Abstract
CD22 is an inhibitory coreceptor of the B-cell receptor (BCR), and plays a critical role in establishing signalling thresholds for B-cell activation. Like other coreceptors, the ability of CD22 to modulate B-cell signalling is critically dependent upon its proximity to the BCR, and this in turn is governed by the binding of its extracellular domain to α2,6-linked sialic acid ligands. Manipulation of CD22 ligand binding in various experimental settings has profound effects on B-cell signalling, but as yet there is no complete model for how ligand binding in vivo controls normal CD22 function. Several elegant studies have recently shed light on this issue, although the results appear to suggest two mutually exclusive models for the role of ligand binding; in either promoting or inhibiting, CD22 function. We shall therefore discuss these results in detail, and suggest possible approaches by which these conflicting experimental findings might be reconciled. We shall also consider a second important issue in CD22 biology, which relates to the role that defects in this receptor might play in mediating autoimmune disease. We review the current evidence for this, and discuss the importance of genetic background in modifying CD22 function and predisposition to autoimmunity.
Keywords: autoimmunity, B cell, CD22, inhibitory receptor, sialic acid
Introduction
CD22 is an inhibitory coreceptor of the B-cell receptor (BCR) and, like other coreceptors, is required to modulate the antigen receptor signal in response to cues from the local microenvironment.1 This is essential to ensure that an appropriate humoral response is mounted against pathogens, but that reactivity to self antigens and autoimmunity is avoided.2–4 To fine-tune the BCR signal, and to integrate an extensive variety of environmental signals, B cells express a broad repertoire of activatory and inhibitory coreceptors on their surface.5,6 These molecules possess immunoreceptor tyrosine-based activation (ITAM) and inhibition (ITIM) motifs in their intracellular domains, which serve as scaffolds to recruit signalling molecules that either augment or inhibit the BCR signal. The extent to which these receptors are associated with the BCR, and hence their capacity to modulate its signal, is governed by the interactions of their extracellular domains with as yet incompletely defined sets of ligands. These ligands can exist in cis on the same cell surface, in trans on adjacent cell surfaces, in a soluble form, or bound to cell-associated antigen, for example immunoglobulin G (IgG) in the case of FcγRIIb (CD32b),7 or complement in the case of the CD21/CD19 coreceptor complex.2
CD22 was originally identified as a B-cell-associated adhesion protein that appeared to function in the regulation of B-cell activation.8–13 It is a member of the sialic acid-binding immunoglobulin-like lectin (Siglec) family of adhesion molecules,14 and binds specifically to glycans that possess sialic acid, attached in α2,6-linkage to an underlying β1,4-linked galactose residue (α2,6Sia).15,16 This is a common structure on N-linked glycans and is abundantly expressed on the surface of many cells, including erythrocytes, monocytes, cytokine-activated endothelial cells, T cells and B cells.17–19 In addition, α2,6Sia residues are present on some soluble plasma proteins such as haptoglobin and IgM, and recombinant CD22 molecules have been reported to bind these glycoproteins.18 Which of these ligands are physiologically important, and how binding to them is transduced to effect changes in BCR signalling, is not yet well understood. The focus of this review is therefore to consider the recent advances that have furthered our understanding of the role that ligand binding plays in controlling CD22 function at a cellular level. We shall also consider the evidence that defects in CD22 mediate autoimmune disease, and the importance of genetic background in modulating these effects.
CD22 structure and function
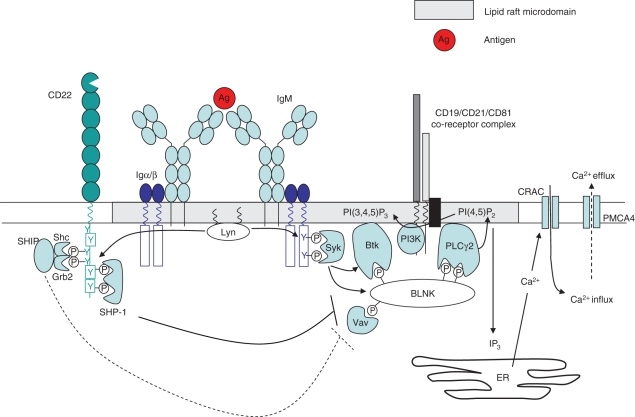
CD22 is a type I membrane protein with molecular weight 140 000 that is expressed at low levels on pre- and immature B cells, maximally on mature B cells,20 and ultimately downregulated on plasma cells.21 The extracellular portion of CD22 comprises seven immunoglobulin domains, the most distal of which is a V-set immunoglobulin domain, and is responsible for binding α2,6Sia ligands.22–24 Within this domain, two arginine residues (R130 and R137 in mouse) are required for α2,6Sia-binding, and mutation of these residues completely abrogates this interaction.25 The intracellular portion of murine CD22 contains six tyrosine residues, three of which (Y762, Y822 and Y842) exist within ITIM motifs.26 Upon cross-linking of the BCR by antigen, the CD22 that is associated with it is rapidly phosphorylated.27 This has been demonstrated to require the activity of Lyn,28,29 an src family protein tyrosine kinase (PTK) that is concentrated in lipid rafts, and is also thought to be in part responsible for phosphorylating the Igα (CD79a) and Igβ (CD79b) chains of the BCR complex.30 Following tyrosine phosphorylation of CD22, docking sites are formed for a number of SH2-domain-containing proteins, including the protein tyrosine phosphatase SHP-1,31 which acts to dephosphorylate components of the BCR signalling cascade to effect a dampening of the BCR signal. Targets of SHP-1 appear to include Vav-1, CD19 and SLP65/BLNK,32–34 all of which are positively involved in Ca2+ signalling (Fig. 1). Another potential target of SHP-1 is the plasma membrane calcium-ATPase (PMCA4), which promotes Ca2+ efflux and attenuation of the BCR signal.35 Both CD22 and SHP-1 are reported to associate with PMCA4 following CD22 phosphorylation, resulting in enhanced PMCA4-mediated Ca2+ efflux, and a further dampening of the BCR signal.
Figure 1.
Upon B-cell receptor (BCR) cross-linking and translocation to lipid rafts, Lyn phosphorylates the immunoreceptor tyrosine-based activation motif tyrosine residues of immunoglobulin α/β. This creates docking sites for other protein tyrosine kinase such as Syk, which in turn phosphorylate and recruit the adaptor molecule BLNK (B-cell linker protein). BLNK forms a scaffold for the association of numerous signalling components, including Vav-1, BTK and phospholipase Cγ2 (PLCγ2). Many of these proteins also contain pleckstrin homology domains, and are stabilized in this complex by interaction with the phospholipid PI(3,4,5)P3 [produced by phosphoinositide 3-kinase (PI3K)]. Calcium induction upon BCR ligation is brought about by PLCγ2, which converts PI(4,5)P2 to I(1,4,5)P3 (IP3) and diacylglycerol (DAG). IP3 promotes calcium release from intracellular stores, which in turn triggers an influx of calcium through the opening of Ca2+-release-activated channels (CRACs), and activation of the nuclear factor κB, nuclear factor of activated T cells and extracellular signal-regulated kinase signalling pathways. Calcium efflux is mediated by the plasma membrane Ca2+-ATPase (PMCA4). CD22 serves to inhibit the BCR signal at several points in this signalling cascade. Following phosphorylation of its immunoreceptor tyrosine-based inhibitory motif residues by Lyn, CD22 provides a docking site for the recruitment of SHP-1. SHP-1 acts to dephosphorylate and inactivate Vav-1, BLNK and CD19 (accompanied by a reduction in PI3K activity) to effect a dampening of the BCR signal. SHP-1 also promotes calcium efflux through activation of PMCA4.
