Abstract
Synthetic anti-idiotypic antibodies represent a potentially valuable tool for the isolation and characterization of B cells that produce xenoantibodies. An anti-idiotypic antibody that binds to a subset of B cells producing antibodies encoded by the variable-region heavy chain 3 (VH3) germline genes DP35 [immunoglobulin variable-region heavy chain 3-11 (IGHV3-11)], DP-53 and DP-54 plus a small number of VH4 gene-encoded antibodies in humans has recently been identified. These germline progenitors also encode xenoantibodies in humans. We tested whether the small, clearly defined group of B cells identified with this anti-idiotypic antibody produce xenoantibodies in non-human primates mounting active immune responses to porcine xenografts. Peripheral blood B cells were sorted by flow cytometry on the basis of phenotype, and cDNA libraries were prepared from each of these sorted groups of cells. Immunoglobulin VH gene libraries were prepared from the sorted cells, and the VH genes expressed in each of the sorted groups were identified by nucleic acid sequencing. Our results indicate that xenoantibody-producing peripheral blood B cells, defined on the basis of binding to fluorescein isothiocyanate (FITC)-conjugated galactose α(1,3) galactose–bovine serum albumin (Gal-BSA) and the anti-idiotypic antibody 2G10, used the IGHV3-11 germline gene to encode xenoantibodies and were phenotypically CD11b+ (Mac-1+) and CD5–. This novel reagent may be used in numerous applications including definition of xenoantibody-producing B-cell subsets in humans and non-human primates and immunosuppression by depletion of B cells producing anti-Gal xenoantibodies.
Keywords: galactose α(1,3) galactose carbohydrate; xenotransplantation; xenoantibody response
Introduction
The ability to use pigs as organ donors for transplantation into humans represents a solution for the increasing shortage of organs for human transplantation. Xenografts, however, are rejected by antibodies that bind to the galactose α(1,3) galactose (Gal) carbohydrate that is present on wild-type pig cells and absent in humans and Old World primates.1–3 Although hyperacute rejection can now be prevented by several methods including depletion of natural antibodies prior to transplantation4–6 and genetic modification of the donor pig organs,7–11 these procedures do not prevent acute humoral xenograft rejection. The nature of the xenoantigenic targets identified by antibodies directed at non-Gal xenoantigens remains controversial.12–15 It has been speculated that xenoantibodies directed at non-Gal xenoantigens may bind to other structurally related carbohydrates that present a barrier to long-term xenotransplantation.12,13
The availability of novel reagents designed to selectively eliminate B lymphocytes producing anticarbohydrate xenoantibodies would significantly impact the field of xenotransplantation. Identification of the appropriate target cell population requires further studies on the phenotypic characteristics of xenoreactive B cells. Studies in galactosyltransferase-deficient mice have shown that peritoneal cavity-derived B cells with anti-Gal receptors are phenotypically B1b-like, defined as CD21–/1ow IgMhigh CD5– CD43+ Mac-1+.16 These peritoneal cavity-derived B cells do not secrete anti-Gal antibodies, but are the precursors of anti-Gal-secreting B cells that can be found in the spleen.17 These cells no longer express Mac-1 (CD11b), but remain CD21–/1ow IgMhigh CD5– CD43+. In baboons and humans, B cells producing anti-Gal xenoantibodies can be identified in the spleen and have been characterized on the basis of binding to CD20, CD138 and anti-immunoglobulin.18 Additional phenotypic markers expressed on these cells have not yet been defined. The use of anti-idiotypic antibodies provides a unique approach to sorting these live cells that is useful for further characterization of their phenotypic features.
Materials and methods
Animals
Nine young adult, captive-bred rhesus monkeys (Macaca mulatta) were obtained from the California National Primate Research Center (CNPRC; University of California, Davis, CA) primate colony, where the animals were housed and all surgical and sampling procedures were conducted. All work was approved by the Animal Care and Use Committee of the CNPRC at the University of California, Davis.
Flow cytometry
Antibody binding to peripheral blood samples obtained from three normal control rhesus monkeys, three monkeys transplanted with neonatal porcine hearts, and three monkeys immunized with fetal pig islet cells at day 66 of gestation were compared. Normal human blood sample controls (n = 6) were obtained from anonymous donors as expired blood that could not be used by the blood bank. The blood samples (approximately 17–20 million Ficoll-separated peripheral blood lymphocytes) were analysed by dual- and tri-colour flow cytometry, performed on a FACSCalibur cytometer using CellQuest software (Becton Dickinson, San Jose, CA). Labels were fluorescein isothiocyanate–galactose–bovine serum albumin (FITC-Gal-BSA), allophycocyanin (APC)-labelled mouse anti-human-CD11b/Mac-1 (#550019; BD Pharmingen, San Diego, CA) and phycoerythrin (PE)-labelled mouse anti-human CD5 antibodies (#R0842; Dako, Carpinteria, CA). FITC was conjugated to either purified Galili pentasaccharide–BSA (V-Laboratories, Covington, LA) or control BSA alone (Sigma, St Louis, MO) using the QuickTag FITC conjugation kit (Roche, Indianapolis, IN) under conditions recommended by the manufacturer. Cells that were labelled with the mouse monoclonal IgG1 2G10 anti-mouse/human antibody were identified using a secondary anti-mouse PE- or FITC-conjugated IgG antibody (Sigma). The 2G10 antibody was a kind gift from Dr F. Shakib (University of Nottingham, Nottingham, UK). Non-specific labelling to Fc receptors was blocked by preincubation with Fc blocker reagents (Sigma) or 2% normal blood type AB Rh factor− (ABRh–) human serum. Labelled cells were identified by setting the gates to exclude non-specific binding to peripheral blood lymphocytes (PBL) which was determined using appropriate isotype-matched antibody controls (BD Pharmingen). Background labelling was subtracted in the determination of the percentage of labelled cells. Labelled cells were collected in bulk and as single cells into 96-well plates (RNeasy 96 with vacuum QIAvac 96; Qiagen, Valencia, CA) for identification of immunoglobulin variable-region heavy chain (IgVH) gene usage in single cells. Sorts were performed using FACSCalibur and FACSVantage flow cytometers (Becton Dickinson).
