Abstract
The stability of the ompA mRNA depends on the bacterial growth rate. The 5′ untranslated region is the stability determinant of this transcript and the target of the endoribonuclease, RNase E, the key player of mRNA degradation. An RNA-binding protein with affinity for the 5′ untranslated region ompA was purified and identified as Hfq, a host factor initially recognized for its function in phage Qβ replication. The ompA RNA-binding activity parallels the amount of Hfq, which is elevated in bacteria cultured at slow growth rate, a condition leading to facilitated degradation of the ompA mRNA. In hfq mutant cells with a deficient Hfq gene product, the RNA-binding activity is missing, and analysis of the ompA mRNA showed that the growth-rate dependence of degradation is lost. Furthermore, the half-life of the ompA mRNA is prolonged in the mutant cells, irrespective of growth rate. Hfq has no affinity for the lpp transcript whose degradation, like that of bulk mRNA, is not affected by bacterial growth rate. Compatible with our results, we found that the intracellular concentration of RNase E and its associated degradosome components is independent of bacterial growth rate. Thus our results suggest a regulatory role for Hfq that specifically facilitates the ompA mRNA degradation in a growth rate-dependent manner.
Keywords: RNA-binding protein/mobility-shift assay/hfq mutants/mRNA half-life
mRNA stability is an important means of cells to control the level of gene expression. An increasing number of examples have been reported where up- or down-regulation of mRNA stability is instrumental for bacteria to meet the needs of a changing environment. The stability of single transcripts or even of entire pools of mRNA has been shown to be regulated by challenges such as antibiotics, nutritional stress, transition of growth stage, and bacterial growth rate (for review see ref. 1). One of the earliest studied examples of alterations in the decay rate was the ompA mRNA in Escherichia coli (coding for the major outer membrane protein A, OmpA). The stability of this transcript depends on the bacterial growth rate (2, 3). In contrast, the decay of most E. coli transcripts seems not to be affected by bacterial growth rate (4). The stability of the ompA mRNA has been assigned to RNase E cleavages that initiate the degradation of this transcript in the 5′ untranslated region (UTR) (1). The ompA 5′ UTR seems to be composed of two stem-loop structures flanked and/or interfered with AU-rich regions (5, 6). The 5′ terminal hairpin is essential for the relatively long half-life of the ompA mRNA, whereas the downstream stem-loop structure has less effect on the stability of the message (7).
RNA decay and processing in E. coli involve the coordinated actions of site-specific endoribonucleases, exoribonucleases, and poly(A) polymerase (for review see ref. 1). The endoribonuclease RNase E has been identified as the key player of mRNA decay (8). This enzyme cleaves RNA in single-stranded AU-rich regions, e.g. in the 5′ UTR of the ompA mRNA and in the 5S ribosomal RNA precursor, 9S RNA (9–11). In E. coli, RNase E seems to be part of a high molecular weight complex, called the degradosome, which contains a 3′–5′ exoribonuclease, polynucleotide phosphorylase (PNPase), a DEAD box helicase, RhlB, and enolase as major components, polyphosphate kinase as a minor component, and, at least facultatively, the protein chaperones DnaK and GroEL (12–14).
In an attempt to identify trans-acting factors that could regulate mRNA stability, three RNA-binding activities (RBAs), named RBA1, RBA2, and RBA3, recently have been discovered by probing cellular extracts with the 5′ UTR of the ompA mRNA in mobility-shift assays (15). RBA1 was found to be up-regulated under conditions of extreme nutritional stress (i.e., culturing bacteria anaerobically at an extremely slow growth rate), leading to an increased stability of the ompA mRNA, as well as of bulk mRNA (16). Thus, RBA1 has been suggested to act as an mRNA decay repressor, and several observations implied that RBA1 contains the chaperone GroEL.
We repeated the previous approach with the 5′ UTR of the ompA mRNA as a target and probed for RBAs that could explain its longevity at fast growth rate. Interestingly, we found an RBA that was increased when the ompA mRNA is destabilized. This finding prompted us to further purify and characterize this RBA and led us to identify it to be Hfq (host factor I), the product of the hfq gene, which is needed for phage Qβ RNA replication (17). Hfq is a heat-stable protein with subunits of 11.2 kDa occurring in hexa- or pentamers (18). The affinity of Hfq for RNA has been documented and its preference for single-stranded AU-rich regions has been recognized (19). It is an abundant protein with 30,000 to 60,000 molecules per cell (20), and its number has been found to increase during stationary phase (21). Recently, an increasing amount of evidence has shown that this factor affects expression of a number of genes involved in stress response, such as the facilitation of the translation of the rpoS mRNA, coding for the stationary phase transcription factor, σs (22, 23). Finally, an involvement of Hfq in negative regulation of mutS, miaA, and hfq mRNA stabilities has been reported (21). Although it is clear that Hfq is able to influence phage Qβ RNA replication, translation, and mRNA degradation, its precise mode of interaction with RNA remains undetermined.
In this study we determine the novel feature of Hfq protein to bind the 5′ UTR of the ompA mRNA. Our data indicate that it participates in the regulation of ompA mRNA stability in response to changes in growth rate. Mutation in the hfq gene leads to an increase of the ompA mRNA half-life, suggesting it plays a role as a destabilizing factor. These results are in agreement with the hypothesis that Hfq functions as an RNA chaperone altering transcript stability and/or translation efficiency.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, Media, and Growth Conditions.