In addition to SHP-1, CD22 recruits a number of other SH2-domain-containing proteins, including Syk, phospholipase Cγ2 (PLCγ2), phosphoinositide 3-kinase (PI3K), and SHIP as a quaternary complex with Grb2 and Shc.36–39 While Grb2 and SHIP may act to further inhibit Ca2+ mobilization by the BCR,40,41 the role of the other molecules that are recruited is unclear. Since these molecules are more typically involved in augmenting the BCR signal, and because CD22 possesses two ITAM motifs in its cytoplasmic tail, it has been suggested that under some circumstances CD22 may provide positive signals and promote B-cell survival.42 Consistent with this prediction, it was observed that a number of anti-CD22 antibodies could augment anti-immunoglobulin-induced proliferation,9 although this is likely to reflect sequestration of CD22 away from the BCR, rather than positive signalling per se.31 More recently, however, it has been observed that even in the absence of BCR ligation, CD22 cross-linking results in c-Jun N-terminal kinase (JNK) signalling and proliferation of human tonsillar B cells.42,43 Therefore, while CD22 appears to inhibit signals that emanate from the BCR, it might also initiate positive signals when ligated to itself, or perhaps other surface receptors.44
In spite of its potential to provide both positive and negative signals, four independent lines of CD22-deficient mice,20,45–47 have demonstrated that CD22 functions predominantly as an inhibitory receptor in vivo. In all of the mouse lines generated, CD22-deficiency resulted in elevated Ca2+ mobilization in response to BCR stimulation in vitro, which is consistent with CD22’s role as an inhibitory coreceptor. Strikingly, while B-cell development and the number of B cells present in the spleens of these mice were normal, peripheral B cells expressed considerably less surface IgM (sIgM) than their wild-type counterparts. This is reminiscent of the anergic phenotype observed in B cells from anti-hen egg lysozyme (anti-HEL) BCR transgenic mice in the presence of soluble HEL,48 and suggests that CD22-deficient B cells have undergone chronic activation in vivo. In support of this notion, CD22-deficient B cells also express high levels of major histocompatibility complex (MHC) class II. In response to BCR stimulation, CD22-deficient B cells predominantly undergo apoptosis,20,49 which explains their reduced proliferation in response to BCR stimulation in vitro, and enhanced turnover in vivo.20,46,49 Interestingly, the effects of CD22 deficiency on proliferation and apoptosis can be rescued by coligation with anti-CD40, suggesting that CD22-deficient cells are not irreversibly programmed to die, and may be rescued by T-cell help.49 Consistent with this, the ability of CD22-deficient mice to mount T-dependent immune responses remains intact, and subsequent germinal centre formation occurs normally.49
In addition to alterations in the peripheral B-cell phenotype, CD22-deficient mice have severely reduced numbers of marginal zone (MZ) B cells and consequently exhibit drastically impaired immune responses to T-independent antigens.20,46,50 It has been suggested that loss of MZ B cells in these mice occurs as a consequence of altered BCR signalling during development because there is good evidence to suggest that the maturation of transitional B cells into one of the three mature B-cell subsets [follicular (B-2) B cells, marginal zone (MZ) B cells, or peritoneal (B-1) B cells] is driven, at least in part, by differences in BCR signal strength (reviewed in detail elsewhere51,52). Some controversy remains as to the exact nature of the signal required to generate the three B-cell subsets,53,54 but evidence from Aiolos−/− mice suggests that a strong BCR/Btk signal disrupts MZ B-cell development.55 In a similar fashion, the enhanced BCR signal afforded by CD22 deficiency might culminate in the loss of MZ B cells. An alternative possibility, however, is that ligand binding by CD22 is required for the homing or retention of MZ B cells in the spleen. This hypothesis is supported by the loss of marginal zone B cells in ligand-deficient mice (discussed below), where BCR signalling is in fact much reduced.56–58
CD22 ligands
The ability of CD22 to inhibit antigen-induced signalling depends upon its proximity to the BCR.31 This is expected to be largely determined by binding of its extracellular domain to α2,6Sia,59 and consequently much effort has gone into identifying the physiologically relevant ligands. CD22 may interact with ligands in cis on the B-cell surface, in trans on the surface of other cells, on soluble glycoproteins, or attached to cell-associated antigen. Since the concentration of α2,6Sia on the B-cell surface is estimated to be in the region of 25–30 mm,60 around 100-fold higher than the Kd of CD22 for α2,6Sia (0·1–0·3 mm15,61,62), most CD22 is predicted to interact with glycoprotein ligands on the same cell. This calculation appears to be correct, because CD22 on most B-cell surfaces is ‘masked’, and unable to bind exogenous sialoside probes unless the cells are first pretreated with sialidase or periodate.63,64 Interestingly, however, under some circumstances CD22 appears to be ‘unmasked’. This has been reported upon cellular activation,64 and on certain B-cell subsets, particularly marginal zone and transitional B cells.65 The significance of this unmasking of CD22 is not yet fully understood, but presumably promotes interaction of CD22 with ligands in trans, or with a distinct subset of cis ligands, perhaps including the BCR.
Consistent with the prediction that CD22 interacts predominantly with cis ligands, the B-cell glycoproteins sIgM and CD45 have been identified by coimmunoprecipitation as prominent CD22 binding partners.23,27,66 However, mutation of the CD22 ligand-binding domain does not affect the interaction of CD22 with these glycoproteins,67 and hence it must be concluded that interaction is mediated by protein–protein contacts. More recently, an elegant approach employing photo-affinity cross-linking of glycan ligands has established that in situ, CD22 exists primarily as homomultimeric complexes in clathrin-coated pit microdomains.68 No discernable binding to CD45, sIgM, PMCA4 or other putative cis ligands was reported, suggesting that if CD22 does interact with these molecules, the interaction is mediated entirely by protein–protein contacts.
In addition to identifying cis ligands of CD22, attempts have been made to establish the physiologically relevant trans ligands. CD22 was first identified as an adhesion molecule,11,17 and COS cells expressing recombinant CD22 bind to lymphocytes and monocytes in a sialic acid-dependent manner.17 Approaches to identify specific ligands have involved immunoprecipitation of T-cell lysates with CD22-Fc fusion proteins, and have identified CD45 as a prominent ligand on T cells.69,70 A more recent study has, however, demonstrated that adhesion of B and T cells, and redistribution of CD22 to sites of cell–cell contact, does not require CD45,60 suggesting that other T-cell ligands may exist. CD22 is also reported to bind soluble proteins, such as the α2,6-sialylated serum glycoproteins IgM and haptoglobin,18 although the physiological relevance of these interactions remains unknown because these glycoconjugates presumably bind CD22 with low avidity, and would be poorly able to compete with clustered cell-associated ligands.
Effects of ligand binding on CD22 function
The cis ligands appear to be important modulators of CD22 activity, as demonstrated by cell lines expressing mutant forms of CD22 that lack sialic-acid-binding activity.71 These cells were hyperresponsive to BCR stimulation, resulting in elevated Ca2+ responses and increased CD22 tyrosine phosphorylation and SHP-1 recruitment. Analogous results were obtained in a different system, which used a synthetic sialoside to abrogate ligand binding in primary cells,72 and suggest that ligand binding in cis is required for the inhibitory function of CD22. A number of studies have also highlighted the importance of interactions that CD22 makes in trans, and their potential to mediate cross-talk between cells. A recent study has demonstrated that B-cell activation by antigen displayed on a target cell is depressed if the target coexpresses α2,6Sia glycoconjugates.73 Since α2,6-sialylation is largely a feature of higher eukaryotes, it has been suggested that interaction of CD22 in this way may serve to dampen the B-cell response to self antigens. Furthermore, trans interactions may have important implications for T-cell signalling because ligation of T cells by CD22–Fc fusion proteins is sufficient to alter CD3-induced T-cell activation,69,74 and CD22 blockade in mixed lymphocyte costimulatory assays results in reduced T-cell proliferation.42 However, it has recently been demonstrated that CD22 is expressed at low, but functionally significant, levels on T cells75 so the interpretation of these studies must now consider the role of T-cell-intrinsic CD22 ligation.
A better established role for CD22 trans interactions appears to be in controlling certain aspects of B-cell adhesion and migration. CD22 ligands are expressed on sinusoidal endothelial cells of murine bone marrow (but not other tissues) and interaction of CD22 has been implicated in the homing of recirculating mature (IgMlo IgDhi) B cells to the bone marrow.76 In support of this hypothesis, CD22-deficient mice have decreased numbers of recirculating B cells,20,47 and decreased numbers of IgM-secreting plasma cells in the bone marrow.76 Repopulation of RAG−/− bone marrow by CD22-deficient B cells was only half as efficient as repopulation with wild-type cells20 and, furthermore, a reduction in recirculating cells can be achieved by blocking CD22-α2,6Sia interactions, either by previous injection with anti-CD22 antibodies, or CD22–Fc fusion proteins.76 Interestingly, subsets of mature B cells with unmasked CD22 are enriched two- to fivefold in the bone marrow,77 suggesting that under certain circumstances, interactions of CD22 in cis may be modulated to promote ligation in trans. While unmasking of CD22 is required for its binding to low-affinity sialoside probes,63 binding of higher-affinity multivalent probes,78 or redistribution of CD22 to sites of cell–cell contact,60 does not require previous unmasking. This therefore suggests the existence of a dynamic equilibrium, in which CD22 can switch readily between cis and trans interactions, depending on their relative affinity/avidity. Nevertheless, deliberate masking/unmasking of CD22 under different physiological circumstances allows this equilibrium to be skewed in favour of one interaction or the other.
To better understand the physiological roles that CD22 ligands might play in modulating its function, it has proved valuable to examine mouse lines that are either deficient in CD22 ligand, or that express mutant forms of CD22 that lack ligand-binding activity. The results and implications of these studies are discussed below, and many of the key phenotypic features of the mouse strains used to study CD22 function are summarized in Table 1.
Table 1.