Preparation of cDNA libraries
PBL were Ficoll-separated from whole blood samples. These cells were labelled for flow cytometry and sorted on the basis of cell surface markers as described above, and cDNA libraries were prepared from the sorted cells. RNA was prepared using the RNeasy Kit (Qiagen) followed by synthesis of cDNA using either Omniscript or Senscript reverse transcriptase (Qiagen). Microcon-100 columns (Amicon; Millipore, Billeria, MA) were used for cDNA purification. The cDNA libraries of genes encoding IgM antibodies were constructed and polymerase chain reaction (PCR) amplifications were performed as previously described.19–21 Purified PCR products were ligated into a pCR®2·1 vector (TA Cloning Kit; Invitrogen, Carlsbad, CA) and INVαF′ One Shot™ competent cells (Invitrogen) were transformed with the ligation reactions and plated onto Xgal-containing Luria Broth (LB) ampicillin plates.
DNA sequence analysis
The DNA sequence was analysed for a total of 100 cDNA clones that were prepared from the VH3 cDNA libraries. The DNA was prepared using the QIAPrep Spin MiniPrep Kit (Qiagen) and was sequenced using the ALFexpress™ automated DNA sequencer and AutoCycle™ Sequencing Kit (Amersham Biosciences, Piscataway, NJ). The results were analysed using omiga software (Oxford Molecular, Madison, WI) and the closest identifiable germline counterparts were identified in GenBank using blast. The sequences reported here have been entered into GenBank as AY986401 to AY986406, and as AY986395 to AY986400.
Results
A subset of B cells expressing antibodies encoded by select VH3 germline genes is identified using the anti-idiotypic antibody 2G10
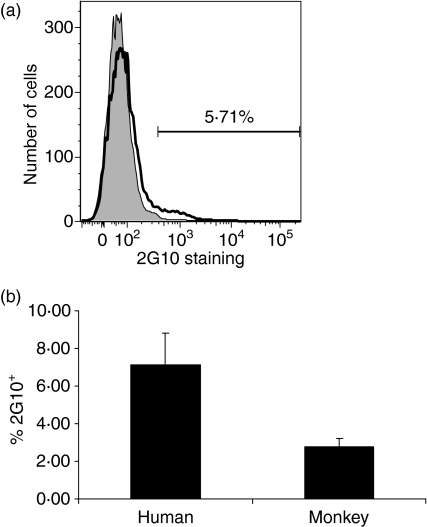
Analysis of labelled peripheral blood cells by flow cytometry indicated that 4–12% of B cells in normal humans and 1·5–5% of B cells in normal rhesus monkeys bound to the anti-idiotypic antibody 2G10 (Fig. 1). It has previously been reported that the 2G10 anti-idiotypic antibody identifies a small subset of B cells and is likely to be directed at sequences encoded by a small number of VH3 and VH4 genes, including DP-35, DP-53 and DP-54.23 In an effort to further characterize the germline progenitors encoding antibodies in B cells identified using anti-idiotypic antibody 2G10, we isolated the 2G10-binding cells by flow cytometry and prepared cDNA libraries of genes encoding immunoglobulins in this population of B cells. The results indicate that human 2G10+ cells produce antibodies encoded by germline progenitors IGHV3-11, IGHV3-74, V3-7, V3-23, V3-21 and V3-30. One hundred clones were sequenced from the VH3 family libraries prepared from the sorted 2G10+ B cells. Twenty per cent of the sequences obtained from the isolated human cells corresponded to antibodies encoded by the IGHV3-11 germline progenitor, 27% were V3-7 and 7% of the sequences were antibodies encoded by the germline gene V3-74. The percentage of cells utilizing the germline progenitors V3-23, V3-21 and V3-30 was less than 1%. The 2G10+ population therefore identifies B cells that express the same VH3-encoded antibodies that are capable of binding to the Gal carbohydrate.19–22 We then confirmed that the subpopulation of 2G10+ B cells expressed antibodies that bind the Gal carbohydrate by dual-colour flow cytometry using FITC-labelled BSA-conjugated Gal carbohydrate. We prepared cDNA libraries from the sorted Gal+ 2G10+ B cells and found that these antibodies were encoded by the IGHV3-11 germline progenitor (Fig. 2).
