.Pearson R, Weston K. c-Myb regulates the proliferation of immature thymocytes following beta-selection. EMBO J. 2000;19:6112–20. doi: 10.1093/emboj/19.22.6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lieu YK, Kumar A, Pajerowski AG, Rogers TJ, Reddy EP. Requirement of c-myb in T cell development and in mature T cell function. Proc Natl Acad Sci USA. 2004;101:14853–60. doi: 10.1073/pnas.0405338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurice D, Hooper J, Lang G, Weston K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J. doi: 10.1038/sj.emboj.7601801. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siu G, Wurster AL, Lipsick JS, Hedrick SM. Expression of the CD4 gene requires a Myb transcription factor. Mol Cell Biol. 1992;12:1592–60. doi: 10.1128/mcb.12.4.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakayama K, Yamamoto R, Ishii S, Nakauchi H. Binding of c-Myb to the core sequence of the CD4 promoter. Int Immunol. 1993;5:817–24. doi: 10.1093/intimm/5.8.817. [DOI] [PubMed] [Google Scholar]
- 10.Wang QF, Lauring J, Schlissel MS. c-Myb binds to a sequence in the proximal region of the RAG-2 promoter and is essential for promoter activity in T-lineage cells. Mol Cell Biol. 2000;20:9203. doi: 10.1128/mcb.20.24.9203-9211.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frampton J, Ramqvist T, Graf T. v-Myb of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis in myeloid cells. Genes Dev. 1996;10:2720–31. doi: 10.1101/gad.10.21.2720. [DOI] [PubMed] [Google Scholar]
- 12.Taylor D, Badiani P, Weston K. A dominant interfering Myb mutant causes apoptosis in T cells. Genes Dev. 1996;10:2732–44. doi: 10.1101/gad.10.21.2732. [DOI] [PubMed] [Google Scholar]
- 13.Salomoni P, Perrotti D, Martinez R, Franceschi C, Calabretta B. Resistance to apoptosis in CTLL-2 cells constitutively expressing c-Myb is associated with induction of BCL-2 expression and Myb-dependent regulation of bcl-2 promoter activity. Proc Natl Acad Sci USA. 1997;94:3296–301. doi: 10.1073/pnas.94.7.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang G, White JR, Argent-Katwala MJ, Allinson CG, Weston K. Myb proteins regulate the expression of diverse target genes. Oncogene. 2005;24:1375–84. doi: 10.1038/sj.onc.1208301. [DOI] [PubMed] [Google Scholar]
- 15.Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–22. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 16.Adam M, Robert F, Larochelle M, Gaudreau L. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol Cell Biol. 2001;21:6270–9. doi: 10.1128/MCB.21.18.6270-6279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–36. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 18.Krogan NJ, Baetz K, Keogh MC, et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci USA. 2004;101:13513–8. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rangasamy D, Greaves I, Tremethick DJ. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat Struct Mol Biol. 2004;11:650–8. doi: 10.1038/nsmb786. [DOI] [PubMed] [Google Scholar]
- 20.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci USA. 2005;102:18385–90. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–48. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–31. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006;20:700–10. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhillon N, Oki M, Szyjka SJ, Aparicio OM, Kamakaka RT. H2A.Z functions to regulate progression through the cell cycle. Mol Cell Biol. 2006;26:489–501. doi: 10.1128/MCB.26.2.489-501.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greaves IK, Rangasamy D, Devoy M, Marshall Graves JA, Tremethick DJ. The X and Y chromosomes assemble into H2A.Z, containing facultative heterochromatin, following meiosis. Mol Cell Biol. 2006;26:5394–405. doi: 10.1128/MCB.00519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keogh MC, Mennella TA, Sawa C, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–72. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–22. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–29. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalton S, Robins AJ, Harvey RP, Wells JR. Transcription from the intron-containing chicken histone H2A.F gene is not S-phase regulated. Nucl Acids Res. 1989;17:1745–50. doi: 10.1093/nar/17.4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatch CL, Bonner WM. The human histone H2A.Z gene. Sequence and regulation. J Biol Chem. 1990;265:15211–220. [PubMed] [Google Scholar]
- 32.Harvey RP, Whiting JA, Coles LS, Krieg PA, Wells JR. H2A.F: an extremely variant histone H2A sequence expressed in the chicken embryo. Proc Natl Acad Sci USA. 1983;80:2819–28. doi: 10.1073/pnas.80.10.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatch CL, Bonner WM. Characterization of the proximal promoter of the human histone H2A.Z. Gene DNA Cell Biol. 1995;14:257–66. doi: 10.1089/dna.1995.14.257. [DOI] [PubMed] [Google Scholar]
- 34.Hatch CL, Bonner WM. An upstream region of the H2AZ gene promoter modulates promoter activity in different cell types. Biochim Biophys Acta. 1996;1305:59–62. doi: 10.1016/0167-4781(95)00223-5. [DOI] [PubMed] [Google Scholar]
- 35.Badiani P, Kioussis D, Swirsky D, Lampert I, Weston K. T-cell lymphomas in v-Myb transgenic mice. Oncogene. 1996;13:2205. [PubMed] [Google Scholar]
- 36.Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 2003;22:4478–85. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shinkai Y, Koyasu S, Nakayama K, Murphy KM, Loh DY, Reinherz EL, Alt FW. Restoration of T cell development in RAG-2-deficient mice by functional TCR transgenes. Science. 1993;259:822–8. doi: 10.1126/science.8430336. [DOI] [PubMed] [Google Scholar]
- 38.Lee PP, Fitzpatrick DR, Beard C, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–9. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 39.de Boer J, Williams A, Skavdis G, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–8. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 40.Weston K. Extension of the DNA binding consensus of the chicken c-Myb and v-Myb proteins. Nucl Acids Res. 1992;20:3043–9. doi: 10.1093/nar/20.12.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 42.Garcia P, Berlanga O, Watson R, Frampton J. Generation of a conditional allele of the B-myb gene. Genesis. 2005;43:189–93. doi: 10.1002/gene.20170. [DOI] [PubMed] [Google Scholar]
- 43.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–8. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 44.Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausio J. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J Biol Chem. 2001;276:41945–54. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- 45.Mo X, Kowenz-Leutz E, Laumonnier Y, Xu H, Leutz A. Histone H3 tail positioning and acetylation by the c-Myb but not the v-Myb DNA-binding SANT domain. Genes Dev. 2005;19:2447–54. doi: 10.1101/gad.355405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanhope-Baker P, Hudson KM, Shaffer AL, Constantinescu A, Schlissel MS. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85:887–97. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- 47.Mathieu N, Hempel WM, Spicuglia S, Verthuy C, Ferrier P. Chromatin remodeling by the T cell receptor (TCR)-beta gene enhancer during early T cell development. Implications for the control of TCR-beta locus recombination. J Exp Med. 2000;192:625–35. doi: 10.1084/jem.192.5.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–511. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- 49.Mauvieux L, Villey I, de Villartay JP. TEA regulates local TCR-Jalpha accessibility through histone acetylation. Eur J Immunol. 2003;33:2216–22. doi: 10.1002/eji.200323867. [DOI] [PubMed] [Google Scholar]