Key phenotypic features of mice with genetically altered CD22 expression and function
| Key phenotypic features | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B-cell populations | B-cell surface phenotype | Signalling (anti-IgM) | Mouse | |||||||||||
| Mouse strain | MZ | B1 | Recirc. | sIgM | MHC II | CD22 | Ca2+ | Prolif. | Apop. | Serum IgM | Td | Ti | AutoAb | Refs |
| CD22−/− (129 × C57BL/6) (derived from 129 ES cells) | ↓ | ↑ | ↓ | ↓ | ↑ | n.a. | ↑ | ↓ | ↑ | ↑ | ↔ | ↓ | Yes (1 of 3 lines) | 45–47 |
| CD22−/− (C57BL/6) (derived from C57BL/6 ES cells) | ↓ | ↔ | ↓ | ↓ | ↑ | n.a. | ↑ | ↔ | ↑ | ↑ (> 5 mo) | ↔ | ↓ | No | 20 |
| CD22Δ1–2 (CD22 lacking the two N-terminalligand-binding Ig domains) | ↓ | ↑ | ↓ | ↓ | ↑ | ↓ | ↔ | ↓ | n.d. | n.d. | n.d. | n.d. | n.d. | 89 |
| CD22AA (CD22 with key ligand-binding Argresidues mutated to Ala) | ↓ | ↑ | ↓ | ↓ | ↑ | ↓ | ↔ | ↓ | n.d. | n.d. | n.d. | n.d. | n.d. | 89 |
| ST6Gal I−/− (cannot produce α2,6Sia ligands) | ↓ | ↔ | ↓ | ↓↓ | ↔ | ↓ | ↓ | ↓ | n.d. | ↓ | ↓ | ↓ | n.d. | 56–58,82 |
| ST6Gal I−/− × CD22−/− | ↓ | ↔ | ↓ | ↓↓ | n.d | n.a. | ↑ | ↓ | n.d. | n.d. | n.d. | n.d. | n.d. | 56,57,82 |
| CD22a.B6 (congenic of CD22a allotype from NZB,on a C57BL/6 background) | ↔ | n.d. | n.d. | ↓ | ↑ | ↓ | ↔ | ↓ | n.d. | n.d. | n.d. | n.d. | Yes (Coombs Ab) | 103,109 |
A summary of mouse lines generated to study CD22 function and their key phenotypic features.
Td, T-dependent immune response; Ti, T-independent immune response; n.a., not applicable; n.d., not done.
Ligand-deficient mice
Ligand-deficient mice are those lacking ST6Gal I sialyltransferase (ST6Gal I), the enzyme that is solely responsible for producing the α2,6Sia terminus on N (and some O) glycans.79,80 Somewhat surprisingly, given that in vitro studies would predict that α2,6Sia is required for CD22-mediated signal inhibition,71,72 these mice have a hyporesponsive B-cell phenotype.58 In response to BCR stimulation, ST6Gal I-deficient B cells exhibit drastically reduced Ca2+ mobilization, and show reduced proliferation in response to CD40, BCR and lipopolysaccharide-induced signals.58 Interestingly, this can be reversed in the presence of interleukin-4, perhaps as the result of the inhibitory effects of this cytokine on CD22 function.81 Similar to CD22-deficient B cells, expression of sIgM is reduced on ST6Gal I-deficient B cells, but this does not appear to be a result of chronic activation because these cells do not upregulate MHC class II, CD86 or other activation markers.58 Rather, this appears to be a consequence of enhanced CD22-dependent sIgM endocytosis,82 which will be discussed in more detail below. B-cell development in ST6Gal I-deficient mice is largely normal, with the exception of MZ B cells, numbers of which are reduced to a similar extent to that seen in CD22-deficient mice.56,57 Consistent with B-cell hyporesponsiveness and a lack of MZ B cells, ST6Gal I-deficient mice are severely impaired in their ability to mount both T-dependent, and T-independent, immune responses.58
Of course, plausible explanations for these results are that sialic acid deficiency affects the function of other glycoproteins, or that other sialic acid-binding proteins, such as Siglec-G,83 might influence the B-cell phenotype. However, in ST6Gal I × CD22 double-deficient mice, BCR-induced signalling and proliferation is restored to CD22-deficient levels,56,57 thereby specifically implicating CD22 in mediating B-cell suppression in the absence of ligand. This enhanced suppression appears to correlate with increased coclustering of CD22 and sIgM, and altered microdomain localization of these two receptors.56,82 In wild-type cells, CD22 exists in clathrin-rich domains, presumably as homomultimers,68 and gives rise to a punctate staining pattern by fluorescence microscopy.56 In ligand-deficient cells, however, its expression pattern is more dispersed, consistent with a shift towards heteroypic, rather than homotypic, interactions.82 Coclustering of CD22 and sIgM is increased twofold in the absence of ligand, and this is accompanied by a significant increase in the association of sIgM with mixed clathrin- and GM1-rich membrane microdomains, and a reduction of sIgM associated with neither marker.56,82 This redistribution is not seen in ST6Gal I × CD22 double-deficient cells, indicating that association of sIgM with mixed clathrin/GM1 domains results from the disruption of CD22 cis interactions.56 Therefore, interaction of CD22 with α2,6Sia ligands not only determines its ability to inhibit BCR signalling, but also influences the localization and trafficking of the BCR itself.
Alterations in the availability of CD22 ligand, and the concomitant changes in sIgM microdomain localization, appear to have at least two important consequences for BCR signalling. The first relates to the fact that in the absence of α2,6Sia, sixfold more SHP-1 is constitutively associated with CD22 in resting cells. This occurs without detectable increases in CD22 phosphorylation56,82 and, more strikingly, occurs even in the absence of Lyn.29 It is not yet clear how disruption of CD22 cis interactions elicits this effect, but one possibility is that enhanced interaction with sIgM alters the conformation of CD22, such that it binds SHP-1 more readily. This increased association of SHP-1 with CD22 correlates with specific reductions in the phosphorylation state of various BCR signalling molecules both before and after BCR stimulation, and hence is likely to represent an important mechanism for B-cell suppression in ST6Gal I-deficient mice.
The second effect of altered microdomain localization is an enhanced rate of sIgM endocytosis,82 which would be expected to attenuate BCR signalling.84 Since sIgM half-life is restored to wild-type levels in ST6Gal I × CD22 double-deficient cells, it appears that CD22 is directly responsible for recruitment of the BCR to clathrin-containing domains for internalization. Consistent with this, CD22 is reported to interact with the clathrin-coated pit adapter protein AP50, and undergoes clathrin-dependent endocytosis when ligated by antibodies, or high-affinity multivalent sialosides.78,85,86 Interaction of CD22 with AP50 occurs via YxxΦ (where Φ is a hydrophobic residue) sorting motifs contained within its ITIM sequences,87 and is abrogated by tyrosine phosphorylation. Interaction of CD22 with AP50 therefore occurs in a reciprocal fashion to SHP-1 binding,88 suggesting a level at which internalization of CD22, and perhaps of sIgM, might be regulated.
Non-ligand-binding CD22 gene-targeted mice
Complementary studies have been conducted using gene-targeted mice that express mutated forms of CD22 that cannot bind ligand: either CD22Δ1-2, which lacks the two N-terminal immunoglobulin domains, or CD22AA, in which the critical ligand-binding residues, R130 and R137, have been replaced by alanine.89 CD22Δ1-2 and CD22AA B cells express lower sIgM and CD22 but, in contrast to ST6Gal I knockout cells, this appears to represent a chronically activated phenotype, because MHC class II expression is also elevated. Consistent with this, CD22Δ1-2 and CD22AA cells show reduced proliferation in response to BCR stimulation in vitro, but higher CD40-induced proliferation, and a higher rate of B-cell turnover in vivo. Therefore, rather than resembling the ST6Gal I-deficient mice, as might have been expected, these gene-targeted mice appear to have a hyperresponsive phenotype that is more reminiscent of the CD22 knockout. In fact, in many regards these gene-targeted mice behave identically to CD22 knockouts, with the exception that calcium signalling and protein tyrosine phosphorylation in response to BCR stimulation are equivalent to wild type. These are interpreted as ‘ligand-independent’ CD22 functions by the authors, but others have suggested that they might merely represent a failure to detect small, but physiologically relevant, differences in calcium flux because of the saturating quantities of anti-IgM used in these in vitro experiments.59 Overall, the evidence therefore suggests that the inhibitory function of CD22 is impaired in CD22AA and CD22Δ1-2 B cells, such that these cells become chronically activated in vivo. This explanation is in agreement with many in vitro studies of CD22 function,71,72 and is consistent with a model in which association of CD22 with the BCR, and subsequent signal inhibition, requires α2,6Sia ligand-binding activity. In support of this hypothesis, it has recently been demonstrated that disruption of α2,6Sia interactions using sialoside inhibitors prevents cocapping of CD22 with the BCR upon B-cell activation.90
A model for CD22 function
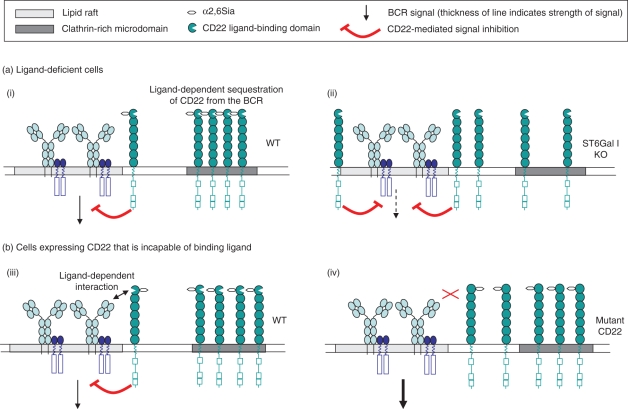
At first glance, the simplest interpretations of the phenotypes of these two mouse models are not easily reconciled. The CD22AA and CD22Δ1-2 mice suggest that ligand binding is required for the inhibitory effects of CD22, while the ST6Gal I knockout would imply that ligand binding actually relieves CD22-mediated inhibition (Fig. 2). In the past, discrepancies between these two models have been partly attributed to the fact that these mice have mixed 129/Sv × C57BL/6 genetic backgrounds, and may therefore carry different epistatic modifiers of CD22 function. While this is undoubtedly an important consideration, it is difficult to envisage how small genetic differences could give rise to two such disparate phenotypes. There are, however, many incompletely understood aspects of CD22 biology that may provide alternative avenues through which these controversies may be reconciled, and these are discussed below.