Figure 1.
Lymphocytes that bind to the anti-idiotypic antibody 2G10 were identified by flow cytometry. (a) The percentage of 2G10-binding human lymphocytes in a representative experiment. (b) The percentage of 2G10+ lymphocytes in normal humans (n = 6) and normal rhesus monkeys (n = 3) ± standard error.
Figure 2.
Cells that bind to the anti-idiotypic antibody 2G10 and to fluorescein isothiocyanate–galactose–bovine serum albumin (Galα1-3Galβ1-4Glc-BSA-FITC) were identified by two-colour flow cytometry and sorted, and cDNA libraries were prepared to identify the immunoglobulin variable-region heavy chain (IgVH) genes encoding these antibodies. The germline progenitor encoding xenoantibodies in this sorted population was IGHV3-11.
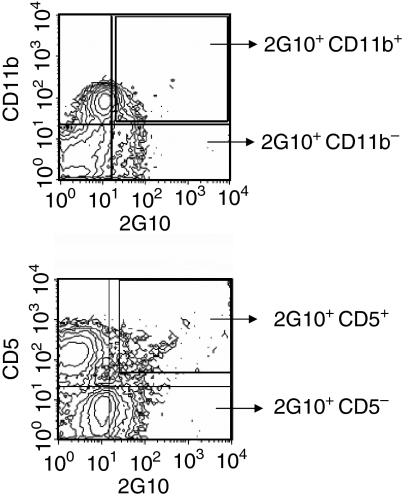
Xenoantibody-producing cells are CD11b+ CD5– 2G10+
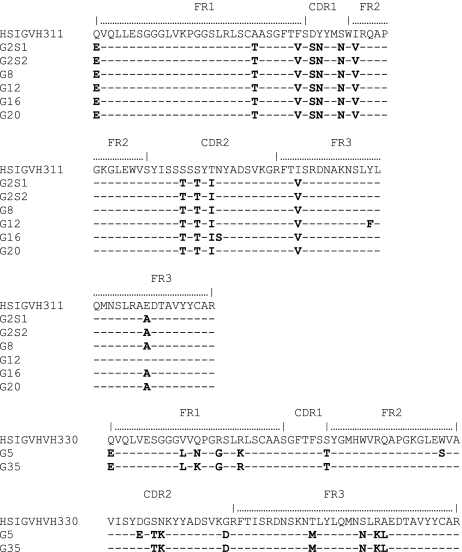
Natural antibody-producing B cells in the peritoneal cavity of mice have been shown to have phenotypic properties of B1b cells (CD21–/low IgMhigh CD5– CD43+ Mac-1+).16 In our study, we isolated B cells from peripheral blood by multicolour flow cytometry on the basis of phenotype. We initially identified groups on the basis of binding to CD11b, CD5 and 2G10 in humans (Fig. 3). The results indicate that most 2G10+ cells were CD5– (only 1·6% were double +). Double-labelling for 2G10 and CD11b demonstrated that 12·5% of the cells defined by 2G10 were CD11b (Mac-1)+. We then sorted cell populations into four different groups: (1) cells that bind to the anti-idiotypic antibody 2G10; (2) CD11b+ CD5– 2G10+ cells; (3) CD11b– CD5– 2G10+ cells, and (4) FITC-Gal+ 2G10+ CD11b+ cells. The populations were sorted using the FACSVantage. Individual cells were collected directly into 96-well plates and a bulk population was also sorted for the preparation of cDNA libraries. Immunoglobulin VH gene libraries were generated from each of these sorted cell groups and the genes encoding antibodies were identified by nucleic acid sequencing, then compared to sequences in the databases and analysed using blast. The results indicated that the CD11b+ CD5– 2G10+ cell population predominantly utilized the IGHV3-11 germline progenitor to encode antibodies and a small population of antibodies were encoded by HSIGVH330 (Fig. 4). These genes utilized the IGHJ4*02 germline gene within the complementarity determining region 3 (CDR3) region (Fig. 5). The CD11b– CD5– sorted cells used various germline genes to encode antibodies. These germline genes were sequenced and identified as VH3-33, DP31, IGHYAAJ, IGH277 and M99651. The antibodies expressed by the human B cells in this group do not bind to the FITC-labelled BSA-conjugated Gal carbohydrate.
Figure 3.
Human peripheral blood B cells were isolated and sorted using anti-idiotypic antibody 2G10, anti-CD5 and anti-CD11b. Dual labelling showed that 12·5% of the cells identified using 2G10 were CD11b+. Most 2G10+ cells were CD5–.
Figure 4.
Amino acid sequence of the antibodies encoded by the immunoglobulin variable-region heavy chain (IgVH) genes expressed in the population of sorted CD11b+ CD5– 2G10+ human B cells. The IGHV3-11cyno gene previously reported by our laboratory to encode xenoantibodies in induced antibody responses to human decay accelerating factor (hDAF) transgenic pig organs in cynomolgus monkeys and in rhesus monkeys with transplants of wild-type organs is identified in this sorted population derived from human peripheral blood. CDR, complementarity-determining region; FR, framework.