Figure 2.
(a) Studies in ligand-deficient mice suggest that CD22 is normally sequestered away from the B-cell receptor (BCR) in clathrin-rich domains (i). Sequestration is mediated by homotypic interactions with other CD22 molecules, via α2,6-linked sialic acid (α2,6Sia) binding. In the absence of ligand, CD22 is released from these constraints, resulting in greater association with the BCR and suppression of signalling (ii). (b) Studies in gene-targeted mice (that express CD22 that cannot bind ligand) suggest that CD22 normally makes α2,6Sia-dependent contacts with the BCR (or other BCR-associated proteins) to mediate signal inhibition (iii). When ligand binding is precluded, CD22 cannot interact with the BCR, and the BCR signal is not effectively inhibited (iv).
It would seem likely that in wild-type cells, α2,6Sia-mediated interactions would work in conjunction with other interactions, such as those mediated by protein–protein contacts. CD22 might interact with other surface proteins, such as CD45 and sIgM,67 or perhaps with intracellular adaptor proteins such as AP50, which might tether CD22 to clathrin-containing regions of the membrane. A further possibility is that CD22 might be targeted to certain membrane domains through interaction with tetraspanins; either directly, or through its reported carbohydrate-independent interaction with members of the Ly-6 superfamily.91 In addition to this complex web of cis interactions, CD22 localization in vivo is likely to be influenced through interaction with trans ligands on neighbouring cells, and so will be exquisitely sensitive to the B cell’s microanatomical environment. These important interactions are disrupted when cells are studied in vitro so the roles of these factors in regulating CD22 activity cannot easily be characterized.
Other complications in interpreting these studies arise from our incomplete understanding of B-cell signalling pathways. First, there are a number of reports to suggest that CD22 elicits both positive and negative signals, and we are therefore likely to observe only the net effect of these. Indeed analysis of the phosphorylation kinetics of various positive and negative signalling molecules is consistent with these dual signalling roles.32,44 Second, differences in B-cell signalling ex vivo presumably reflect adaptations to signals that have been received during B-cell ontogeny. In the CD22AA and CD22α1-2 mice, for example, we are studying a post-activation phenotype, in which many alterations in CD22 distribution and function may have already occurred to compensate for enhanced signalling. Furthermore, it is likely that other BCR signalling components will have been up- or downregulated to compensate for altered signalling thresholds, and therefore functional studies in vitro are unlikely to be a true reflection of the situation in vivo.
A final outstanding controversy in understanding the function of CD22 relates to its ability to regulate BCRs of different isotypes. Early studies of chimeric BCRs expressed in B-cell lines suggested that CD22 could not inhibit signals from the IgG BCR, and that this might explain increased B-cell proliferation in the memory response.92,93 More recently, however, studies in primary B cells have demonstrated that CD22 can regulate IgG BCR signalling, highlighting the potential importance of this coreceptor in regulating not only the primary response, but also memory responses.94,95
In summary, while many of the intricacies of the role of CD22 in vivo await further investigation, it is clear that this receptor plays an important role in establishing thresholds for B-cell activation, and that alterations in α2,6Sia-binding have profound effects on B-cell phenotypes.
Autoimmunity
Given its role as a negative regulator of BCR signalling, it seems likely that defects in CD22 might predispose to autoimmunity, perhaps in a similar fashion to FcγRIIb.6 While CD22-deficient mice do not develop overt autoimmune disease, one line of CD22-deficient mice is reported to develop autoantibodies; initially anti-DNA antibodies of the IgM isotype at around 5 months of age, and then high-affinity, isotype-switched autoantibodies by 8 months of age.45,96 Furthermore, it has been demonstrated that in the presence of the Y-linked autoimmune accelerator (Yaa) locus, CD22 heterozygosity is sufficient to promote the generation of IgG anti-DNA antibodies.97 However, because spontaneous autoantibody development has so far only been observed in one line of CD22-deficient mice, it is likely that loss of tolerance through CD22 deficiency is subject to modulation by other loci, which differ according to the genetic background on which the mice were derived. The CD22-deficient line in question was generated using 129/Sv (129) embryonic stem cells and, despite extensive backcrossing, still retains a significant portion of 129 chromosome 7 in the region surrounding Cd22.97 It is pertinent to note, therefore, that mice derived directly from C57BL/6 (B6) embryonic stem cells do not generate autoantibodies,20 and it is therefore likely that an important 129-derived modifying locus lies close to Cd22 on proximal chromosome 7. It has recently been demonstrated that a number of 129-derived chromosome intervals predispose to the development of autoimmunity when expressed on a B6 genetic background.98,99 For example, much of the autoimmune-prone phenotype of C1q and SAP (serum amyloid P component) knockout mice can be attributed not to these targeted mutations, but to the presence of a 129-derived interval that surrounds these genes on distal chromosome 1.100,101 Recently, a second interval has been identified on proximal chromosome 7, which predisposes to the development of glomerulonephritis in C1q-deficient mice.99 Since a considerable portion of this interval is also present in 129 × B6 CD22-deficient mice, it may synergize with the effects of CD22 deficiency, or indeed act alone, to promote autoantibody generation in these mice.
On a genetic background where other loci predispose to autoimmunity, one might predict that B-cell dysregulation through defects in CD22 would promote the development of autoantibodies and accelerate disease progression. It is interesting, therefore, that a number of autoimmune-prone strains of mice possess CD22 polymorphisms, and that several studies have associated the region of chromosome 7 that contains CD22 with the development of autoantibodies, lupus-like glomerulonephritis and autoimmune haemolytic anaemia.102–107 Three major polymorphic forms of Cd22 (Cd22a, -b and -c) have been identified by comparing inbred mouse strains. In general, the Cd22b form is associated with non-autoimmune-prone mice, whereas the Cd22a (and closely related Cd22c) form is present in lupus-prone strains.10,108 The CD22a protein differs from CD22b in that it carries a six-amino-acid deletion, plus several point mutations, in the N-terminal immunoglobulin domain. These mutations map to a region that is distinct from the ligand-binding site,25 so are unlikely to affect this function, but result in the loss of an N-linked glycosylation site, which may adversely affect homodimerization.108 In addition to these alterations in the CD22-coding sequence, expression of the Cd22a form is dysregulated as a result of a short interspersed nucleotide element insertion in the second intron.97 This results in abnormal processing of Cd22 mRNA, producing aberrant CD22 molecules with substantial deletions in the ligand-binding domain, which have recently been demonstrated to reduce ligand binding.109 Furthermore, while CD22 is normally upregulated on LPS stimulation, this occurs only half as efficiently in Cd22a mice, because of the production of aberrant transcripts containing premature stop codons.97
The effects of the Cd22a polymorphism have been neatly dissected from an autoimmune-prone genetic background by the generation of a B6.Cd22a congenic mouse.109 Compared to Cd22b cells, Cd22a cells exhibit a chronically activated phenotype, much like the gene-targeted mice (CD22AA and CD22Δ1-2) in which CD22 ligand binding is abolished.89 Like CD22AA and CD22Δ1-2 B cells, Cd22a cells have low levels of sIgM, elevated MHC class II, and reduced BCR-induced proliferation. Interestingly, despite this chronically activated phenotype, no difference was observed in BCR-induced Ca2+ flux, exactly paralleling the gene-targeted mice.
In the situations described above, loss of CD22 function is associated with autoimmunity, presumably as a consequence of altered BCR signalling thresholds and B-cell dysregulation. In lyn-deficient mice, reduced BCR signalling thresholds and B-cell hyperactivity predispose to the development of autoimmunity through spontaneous B-cell activation, class switch recombination and antibody hypersecretion.110,111 In a similar fashion, altered BCR signalling in CD22-deficient mice might have similar effects, especially when combined with defects in other components of the inhibitory pathway, for example haploinsufficiency in Lyn and SHP-1,28 or with as yet unidentified 129-derived genes.49,89 Altered BCR signalling in CD22-deficient mice might also predispose to autoimmunity by promoting the generation of B-1 B cells, which appear to require strong BCR signals for their development.52 B-1 B cells have been implicated in the pathogenesis of a number of autoimmune diseases,112 and are found in increased numbers in two lines of CD22-deficient mice.45,47
As yet, there is no convincing link between CD22 polymorphisms in humans, and the development of autoimmunity. One study has identified an association of the Q152E polymorphism with systemic lupus erythematosus, but this result was statistically insignificant.113 Another study links reduced CD22 expression on B cells with cutaneous systemic sclerosis,114 similar to the flaky-skin mouse model of this disease.115
Conclusions
Little doubt now remains that CD22 acts predominantly as an inhibitory coreceptor of the BCR, but much controversy still surrounds the mechanisms that modulate its function, and the role of α2,6Sia-binding activity. Nevertheless, it is clear that CD22 plays a key part in establishing the BCR signalling threshold, and alterations in CD22 activity have profound implications for the generation of B-cell hypo- or hyper-activity. In turn, changes in CD22 function appear to influence predisposition to autoimmunity, although the effects of CD22 can be modulated by other genetic loci. Loci that modulate, or exacerbate, the effects of CD22 deficiency might be predicted to affect B-cell signalling, the induction of tolerance, or even the degree to which T cells provide help to overcome chronic BCR stimulation. A challenge for the future therefore lies in identifying these epistatic loci, which will bring a better understanding of the molecular mechanisms involved in the control of signalling thresholds, and the generation of B-cell tolerance.
Abbreviations
- α2,6Sia
α2,6-linked sialic acid
- sIgM
cell surface immunoglobulin M
- HEL
hen egg lysozyme
References
- 1.Cyster JG, Goodnow CC. Tuning antigen receptor signaling by CD22: integrating cues from antigens and the microenvironment. Immunity. 1997;6:509–17. doi: 10.1016/s1074-7613(00)80339-8. [DOI] [PubMed] [Google Scholar]
- 2.Fearon DT, Carroll MC. Regulation of B lymphocyte responses to foreign and self-antigens by the CD19/CD21 complex. Annu Rev Immunol. 2000;18:393–422. doi: 10.1146/annurev.immunol.18.1.393. [DOI] [PubMed] [Google Scholar]
- 3.Rickert RC. Regulation of B lymphocyte activation by complement C3 and the B cell coreceptor complex. Curr Opin Immunol. 2005;17:237–43. doi: 10.1016/j.coi.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Grimaldi CM, Hicks R, Diamond B. B cell selection and susceptibility to autoimmunity. J Immunol. 2005;174:1775–81. doi: 10.4049/jimmunol.174.4.1775. [DOI] [PubMed] [Google Scholar]
- 5.Poe JC, Hasegawa M, Tedder TF. CD19, CD21, and CD22: multifaceted response regulators of B lymphocyte signal transduction. Int Rev Immunol. 2001;20:739–62. doi: 10.3109/08830180109045588. [DOI] [PubMed] [Google Scholar]
- 6.Pritchard NR, Smith KGC. B cell inhibitory receptors and autoimmunity. Immunology. 2003;108:263–73. doi: 10.1046/j.1365-2567.2003.01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malbec O, Fridman WH, Daeron M. Negative regulation of hematopoietic cell activation and proliferation by Fc gamma RIIB. Curr Top Microbiol Immunol. 1999;244:13–27. [PubMed] [Google Scholar]
- 8.Pezzutto A, Dorken B, Moldenhauer G, Clark EA. Amplification of human B cell activation by a monoclonal antibody to the B cell-specific antigen CD22, Bp 130/140. J Immunol. 1987;138:98–103. [PubMed] [Google Scholar]
- 9.Pezzutto A, Rabinovitch PS, Dorken B, Moldenhauer G, Clark EA. Role of the CD22 human B cell antigen in B cell triggering by anti-immunoglobulin. J Immunol. 1988;140:1791–5. [PubMed] [Google Scholar]
- 10.Law CL, Torres RM, Sundberg HA, Parkhouse RM, Brannan CI, Copeland NG, Jenkins NA, Clark EA. Organization of the murine Cd22 locus. Mapping to chromosome 7 and characterization of two alleles. J Immunol. 1993;151:175–87. [PubMed] [Google Scholar]
- 11.Stamenkovic I, Seed B. The B-cell antigen CD22 mediates monocyte and erythrocyte adhesion. Nature. 1990;345:74–7. doi: 10.1038/345074a0. [DOI] [PubMed] [Google Scholar]
- 12.Torres RM, Law CL, Santos-Argumedo L, Kirkham PA, Grabstein K, Parkhouse RM, Clark EA. Identification and characterization of the murine homologue of CD22, a B lymphocyte-restricted adhesion molecule. J Immunol. 1992;149:2641–9. [PubMed] [Google Scholar]
- 13.Wilson GL, Fox CH, Fauci AS, Kehrl JH. cDNA cloning of the B cell membrane protein CD22: a mediator of B–B cell interactions. J Exp Med. 1991;173:137–46. doi: 10.1084/jem.173.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sgroi D, Varki A, Braesch-Andersen S, Stamenkovic I. CD22, a B cell-specific immunoglobulin superfamily member, is a sialic acid-binding lectin. J Biol Chem. 1993;268:7011–8. [PubMed] [Google Scholar]
- 15.Powell LD, Jain RK, Matta KL, Sabesan S, Varki A. Characterization of sialyloligosaccharide binding by recombinant soluble and native cell-associated CD22. Evidence for a minimal structural recognition motif and the potential importance of multisite binding. J Biol Chem. 1995;270:7523–32. doi: 10.1074/jbc.270.13.7523. [DOI] [PubMed] [Google Scholar]
- 16.Powell LD, Sgroi D, Sjoberg ER, Stamenkovic I, Varki A. Natural ligands of the B cell adhesion molecule CD22 beta carry N-linked oligosaccharides with alpha-2,6-linked sialic acids that are required for recognition. J Biol Chem. 1993;268:7019–27. [PubMed] [Google Scholar]
- 17.Engel P, Nojima Y, Rothstein D, Zhou LJ, Wilson GL, Kehrl JH, Tedder TF. The same epitope on CD22 of B lymphocytes mediates the adhesion of erythrocytes, T and B lymphocytes, neutrophils, and monocytes. J Immunol. 1993;150:4719–32. [PubMed] [Google Scholar]
- 18.Hanasaki K, Powell LD, Varki A. Binding of human plasma sialoglycoproteins by the B cell-specific lectin CD22. Selective recognition of immunoglobulin M and haptoglobin. J Biol Chem. 1995;270:7543–50. doi: 10.1074/jbc.270.13.7543. [DOI] [PubMed] [Google Scholar]
- 19.Hanasaki K, Varki A, Powell LD. CD22-mediated cell adhesion to cytokine-activated human endothelial cells. Positive and negative regulation by alpha 2-6-sialylation of cellular glycoproteins. J Biol Chem. 1995;270:7533–42. doi: 10.1074/jbc.270.13.7533. [DOI] [PubMed] [Google Scholar]
- 20.Nitschke L, Carsetti R, Ocker B, Kohler G, Lamers MC. CD22 is a negative regulator of B-cell receptor signalling. Curr Biol. 1997;7:133–43. doi: 10.1016/s0960-9822(06)00057-1. [DOI] [PubMed] [Google Scholar]
- 21.Smith KGC, Hewitson TD, Nossal GJ, Tarlinton DM. The phenotype and fate of the antibody-forming cells of the splenic foci. Eur J Immunol. 1996;26:444–8. doi: 10.1002/eji.1830260226. [DOI] [PubMed] [Google Scholar]
- 22.Engel P, Wagner N, Miller AS, Tedder TF. Identification of the ligand-binding domains of CD22, a member of the immunoglobulin superfamily that uniquely binds a sialic acid-dependent ligand. J Exp Med. 1995;181:1581–6. doi: 10.1084/jem.181.4.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Law CL, Aruffo A, Chandran KA, Doty RT, Clark EA. Ig domains 1 and 2 of murine CD22 constitute the ligand-binding domain and bind multiple sialylated ligands expressed on B and T cells. J Immunol. 1995;155:3368–76. [PubMed] [Google Scholar]