Figure 5.
Nucleotide (a) and amino acid (b) sequences of the complementarity determining region 3 (CDR3) region of immunoglobulin genes encoding antibodies in the isolated human CD11b+ CD5– 2G10+ peripheral blood cells.
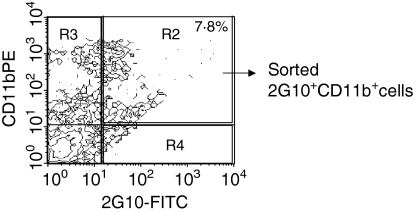
Non-human primates with transplants of porcine xenografts demonstrate an increase in the number of CD11b+ 2G10+ cells
We have recently shown that xenoantibodies in humans and in non-human primates are encoded by a restricted group of germline genes.20,21 The IGHV3-11 germline progenitor and a closely related gene (IGHV3-11cyno) in non-human primates encode xenoantibodies that have been identified by the preparation of cDNA libaries from PBL of humans, rhesus monkeys and cynomolgus monkeys producing high levels of xenoantibodies following exposure to porcine xenografts. In this study, we took a different approach with the intent of identifying the small group of B cells producing xenoantibodies by isolation of the CD11b+ 2G10+ cells from the peripheral blood of rhesus monkeys at 11 days postexposure to porcine xenoantigens. We sought to determine whether this subpopulation of B cells produce xenoantibodies and if they were expanded following transplantation with pig xenografts. Nine monkeys in total were used for the determination of subpopulations of cells from the peripheral blood pre- and/or postexposure to pig cells. In three monkeys with transplants of porcine hearts, the percentage of CD11b+ cells nearly doubled after organ transplantation, rising from 17 to 33% at day 11 post-transplant, and approximately 6–8% of circulating cells were CD11b+ 2G10+ post-transplant (Fig. 6). Exposure to single cells expressing the Gal carbohydrate (porcine islets), rather than solid organs, similarly resulted in a 22% increase in the number of circulating CD11b+ 2G10+ cells, as determined by flow cytometry.
Figure 6.
In rhesus monkeys with transplants of porcine hearts, the percentage of peripheral blood CD11b+ 2G10+ cells was 6–8% (7·8% in this representative animal) as determined by flow cytometry. FITC, fluorescein isothiocyanate; PE, phycoerythrin.
The CD11b+ 2G10+ FITC-Gal+ B cells in rhesus monkeys produce antibodies encoded by IGHV3-11
To confirm that CD11b+ 2G10+ rhesus B cells encoded xenoantibodies, the cells were further defined on the basis of their ability to bind to FITC-Gal. Xenoreactive antibodies that have the ability to reject wild-type porcine xenografts include antibodies that bind to the Gal carbohydrate, and we have previously shown that the IGHV3-11 germline gene encodes xenoantibodies in humans and non-human primates mounting active xenoantibody responses.19–21 The 2G10+ CD11b+ FITC-Gal+ xenoantibody-producing B cells were sorted, and their RNA used for the construction of cDNA libraries. The IGHV3-11 germline progenitor was identified as the gene used to encode xenoantibodies in rhesus peripheral blood B cells that are FITC-Gal+ CD11b+ and 2G10+ using tri-colour flow cytometry (Fig. 7). The amino acid sequences of the genes encoding xenoantibodies produced by the population of expanded B cells following in vivo transplantation of porcine hearts and porcine islets were 96·7 and 98·9% similar, respectively, to the amino acid sequence of genes encoding xenoantibodies previously identified in cDNA libraries generated from the peripheral blood of rhesus xenograft recipients and humans exposed to porcine cells in a bioartificial liver device (Fig. 8).
Figure 7.
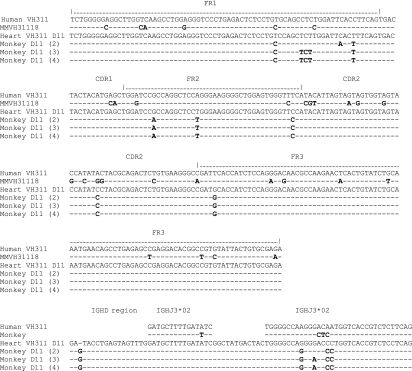
Nucleic acid sequences of genes encoding antibodies identified in rhesus cells sorted by flow cytometry as 2G10+ Gal+ and CD11b+ are shown as sequences 2, 3, and 4. The nucleotide sequence of xenoantibodies identified in the cDNA libraries constructed from peripheral blood cells of the rhesus monkeys with transplants of porcine hearts is shown as ‘Heart VH311’. The cells were isolated at day 11 (D11) post-transplantation. The sequences have been aligned with the closest germline progenitors (identified as the IGHV3-11 germline gene) in human (VH311) and non-human primates (MMVH31118). CDR, complementarity determining region; FR, framework.
Figure 8.