- 24.Nath D, van der Merwe PA, Kelm S, Bradfield P, Crocker PR. The amino-terminal immunoglobulin-like domain of sialoadhesin contains the sialic acid binding site. Comparison with CD22. J Biol Chem. 1995;270:26184–91. doi: 10.1074/jbc.270.44.26184. [DOI] [PubMed] [Google Scholar]
- 25.van der Merwe PA, Crocker PR, Vinson M, Barclay AN, Schauer R, Kelm S. Localization of the putative sialic acid-binding site on the immunoglobulin superfamily cell-surface molecule CD22. J Biol Chem. 1996;271:9273–80. [PubMed] [Google Scholar]
- 26.Fujimoto M, Kuwano Y, Watanabe R, et al. B cell antigen receptor and CD40 differentially regulate CD22 tyrosine phosphorylation. J Immunol. 2006;176:873–9. doi: 10.4049/jimmunol.176.2.873. [DOI] [PubMed] [Google Scholar]
- 27.Leprince C, Draves KE, Geahlen RL, Ledbetter JA, Clark EA. CD22 associates with the human surface IgM–B-cell antigen receptor complex. Proc Natl Acad Sci U S A. 1993;90:3236–40. doi: 10.1073/pnas.90.8.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cornall RJ, Cyster JG, Hibbs ML, Dunn AR, Otipoby KL, Clark EA, Goodnow CC. Polygenic autoimmune traits: Lyn, CD22, and SHP-1 are limiting elements of a biochemical pathway regulating BCR signaling and selection. Immunity. 1998;8:497–508. doi: 10.1016/s1074-7613(00)80554-3. [DOI] [PubMed] [Google Scholar]
- 29.Smith KGC, Tarlinton DM, Doody GM, Hibbs ML, Fearon DT. Inhibition of the B cell by CD22: a requirement for Lyn. J Exp Med. 1998;187:807–11. doi: 10.1084/jem.187.5.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y, Harder KW, Huntington ND, Hibbs ML, Tarlinton DM. Lyn tyrosine kinase: accentuating the positive and the negative. Immunity. 2005;22:9–18. doi: 10.1016/j.immuni.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Doody GM, Justement LB, Delibrias CC, Matthews RJ, Lin J, Thomas ML, Fearon DT. A role in B cell activation for CD22 and the protein tyrosine phosphatase SHP. Science. 1995;269:242–4. doi: 10.1126/science.7618087. [DOI] [PubMed] [Google Scholar]
- 32.Sato S, Jansen PJ, Tedder TF. CD19 and CD22 expression reciprocally regulates tyrosine phosphorylation of Vav protein during B lymphocyte signaling. Proc Natl Acad Sci U S A. 1997;94:13158–62. doi: 10.1073/pnas.94.24.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujimoto M, Bradney AP, Poe JC, Steeber DA, Tedder TF. Modulation of B lymphocyte antigen receptor signal transduction by a CD19/CD22 regulatory loop. Immunity. 1999;11:191–200. doi: 10.1016/s1074-7613(00)80094-1. [DOI] [PubMed] [Google Scholar]
- 34.Gerlach J, Ghosh S, Jumaa H, Reth M, Wienands J, Chan AC, Nitschke L. B cell defects in SLP65/BLNK-deficient mice can be partially corrected by the absence of CD22, an inhibitory coreceptor for BCR signaling. Eur J Immunol. 2003;33:3418–26. doi: 10.1002/eji.200324290. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, McLean PA, Neel BG, Okunade G, Shull GE, Wortis HH. CD22 attenuates calcium signaling by potentiating plasma membrane calcium-ATPase activity. Nat Immunol. 2004;5:651–7. doi: 10.1038/ni1072. [DOI] [PubMed] [Google Scholar]
- 36.Wienands J, Freuler F, Baumann G. Tyrosine-phosphorylated forms of Ig beta, CD22, TCR zeta and HOSS are major ligands for tandem SH2 domains of Syk. Int Immunol. 1995;7:1701–8. doi: 10.1093/intimm/7.11.1701. [DOI] [PubMed] [Google Scholar]
- 37.Law CL, Sidorenko SP, Chandran KA, Zhao Z, Shen SH, Fischer EH, Clark EA. CD22 associates with protein tyrosine phosphatase 1C, Syk, and phospholipase C-gamma(1) upon B cell activation. J Exp Med. 1996;183:547–60. doi: 10.1084/jem.183.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tuscano JM, Engel P, Tedder TF, Agarwal A, Kehrl JH. Involvement of p72syk kinase, p53/56lyn kinase and phosphatidyl inositol-3 kinase in signal transduction via the human B lymphocyte antigen CD22. Eur J Immunol. 1996;26:1246–52. doi: 10.1002/eji.1830260610. [DOI] [PubMed] [Google Scholar]
- 39.Poe JC, Fujimoto M, Jansen PJ, Miller AS, Tedder TF. CD22 forms a quaternary complex with SHIP, Grb2, and Shc. A pathway for regulation of B lymphocyte antigen receptor-induced calcium flux. J Biol Chem. 2000;275:17420–7. doi: 10.1074/jbc.M001892200. [DOI] [PubMed] [Google Scholar]
- 40.Brauweiler AM, Tamir I, Cambier JC. Bilevel control of B-cell activation by the inositol 5-phosphatase SHIP. Immunol Rev. 2000;176:69–74. doi: 10.1034/j.1600-065x.2000.00612.x. [DOI] [PubMed] [Google Scholar]
- 41.Stork B, Engelke M, Frey J, Horejsi V, Hamm-Baarke A, Schraven B, Kurosaki T, Wienands J. Grb2 and the non-T cell activation linker NTAL constitute a Ca2+-regulating signal circuit in B lymphocytes. Immunity. 2004;21:681–91. doi: 10.1016/j.immuni.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 42.Tuscano J, Engel P, Tedder TF, Kehrl JH. Engagement of the adhesion receptor CD22 triggers a potent stimulatory signal for B cells and blocking CD22/CD22L interactions impairs T-cell proliferation. Blood. 1996;87:4723–30. [PubMed] [Google Scholar]
- 43.Tuscano JM, Riva A, Toscano SN, Tedder TF, Kehrl JH. CD22 cross-linking generates B-cell antigen receptor-independent signals that activate the JNK/SAPK signaling cascade. Blood. 1999;94:1382–92. [PubMed] [Google Scholar]
- 44.Sato S, Tuscano JM, Inaoki M, Tedder TF. CD22 negatively and positively regulates signal transduction through the B lymphocyte antigen receptor. Semin Immunol. 1998;10:287–97. doi: 10.1006/smim.1998.0121. [DOI] [PubMed] [Google Scholar]
- 45.O’Keefe TL, Williams GT, Davies SL, Neuberger MS. Hyperresponsive B cells in CD22-deficient mice. Science. 1996;274:798–801. doi: 10.1126/science.274.5288.798. [DOI] [PubMed] [Google Scholar]
- 46.Otipoby KL, Andersson KB, Draves KE, et al. CD22 regulates thymus-independent responses and the lifespan of B cells. Nature. 1996;384:634–7. doi: 10.1038/384634a0. [DOI] [PubMed] [Google Scholar]
- 47.Sato S, Miller AS, Inaoki M, Bock CB, Jansen PJ, Tang ML, Tedder TF. CD22 is both a positive and negative regulator of B lymphocyte antigen receptor signal transduction: altered signaling in CD22-deficient mice. Immunity. 1996;5:551–62. doi: 10.1016/s1074-7613(00)80270-8. [DOI] [PubMed] [Google Scholar]
- 48.Goodnow CC, Crosbie J, Adelstein S, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334:676–82. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 49.Poe JC, Haas KM, Uchida J, Lee Y, Fujimoto M, Tedder TF. Severely impaired B lymphocyte proliferation, survival, and induction of the c-Myc:Cullin 1 ubiquitin ligase pathway resulting from CD22 deficiency on the C57BL/6 genetic background. J Immunol. 2004;172:2100–10. doi: 10.4049/jimmunol.172.4.2100. [DOI] [PubMed] [Google Scholar]
- 50.Samardzic T, Marinkovic D, Danzer CP, Gerlach J, Nitschke L, Wirth T. Reduction of marginal zone B cells in CD22-deficient mice. Eur J Immunol. 2002;32:561–7. doi: 10.1002/1521-4141(200202)32:2<561::AID-IMMU561>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 51.Pillai S, Cariappa A, Moran ST. Marginal zone B cells. Annu Rev Immunol. 2005;23:161–96. doi: 10.1146/annurev.immunol.23.021704.115728. [DOI] [PubMed] [Google Scholar]
- 52.Niiro H, Clark EA. Regulation of B-cell fate by antigen–receptor signals. Nat Rev Immunol. 2002;2:945–56. doi: 10.1038/nri955. [DOI] [PubMed] [Google Scholar]