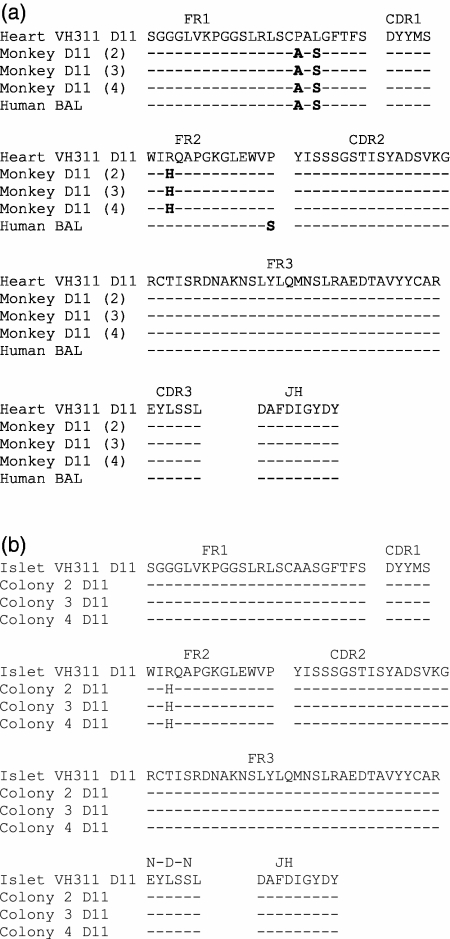
Amino acid sequences of genes encoding antibodies in the sorted 2G10+ Gal+ and CD11b+ population of rhesus monkeys mounting an active immune response to porcine hearts (monkey D11 sequences 2, 3 and 4) are compared with the sequence of immunoglobulin variable-region heavy chain (IgVH) genes encoding xenoantibodies identified in cDNA libraries made from peripheral blood B cells at day 11 (heart VH311 D11) post-transplantation with porcine heart (a) and porcine islet cells (b).20 The sequence of genes encoding xenoantibodies in the peripheral blood of humans exposed to porcine cells in a bioartificial liver is shown as human BAL.19 CDR, complementarity determining region; FR, framework.
Discussion
It has been reported that B cells with anti-Gal receptors are phenotypically like B-1b cells in mice.16 B-1 cells comprise the majority of B cells within the peritoneal and pleural cavities and are characterized by their IgMhi IgD1ow CD11b+ phenotype.23–25 They differ in distribution, function and phenotype when compared with B-2 B cells in secondary lymphoid organs which play a key role in adaptive immune responses. Within the B-1 population, B-1a cells are distinguished from B-1b cells by the presence of CD5 on the B-1a population.26 It has been shown that natural antibody-producing cells that bind to the Gal carbohydrate in the peritoneal cavity are phenotypically CD21– CD23– CD5– CD43+ CD11b+ CD138–, whereas those identified in the spleen did not express CD11b.16 In galactosyltransferase-deficient mice, the peritoneal cavity B cells that produce natural antibodies are the precursors of splenic IgM natural antibody-producing cells.17 In humans and non-human primates, the phenotypic characteristics of the subpopulations of B cells producing xenoantibodies have not yet been clearly defined, but it has been speculated that B1b cells produce xenoantibodies in higher species.18 In this paper, we have further characterized the lymphocyte subpopulations that are responsible for the production of xenoantibodies using both cell-type specific surface markers and an anti-idiotypic antibody (2G10). Our results are consistent with previous work in mice, suggesting that B1b cells produce xenoantibodies in humans and non-human primates. The current study further demonstrates that antibodies expressed by this small population of cells are encoded by a restricted number of germline genes in higher mammals.
The rationale for including this particular anti-idiotypic antibody (2G10) in our experimental design was the fact that it binds to antibodies that are encoded by a small number of VH3 and VH4 germline genes, as demonstrated by inhibition enzyme-linked immunosorbent assay (ELISA).27 The ability of this reagent to identify B cells producing VH3 gene-encoded antibodies such as DP-35 (IGHV3-11), DP-53, DP-54 and V3-74 suggested to us that the specificity of this antibody coincidentally included the germline progenitors that encode xenoantibodies in humans and non-human primates.19–22 The antibody might therefore be useful in the identification of B cells producing xenoantibodies. We examined this issue further by isolating 2G10-binding cells by flow cytometry and defining the IgVH gene usage in the 2G10+ cell population, as well as in phenotypically defined subsets of this group. Peripheral blood B cells producing xenoantibodies were found to be 2G10+ CD11b+ CD5–. We found that the population of CD11b+ 2G10+ cells expands in primates mounting active immune responses after exposure to porcine xenografts and/or Gal+ cells, and the germline progenitor encoding antibodies in these cells was identified as IGHV3-11. The sequences of the genes encoding antibodies in B cells sorted on the basis of phenotype were 99% similar to the previously reported sequence of genes encoding xenoantibodies directed at porcine hearts and islet cells in rhesus monkeys.20 Although the restriction in the usage of select IgVH genes encoding xenoantibodies has been well documented in multiple settings, the ability to sort and define the small group of B cells producing these antibodies reported here is novel. Of interest is the finding that a restricted VH gene usage associated with selection of a particular CDR3 structure is seen in the B-1 cells that were sorted on the basis of phenotype in this B-1b population of cells in humans and non-human primates. Our results are strikingly consistent with previous work in mice demonstrating a restriction in IgVH gene usage in the B-1b cell population and a close association between CDR3 structure and specific VH gene families.28 These findings support the dual model concept whereby VH gene usage and repertoire-shaping mechanisms differ between B-1 and B-2 cells. Germline VH genes may play a role in antibody specificity in B-1 cells, allowing pre-existing innate antibodies to respond to carbohydrate antigens on infectious agents with a select VH gene usage and a restricted CDR3, whereas B-2 responses mediated by encounter with antigen involve somatic mutation and selection to confer specificity.28
The use of this information to deplete xenoreactive B cells and/or alter immune responses to porcine xenografts could be extremely valuable for controlling xenograft rejection. Clinically acceptable immunosuppressive reagents that can effectively control humoral rejection in primates transplanted with porcine xenografts need to be developed. Depletion of anti-α1,3galactosyl antibodies by extracorporeal absorption or administration of synthetic α-Gal-specific immunoabsorbants does not completely remove these xenoantibodies, and they tend to return to reject the grafts.29–32 Various approaches to achieve direct elimination of B cells producing xenoantibodies have also been developed, including depletion using anti-μ monoclonal antibodies, and the development of anti-idiotypic antibodies directed at xenoantibody-producing B cells.33–39 The identification of B cells producing xenoantibodies directed at both Gal and non-Gal determinants would be of benefit, as acute xenograft rejection still occurs in baboons transplanted with Gal knockout pig organs.15 Administration of polyclonal anti-idiotypic antibodies against human anti-Gal antibodies has been shown to reduce serum cytotoxicity to pig cells in baboons by the induction of non-cytotoxic anti-Gal IgG antibodies in vivo.35,36 Monoclonal anti-idiotypic antibodies have been shown to neutralize the cytotoxicity of baboon serum to pig cell lines in vitro as well as reduce the cytotoxicity of baboon serum following in vivo administration.37 The beneficial effects of these treatments, however, have been short-lived and/or ineffective unless combined with other reagents and/or procedures. In this paper, we phenotypically define the small group of B cells encoding antibodies that play a role in antibody-mediated rejection of porcine xenografts in both humans and non-human primates in vivo. Additional studies are ongoing to test the efficacy of depletion of these B cells as an adjunct therapeutic that may enhance the survival of porcine xenografts.
Acknowledgments
We thank Natasha Barteneva, Lora Barsky and Ewa Zielinska for their assistance with the cell sorting, Dr Clare Gregory for performing the islet transplantation in the monkeys and Drs Clare Gregory and Dominic Borie for performing the heart transplantation. We would also like to thank the staff of the University of California National Primate Research Center for taking care of the animals. This work was supported by NIH grant RO1AI52079 (to MKJ).
Abbreviations
- BAL
bioartificial liver device
- CDR
complementarity determining region
- FR
framework
- Gal
galactose α(1,3) galactose
- VH
variable-region heavy chain.
References
- 1.Sandrin MS, Vaughan HA, Dabkowski PL, McKenzie IFC. Anti-pig IgM antibodies in human serum react predominantly with Galα (1–3) Gal epitopes. Proc Natl Acad Sci USA. 1993;90:11391–5. doi: 10.1073/pnas.90.23.11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Platt JL, Fischel RJ, Matas AJ, Reif SA, Bolman RM, Bach FH. Immunopathology of hyperacute xenograft rejection in a swine-to-primate model. Transplantation. 1991;52:214–20. doi: 10.1097/00007890-199108000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Galili U, Shohet SB, Kobrin E, Stults CLM, Macher BA. Man, apes, and old world monkeys differ from other mammals in the expression of α-galactosyl epitopes on nucleated cells. J Biol Chem. 1988;263:17755–62. [PubMed] [Google Scholar]
- 4.Simon PM, Neethling FA, Taniguchi S, Hancock WW, Zopf D, Cooper DK. Intravenous infusion of Galα1–3Gal oligosaccharides in baboons delays hyperacute rejection of porcine heart xenografts. Transplantation. 1998;65:346–53. doi: 10.1097/00007890-199802150-00009. [DOI] [PubMed] [Google Scholar]