- 53.Pillai S. Two lymphoid roads diverge – but does antigen bade B cells to take the road less traveled? Immunity. 2005;23:242–4. doi: 10.1016/j.immuni.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Wen L, Brill-Dashoff J, Shinton SA, Asano M, Hardy RR, Hayakawa K. Evidence of marginal-zone B cell-positive selection in spleen. Immunity. 2005;23:297–308. doi: 10.1016/j.immuni.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 55.Cariappa A, Tang M, Parng C, Nebelitskiy E, Carroll M, Georgopoulos K, Pillai S. The follicular versus marginal zone B lymphocyte cell fate decision is regulated by Aiolos, Btk, and CD21. Immunity. 2001;14:603–15. doi: 10.1016/s1074-7613(01)00135-2. [DOI] [PubMed] [Google Scholar]
- 56.Collins BE, Smith BA, Bengtson P, Paulson JC. Ablation of CD22 in ligand-deficient mice restores B cell receptor signaling. Nat Immunol. 2006;7:199–206. doi: 10.1038/ni1283. [DOI] [PubMed] [Google Scholar]
- 57.Ghosh S, Bandulet C, Nitschke L. Regulation of B cell development and B cell signalling by CD22 and its ligands alpha2,6-linked sialic acids. Int Immunol. 2006;18:603–11. doi: 10.1093/intimm/dxh402. [DOI] [PubMed] [Google Scholar]
- 58.Hennet T, Chui D, Paulson JC, Marth JD. Immune regulation by the ST6Gal sialyltransferase. Proc Natl Acad Sci U S A. 1998;95:4504–9. doi: 10.1073/pnas.95.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nitschke L. The role of CD22 and other inhibitory co-receptors in B-cell activation. Curr Opin Immunol. 2005;17:290–7. doi: 10.1016/j.coi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 60.Collins BE, Blixt O, DeSieno AR, Bovin N, Marth JD, Paulson JC. Masking of CD22 by cis ligands does not prevent redistribution of CD22 to sites of cell contact. Proc Natl Acad Sci U S A. 2004;101:6104–9. doi: 10.1073/pnas.0400851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bakker TR, Piperi C, Davies EA, Merwe PA. Comparison of CD22 binding to native CD45 and synthetic oligosaccharide. Eur J Immunol. 2002;32:1924–32. doi: 10.1002/1521-4141(200207)32:7<1924::AID-IMMU1924>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 62.Blixt O, Collins BE, van den Nieuwenhof IM, Crocker PR, Paulson JC. Sialoside specificity of the siglec family assessed using novel multivalent probes: identification of potent inhibitors of myelin-associated glycoprotein. J Biol Chem. 2003;278:31007–19. doi: 10.1074/jbc.M304331200. [DOI] [PubMed] [Google Scholar]
- 63.Razi N, Varki A. Masking and unmasking of the sialic acid-binding lectin activity of CD22 (Siglec-2) on B lymphocytes. Proc Natl Acad Sci U S A. 1998;95:7469–74. doi: 10.1073/pnas.95.13.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Razi N, Varki A. Cryptic sialic acid binding lectins on human blood leukocytes can be unmasked by sialidase treatment or cellular activation. Glycobiology. 1999;9:1225–34. doi: 10.1093/glycob/9.11.1225. [DOI] [PubMed] [Google Scholar]
- 65.Danzer CP, Collins BE, Blixt O, Paulson JC, Nitschke L. Transitional and marginal zone B cells have a high proportion of unmasked CD22: implications for BCR signaling. Int Immunol. 2003;15:1137–47. doi: 10.1093/intimm/dxg114. [DOI] [PubMed] [Google Scholar]
- 66.Peaker CJ, Neuberger MS. Association of CD22 with the B cell antigen receptor. Eur J Immunol. 1993;23:1358–63. doi: 10.1002/eji.1830230626. [DOI] [PubMed] [Google Scholar]
- 67.Zhang M, Varki A. Cell surface sialic acids do not affect primary CD22 interactions with CD45 and surface IgM nor the rate of constitutive CD22 endocytosis. Glycobiology. 2004;14:939–49. doi: 10.1093/glycob/cwh126. [DOI] [PubMed] [Google Scholar]
- 68.Han S, Collins BE, Bengtson P, Paulson JC. Homomultimeric complexes of CD22 in B cells revealed by protein–glycan cross-linking. Nat Chem Biol. 2005;1:93–7. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- 69.Aruffo A, Kanner SB, Sgroi D, Ledbetter JA, Stamenkovic I. CD22-mediated stimulation of T cells regulates T-cell receptor/CD3-induced signaling. Proc Natl Acad Sci U S A. 1992;89:10242–6. doi: 10.1073/pnas.89.21.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stamenkovic I, Sgroi D, Aruffo A, Sy MS, Anderson T. The B lymphocyte adhesion molecule CD22 interacts with leukocyte common antigen CD45RO on T cells and alpha 2-6 sialyltransferase, CD75, on B cells. Cell. 1991;66:1133–44. doi: 10.1016/0092-8674(91)90036-x. [DOI] [PubMed] [Google Scholar]
- 71.Jin L, McLean PA, Neel BG, Wortis HH. Sialic acid binding domains of CD22 are required for negative regulation of B cell receptor signaling. J Exp Med. 2002;195:1199–205. doi: 10.1084/jem.20011796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kelm S, Gerlach J, Brossmer R, Danzer CP, Nitschke L. The ligand-binding domain of CD22 is needed for inhibition of the B cell receptor signal, as demonstrated by a novel human CD22-specific inhibitor compound. J Exp Med. 2002;195:1207–13. doi: 10.1084/jem.20011783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lanoue A, Batista FD, Stewart M, Neuberger MS. Interaction of CD22 with alpha2,6-linked sialoglycoconjugates: innate recognition of self to dampen B cell autoreactivity? Eur J Immunol. 2002;32:348–55. doi: 10.1002/1521-4141(200202)32:2<348::AID-IMMU348>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 74.Sgroi D, Koretzky GA, Stamenkovic I. Regulation of CD45 engagement by the B-cell receptor CD22. Proc Natl Acad Sci U S A. 1995;92:4026–30. doi: 10.1073/pnas.92.9.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sathish JG, Walters J, Luo JC, et al. CD22 is a functional ligand for SH2 domain-containing protein-tyrosine phosphatase-1 in primary T cells. J Biol Chem. 2004;279:47783–91. doi: 10.1074/jbc.M402354200. [DOI] [PubMed] [Google Scholar]
- 76.Nitschke L, Floyd H, Ferguson DJ, Crocker PR. Identification of CD22 ligands on bone marrow sinusoidal endothelium implicated in CD22-dependent homing of recirculating B cells. J Exp Med. 1999;189:1513–8. doi: 10.1084/jem.189.9.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Floyd H, Nitschke L, Crocker PR. A novel subset of murine B cells that expresses unmasked forms of CD22 is enriched in the bone marrow: implications for B-cell homing to the bone marrow. Immunology. 2000;101:342–7. doi: 10.1046/j.1365-2567.2000.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Collins BE, Blixt O, Han S, Duong B, Li H, Nathan JK, Bovin N, Paulson JC. High-affinity ligand probes of CD22 overcome the threshold set by cis ligands to allow for binding, endocytosis, and killing of B cells. J Immunol. 2006;177:2994–3003. doi: 10.4049/jimmunol.177.5.2994. [DOI] [PubMed] [Google Scholar]
- 79.Harduin-Lepers A, Recchi MA, Delannoy P. 1994, the year of sialyltransferases. Glycobiology. 1995;5:741–58. doi: 10.1093/glycob/5.8.741. [DOI] [PubMed] [Google Scholar]
- 80.Weinstein J, Lee EU, McEntee K, Lai PH, Paulson JC. Primary structure of beta-galactoside alpha 2,6-sialyltransferase. Conversion of membrane-bound enzyme to soluble forms by cleavage of the NH2-terminal signal anchor. J Biol Chem. 1987;262:17735–43. [PubMed] [Google Scholar]
- 81.Rudge EU, Cutler AJ, Pritchard NR, Smith KGC. Interleukin 4 reduces expression of inhibitory receptors on B cells and abolishes CD22 and Fc gamma RII-mediated B cell suppression. J Exp Med. 2002;195:1079–85. doi: 10.1084/jem.20011435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grewal PK, Boton M, Ramirez K, et al. ST6Gal-I restrains CD22-dependent antigen receptor endocytosis and Shp-1 recruitment in normal and pathogenic immune signaling. Mol Cell Biol. 2006;26:4970–81. doi: 10.1128/MCB.00308-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hoffmann A, Kerr S, Jellusova J, et al. Siglec-G is a B1 cell-inhibitory receptor that controls expansion and calcium signaling of the B1 cell population. Nat Immunol. 2007;8:695–704. doi: 10.1038/ni1480. [DOI] [PubMed] [Google Scholar]
- 84.Stoddart A, Jackson AP, Brodsky FM. Plasticity of B cell receptor internalization upon conditional depletion of clathrin. Mol Biol Cell. 2005;16:2339–48. doi: 10.1091/mbc.E05-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shan D, Press OW. Constitutive endocytosis and degradation of CD22 by human B cells. J Immunol. 1995;154:4466–75. [PubMed] [Google Scholar]
- 86.Tateno H, Li H, Schur MJ, Bovin N, Crocker PR, Wakarchuk WW, Paulson JC. Distinct endocytic mechanisms of CD22 (Siglec-2) and Siglec-F reflect roles in cell-signaling and innate immunity. Mol Cell Biol. 2007;27:5699–710. doi: 10.1128/MCB.00383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.John B, Herrin BR, Raman C, Wang YN, Bobbitt KR, Brody BA, Justement LB. The B cell coreceptor CD22 associates with AP50, a clathrin-coated pit adapter protein, via tyrosine-dependent interaction. J Immunol. 2003;170:3534–43. doi: 10.4049/jimmunol.170.7.3534. [DOI] [PubMed] [Google Scholar]
- 88.Yohannan J, Wienands J, Coggeshall KM, Justement LB. Analysis of tyrosine phosphorylation-dependent interactions between stimulatory effector proteins and the B cell co-receptor CD22. J Biol Chem. 1999;274:18769–76. doi: 10.1074/jbc.274.26.18769. [DOI] [PubMed] [Google Scholar]
- 89.Poe JC, Fujimoto Y, Hasegawa M, et al. CD22 regulates B lymphocyte function in vivo through both ligand-dependent and ligand-independent mechanisms. Nat Immunol. 2004;5:1078–87. doi: 10.1038/ni1121. [DOI] [PubMed] [Google Scholar]
- 90.Yu J, Sawada T, Adachi T, et al. Synthetic glycan ligand excludes CD22 from antigen receptor-containing lipid rafts. Biochem Biophys Res Commun. 2007;360:759–64. doi: 10.1016/j.bbrc.2007.06.110. [DOI] [PubMed] [Google Scholar]
- 91.Pflugh DL, Maher SE, Bothwell AL. Ly-6 superfamily members Ly-6A/E, Ly-6C, and Ly-6I recognize two potential ligands expressed by B lymphocytes. J Immunol. 2002;169:5130–6. doi: 10.4049/jimmunol.169.9.5130. [DOI] [PubMed] [Google Scholar]
- 92.Wakabayashi C, Adachi T, Wienands J, Tsubata T. A distinct signaling pathway used by the IgG-containing B cell antigen receptor. Science. 2002;298:2392–5. doi: 10.1126/science.1076963. [DOI] [PubMed] [Google Scholar]
- 93.Silver K, Cornall RJ. Isotype control of B cell signaling. Sci STKE. 2003;2003 doi: 10.1126/stke.2003.184.pe21. [DOI] [PubMed] [Google Scholar]
- 94.Horikawa K, Martin SW, Pogue SL, Silver K, Peng K, Takatsu K, Goodnow CC. Enhancement and suppression of signaling by the conserved tail of IgG memory-type B cell antigen receptors. J Exp Med. 2007;204:759–69. doi: 10.1084/jem.20061923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Waisman A, Kraus M, Seagal J, et al. IgG1 B cell receptor signaling is inhibited by CD22 and promotes the development of B cells whose survival is less dependent on Ig alpha/beta. J Exp Med. 2007;204:747–58. doi: 10.1084/jem.20062024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.O’Keefe TL, Williams GT, Batista FD, Neuberger MS. Deficiency in CD22, a B cell-specific inhibitory receptor, is sufficient to predispose to development of high affinity autoantibodies. J Exp Med. 1999;189:1307–13. doi: 10.1084/jem.189.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mary C, Laporte C, Parzy D, et al. Dysregulated expression of the Cd22 gene as a result of a short interspersed nucleotide element insertion in Cd22a lupus-prone mice. J Immunol. 2000;165:2987–96. doi: 10.4049/jimmunol.165.6.2987. [DOI] [PubMed] [Google Scholar]
- 98.Bygrave AE, Rose KL, Cortes-Hernandez J, et al. Spontaneous autoimmunity in 129 and C57BL/6 mice – implications for autoimmunity described in gene-targeted mice. PLoS Biol. 2004;2:E243. doi: 10.1371/journal.pbio.0020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heidari Y, Bygrave AE, Rigby RJ, Rose KL, Walport MJ, Cook HT, Vyse TJ, Botto M. Identification of chromosome intervals from 129 and C57BL/6 mouse strains linked to the development of systemic lupus erythematosus. Genes Immun. 2006;7:592–9. doi: 10.1038/sj.gene.6364335. [DOI] [PubMed] [Google Scholar]
- 100.Mitchell DA, Pickering MC, Warren J, Fossati-Jimack L, Cortes-Hernandez J, Cook HT, Botto M, Walport MJ. C1q deficiency and autoimmunity: the effects of genetic background on disease expression. J Immunol. 2002;168:2538–43. doi: 10.4049/jimmunol.168.5.2538. [DOI] [PubMed] [Google Scholar]
- 101.Gillmore JD, Hutchinson WL, Herbert J, et al. Autoimmunity and glomerulonephritis in mice with targeted deletion of the serum amyloid P component gene: SAP deficiency or strain combination? Immunology. 2004;112:255–64. doi: 10.1111/j.1365-2567.2004.01860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kikuchi S, Amano H, Amano E, et al. Identification of 2 major loci linked to autoimmune hemolytic anemia in NZB mice. Blood. 2005;106:1323–9. doi: 10.1182/blood-2005-02-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kikuchi S, Fossati-Jimack L, Moll T, et al. Differential role of three major New Zealand Black-derived loci linked with Yaa-induced murine lupus nephritis. J Immunol. 2005;174:1111–7. doi: 10.4049/jimmunol.174.2.1111. [DOI] [PubMed] [Google Scholar]
- 104.Kono DH, Burlingame RW, Owens DG, Kuramochi A, Balderas RS, Balomenos D, Theofilopoulos AN. Lupus susceptibility loci in New Zealand mice. Proc Natl Acad Sci U S A. 1994;91:10168–72. doi: 10.1073/pnas.91.21.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morel L, Rudofsky UH, Longmate JA, Schiffenbauer J, Wakeland EK. Polygenic control of susceptibility to murine systemic lupus erythematosus. Immunity. 1994;1:219–29. doi: 10.1016/1074-7613(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 106.Santiago ML, Mary C, Parzy D, et al. Linkage of a major quantitative trait locus to Yaa gene-induced lupus-like nephritis in (NZW × C57BL/6)F1 mice. Eur J Immunol. 1998;28:4257–67. doi: 10.1002/(SICI)1521-4141(199812)28:12<4257::AID-IMMU4257>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 107.Morel L, Mohan C, Yu Y, Croker BP, Tian N, Deng A, Wakeland EK. Functional dissection of systemic lupus erythematosus using congenic mouse strains. J Immunol. 1997;158:6019–28. [PubMed] [Google Scholar]
- 108.Lajaunias F, Ibnou-Zekri N, Fossati Jimack L, et al. Polymorphisms in the Cd22 gene of inbred mouse strains. Immunogenetics. 1999;49:991–5. doi: 10.1007/s002510050584. [DOI] [PubMed] [Google Scholar]
- 109.Nitschke L, Lajaunias F, Moll T, et al. Expression of aberrant forms of CD22 on B lymphocytes in Cd22a lupus-prone mice affects ligand binding. Int Immunol. 2006;18:59–68. doi: 10.1093/intimm/dxh349. [DOI] [PubMed] [Google Scholar]
- 110.Hibbs ML, Tarlinton DM, Armes J, Grail D, Hodgson G, Maglitto R, Stacker SA, Dunn AR. Multiple defects in the immune system of Lyn-deficient mice, culminating in autoimmune disease. Cell. 1995;83:301–11. doi: 10.1016/0092-8674(95)90171-x. [DOI] [PubMed] [Google Scholar]
- 111.Silver K, Bouriez-Jones T, Crockford T, Ferry H, Tang HL, Cyster JG, Cornall RJ. Spontaneous class switching and B cell hyperactivity increase autoimmunity against intracellular self antigen in Lyn-deficient mice. Eur J Immunol. 2006;36:2920–7. doi: 10.1002/eji.200636462. [DOI] [PubMed] [Google Scholar]
- 112.Duan B, Morel L. Role of B-1a cells in autoimmunity. Autoimmun Rev. 2006;5:403–8. doi: 10.1016/j.autrev.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 113.Hatta Y, Tsuchiya N, Matsushita M, Shiota M, Hagiwara K, Tokunaga K. Identification of the gene variations in human CD22. Immunogenetics. 1999;49:280–6. doi: 10.1007/s002510050494. [DOI] [PubMed] [Google Scholar]
- 114.Hitomi Y, Tsuchiya N, Hasegawa M, Fujimoto M, Takehara K, Tokunaga K, Sato S. Association of CD22 gene polymorphism with susceptibility to limited cutaneous systemic sclerosis. Tissue Antigens. 2007;69:242–9. doi: 10.1111/j.1399-0039.2007.00801.x. [DOI] [PubMed] [Google Scholar]
- 115.Mattsson N, Duzevik EG, Pelsue SC. Expansion of CD22lo B cells in the spleen of autoimmune-prone flaky skin mice. Cell Immunol. 2005;234:124–32. doi: 10.1016/j.cellimm.2005.06.005. [DOI] [PubMed] [Google Scholar]