- 5.Lam TT, Hausen B, Boeke-Purkis K, et al. Hyperacute rejection of hDAF-transgenic pig organ xenografts in cynomolgus monkeys: influence of pre-existing anti-pig antibodies and prevention by the alpha GAL glycoconjugate GAS914. Xenotransplantation. 2004;11:517–24. doi: 10.1111/j.1399-3089.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen G, Sun H, Yang H, et al. The role of anti-non-Gal antibodies in the development of acute humoral xenograft rejection of hDAF transgenic porcine kidneys in baboons receiving anti-Gal antibody neutralization therapy. Transplantation. 2006;81:273–83. doi: 10.1097/01.tp.0000188138.53502.de. [DOI] [PubMed] [Google Scholar]
- 7.Lam TT, Paniagua R, Shivaram G, Schuurman HJ, Borie DC, Morris RE. Anti-non-Gal porcine endothelial cell antibodies in acute humoral xenograft rejection of hDAF-transgenic porcine hearts in cynomolgus monkeys. Xenotransplantation. 2004;11:531–5. doi: 10.1111/j.1399-3089.2004.00175.x. [DOI] [PubMed] [Google Scholar]
- 8.Ramirez P, Montoya MJ, Rios A, et al. Prevention of hyperacute rejection in a model of orthotopic liver xenotransplantation from pig to baboon using polytransgenic pig livers (CD55, CD59, and H-transferase) Transplant Proc. 2005;37:4103–6. doi: 10.1016/j.transproceed.2005.09.186. [DOI] [PubMed] [Google Scholar]
- 9.Phelps CJ, Koike C, Vaught TD, et al. Production of alpha 1,3-galactosyltransferase-deficient pigs. Science. 2003;299:411–4. doi: 10.1126/science.1078942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai L, Kolber-Simonds D, Park KW, et al. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science. 2002;295:1089–92. doi: 10.1126/science.1068228. [DOI] [PubMed] [Google Scholar]
- 11.Harrison SJ, Guidolin A, Faast R, Crocker LA, Giannakis C, D'Apice AJ, Nottle MB, Lyons I. Efficient generation of alpha(1,3) galactosyltransferase knockout porcine fetal fibroblasts for nuclear transfer. Transgenic Res. 2002;11:143–50. doi: 10.1023/a:1015262108526. [DOI] [PubMed] [Google Scholar]
- 12.Milland J, Christiansen D, Sandrin MS. Alpha1,3-galactosyltransferase knockout pigs are available for xenotransplantation: are glycosyltransferases still relevant? Immunol Cell Biol. 2005;83:687–93. doi: 10.1111/j.1440-1711.2005.01398.x. [DOI] [PubMed] [Google Scholar]
- 13.Milland J, Christiansen D, Lazarus BD, Taylor SG, Xing PX, Sandrin MS. The molecular basis for gal alpha(1,3)Gal expression in animals with a deletion of the alpha1,3galactosyltransferase gene. J Immunol. 2006;176:2448–54. doi: 10.4049/jimmunol.176.4.2448. [DOI] [PubMed] [Google Scholar]
- 14.Sharma A, Naziruddin B, Cui C, et al. Pig cells that lack the gene for alpha1–3 galactosyltransferase express low levels of the Gal antigen. Transplantation. 2003;75:430–6. doi: 10.1097/01.TP.0000053615.98201.77. [DOI] [PubMed] [Google Scholar]
- 15.Chen G, Qian H, Starzl T, et al. Acute rejection is associated with antibodies to non-Gal antigens in baboons using Gal-knockout pig kidneys. Nat Med. 2005;11:1295–8. doi: 10.1038/nm1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohdan H, Swenson KG, Kruger Gray HS, Yong-Guang Y, Xu Y, Thall A, Sykes M. Mac-1 negative B-1b phenotype of natural antibody-producing cells, including those responding to gal α1,3 galactosyltransferase-deficient mice. J Immunol. 2000;165:5518–29. doi: 10.4049/jimmunol.165.10.5518. [DOI] [PubMed] [Google Scholar]
- 17.Kawahara T, Ohdan H, Zhao G, Yong-Guang Y, Sykes M. Peritoneal cavity B cells are precursors of splenic IgM natural antibody-producing cells. J Immunol. 2003;171:5406–14. doi: 10.4049/jimmunol.171.10.5406. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y, Yong-Guang Y, Ohdan H, et al. Characterization of anti-Gal antibody-producing cells of baboons and humans. Transplantation. 2006;81:940–8. doi: 10.1097/01.tp.0000203300.87272.a3. [DOI] [PubMed] [Google Scholar]
- 19.Kearns-Jonker M, Swensson J, Ghiuzeli C, Chu W, Osame Y, Baquerizo A, Demetriou A, Cramer DV. The human antibody response to porcine xenoantigens is encoded by IGHV3-11 and IGHV3-74 IgVH germline progenitors. J Immunol. 1999;163:4399–412. [PubMed] [Google Scholar]
- 20.Kleihauer A, Gregory CR, Borie D, et al. Identification of the VH genes encoding xenoantibodies in non-immunosuppressed rhesus monkeys. Immunology. 2005;116:89–102. doi: 10.1111/j.1365-2567.2005.02204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zahorsky-Reeves J, Gregory C, Cramer DV, Kyles AE, Borie DC, Christe KL, Starnes VA, Kearns-Jonker M. Similarities in the immunoglobulin response and VH gene usage in rhesus monkeys and humans exposed to porcine hepatocytes. BMC Immunol. 2006;7:3. doi: 10.1186/1471-2172-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Radic MZ, Galili U. Human anti-Gal heavy chain genes. Preferential use of VH3 and the presence of somatic mutations. J Immunol. 1995;155:1276–85. [PubMed] [Google Scholar]
- 23.Hayakawa K, Hardy RR, Herzenberg LA. Progenitors for Ly-1 B cells are distinct from progenitors for other B cells. J Exp Med. 1985;161:1554–68. doi: 10.1084/jem.161.6.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalor PA, Stall AM, Adams S, Herzenberg LA. Permanent alteration of the murine Ly-1B repertoire due to selective depletion of Ly-1 B cells in neonatal animals. Eur J Immunol. 1989;19:501–6. doi: 10.1002/eji.1830190314. [DOI] [PubMed] [Google Scholar]
- 25.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 b cell-specified progenitor. Nature Immunol. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 26.Kantor A, Herzenberg LA. Origin of murine B cell lineages. Annu Rev Immunol. 1993;11:501–38. doi: 10.1146/annurev.iy.11.040193.002441. [DOI] [PubMed] [Google Scholar]
- 27.McElveen JE, Furtado PB, Smith SJ, Clark MR, Spendlove I, Sewell HF, Shakib F. Characterisation of a mouse monoclonal anti-idiotype reactive with a V region sequence commonly used by human immunoglobulins. Mol Pathol. 2000;53:77–82. doi: 10.1136/mp.53.2.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seidl K, Wilshire JA, MacKenzie JD, Kantor A, Herzenberg L, Herzenberg L. Predominant VH genes expressed in innate antibodies are associated with distinctive antigen binding sites. Proc Natl Acad Sci USA. 1999;96:2262–7. doi: 10.1073/pnas.96.5.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alwayn IP, Basker M, Buhler L, Cooper DK. The problem of anti-pig antibodies in pig-to-primate xenografting: current and novel methods of depletion and/or suppression of production of anti-pig antibodies. Xenotransplantation. 1999;6:157–68. doi: 10.1034/j.1399-3089.1999.00030.x. [DOI] [PubMed] [Google Scholar]
- 30.Watts A, Foley A, Awwad M, et al. Plasma perfusion by apheresis through a Gal immunoaffinity column successfully depletes anti-Gal antibody: experience with 320 aphereses in baboons. Xenotransplantation. 2000;7:181–5. doi: 10.1034/j.1399-3089.2000.00068.x. [DOI] [PubMed] [Google Scholar]
- 31.Gerber B, Tinguely C, Bovin NV, Rieben R, Carrel T, Nydegger UE. Differences between synthetic oliogosaccharide immunoabsorbents in depletion capacity for xenoreactive anti-Gal α1,3Gal antibodies from human serum. Xenotransplantation. 2001;8:106–14. doi: 10.1034/j.1399-3089.2001.00064.x. [DOI] [PubMed] [Google Scholar]
- 32.Weintraub LJ, Holgersson J. Multivalent Gal α1,3 Gal substitution makes recombinant mucin-immunoglobulins efficient absorbers of anti-pig antibodies. Xenotransplantation. 2003;10:149–63. doi: 10.1034/j.1399-3089.2003.01144.x. [DOI] [PubMed] [Google Scholar]
- 33.Dehoux JP, Hori S, Talpe S, Bazin H, Latinne D, Soares MP, Gianello P. Specific depletion of preformed IgM natural antibodies by administration of anti-mu monoclonal antibody suppresses hyperacute rejection of pig to baboon renal xenografts. Transplantation. 2000;70:935–46. doi: 10.1097/00007890-200009270-00011. [DOI] [PubMed] [Google Scholar]
- 34.Sato K, Takigami K, Miyatake T, Czismadia E, Latinne D, Bazin H, Bach FH, Soares MP. Suppression of delayed xenograft rejection by specific depletion of elicited antibodies of the IgM isotype. Transplantation. 1999;68:844–54. doi: 10.1097/00007890-199909270-00018. [DOI] [PubMed] [Google Scholar]
- 35.McMorrow IM, Buhler L, Treter S, et al. Modulation of the in vivo primate anti-Gal response through administration of anti-idiotypic antibodies. Xenotransplantation. 2002;9:106–14. doi: 10.1034/j.1399-3089.2002.1o028.x. [DOI] [PubMed] [Google Scholar]
- 36.Buhler L, Treter S, McMorrow I, et al. Injection of porcine anti-idiotypic antibodies to primate anti-Gal antibodies leads to active inhibition of serum cytotoxicity in a baboon. Transplant Proc. 2000;32:1102. doi: 10.1016/s0041-1345(00)01144-1. [DOI] [PubMed] [Google Scholar]
- 37.Koren E, Milotec F, Neethling F, Mirna F, Kobayashi R, Taniguchi S, Cooper DKC. Monoclonal antiidiotypic antibodies neutralize cytotoxic effects of anti-Gal antibodies. Transplantation. 1996;62:837–43. doi: 10.1097/00007890-199609270-00023. [DOI] [PubMed] [Google Scholar]
- 38.Schussler O, Genevaz D, Latremouille C, Goussev N, Kaveri S, Glotz D. Intravenous immunoglobulins for therapeutic use contain anti-idiotypes against xenophile antibodies and prolong discordant graft survival. Clin Immunol Immunopathol. 1998;86:183–91. doi: 10.1006/clin.1997.4484. [DOI] [PubMed] [Google Scholar]
- 39.Lang J, Zhan J, Xu L, Yan Z. Identification of peptide mimetics of xenoreactive alpha-Gal antigenic epitope by phage display. Biochem Biophys Res Commun. 2006;344:214–20. doi: 10.1016/j.bbrc.2006.03.112. [DOI] [PubMed] [Google Scholar]