In this study we used the following E. coli strains: parental strain MC4100 [F− Δ(argF lac) U169 araD139 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR] (24) and its derivatives AM111(hfq1) [hfq∷Ω, BclI], AM112 (hfq2) (hfq∷Ω, KpnI) (22); BL21(DE3) [F− hshS gal λ imm21 Δnin5 int∷T7 gene 1] (25); SBS1936 [hfrC relA pit10 tonA22] and its mutant SBS1935 (rpoS190) (26); MG1655 and its derivative CF1678 [ΔrelA251∷kan ΔspoT209∷cat] (27). Plasmid pHFQ607 containing the hfq chromosomal locus under the control of the T7 RNA-polymerase promoter is described (28).
Cells were grown in Luria–Bertani medium (LB) or Mops (morpholinepropanesulfonic acid) medium supplemented with 0.2% (wt/vol) sodium acetate at 30°C until middle log-phase, as described (2). For overexpression of the hfq gene, BL21(DE3) exponentially growing cells provided with the plasmid pHFQ607 were incubated for 1 hr in the presence of 1 mM isopropyl β-d-thio-galactopyranoside.
Preparation of cell extracts were performed at 4°C. Frozen cells (5 g wet weight) were resuspended in 5 ml of buffer A [20 mM Tris⋅HCl, pH 7.8/5 mM MgCl2/0.1 mM EDTA/0.1 mM DTT/5% (wt/vol) glycerol] supplemented with 1 mM phenylmethylsulfonyl fluoride and 2 μg/ml of lysozyme. The cells were lyzed by ultrasonication and centrifuged at 30,000 g for 30 min, and the supernatant (S30) was treated with 0.05 mg/ml of DNase I (RNase free) for 30 min.
Purification and Sequence Analysis of the RBA.
The S30 extract of MC4100 cells was used for the further purification of the RBA. The protein was precipitated with 35–55% saturated (NH4)2SO4 and resuspended in and dialyzed against buffer A. This fraction was loaded onto a column of DEAE-cellulose previously equilibrated with the same buffer. A linear gradient of 0–700 mM KCl in buffer A was applied, and most of the RBA was eluted in 200–300 mM KCl. The fractions containing RBA were applied to a heparin-agarose column equilibrated with buffer A/200 mM KCl. RBA was found in a single peak eluted between 450 and 550 mM KCl; the bulk of the contaminating proteins flow through the column. The further purification takes advantage of the 5′ UTR ompA mRNA-binding properties of the protein. The 5′ biotinylated oligomer complementary to the 3′ end of target ompA RNA was annealed to in vitro-transcribed ompA substrate, and the resultant RNA⋅DNA duplex was coupled to streptavidin magnetic beads according to the instruction of the manufacturer (Dynal). The active fraction eluted from a heparin-agarose column was added to the beads resuspended in buffer B [10 mM Tris⋅HCl, pH 7.5/5 mM NH4Cl/10 mM EDTA/5 mM DTT/10% (vol/vol) glycerol] and incubated for 15–30 min at 4°C. Unbound proteins were washed out with buffer B. To elute bound protein, the bead suspension was heated up to 65°C for 3 min. The purity of protein obtained was above 95%. The excised Coomassie-stained protein band was in-gel digested (29, 30). Extracted peptides were purified on a R2 Poros microcolumn and eluted in 1 μl of 60% methanol/5% formic acid into a nanoelectrospray capillary. Mass spectrometry was done on an API III triple quadropole instrument (PE-Sciex, Ontario) equipped with an upgraded collision cell and a nanoelectrospray source (31). The sequence tag (733.5)PDQ(1073.6) from the fragment spectrum of a peptide (Mr 1272.7) identified the peptide comprising amino acids 5–15 of the host factor I (SwissProt accession no. P25521) in a nonredundant database. Three other host factor peptides were sequenced from the same digest comprising amino acids 17–24, 17–30, and 19–24.
Western Blot Analysis.
Lysates, prepared by boiling of cells in Laemmli loading buffer, or cell extracts (S30) were separated on SDS-polyacrylamide gels. After protein blotting, nitrocellulose membranes were blocked in 10% dry milk suspension and developed with antibodies against RNase E, PNPase, RhlB helicase (from A. J. Carpousis, Centre National de la Recherche Scientifique, Toulouse, France), Hfq (from A. Ishihama, National Institute of Genetics, Mishima, Shizuoka, Japan), and GroEL (StressGen, Biotechnologies, Victoria, Canada), using the ECL detection kit (Amersham).
Preparation of Labeled RNA Substrates and Mobility-Shift Assays.
32P-labeled ompA mRNA was prepared by in vitro transcription as described (15). The resulting RNA was 203 nt in length and contained the ompA 5′ UTR, a part of the coding region, and an additional hairpin in the 3′ end to protect it from exoribonucleases. Labeled lpp mRNA substrate (245 nt) was transcribed from the T7 RNA-polymerase promoter of the DNA fragment that contained the 5′ UTR and a part of the coding region of the lpp gene. Mobility-shift assays were performed as described (15). S30 extracts (10–30 μg) or 1 ng of purified Hfq were preincubated in buffer B with 5 μg of heparin and 103–104 cpm of 32P-labeled transcripts, and RNA-protein complexes were resolved on 4% native polyacrylamide gels. For autoradiography, gels were dried and exposed to x-ray films.
Determination of mRNA Half-Life.
The RNA isolation and determination of mRNA half-lives by Northern blot analysis were performed as described (3). A 32P-labeled PstI–PstI fragment from pSPomp8 plasmid (9) containing the central part of the ompA gene was used for ompA mRNA detection, whereas the 5′ end and a part of the coding region of the lpp gene (260 nt in length) was applied for lpp mRNA half-life measurement.
RESULTS
The Key Enzymes of RNA Decay Are Not Regulated in a Growth Rate-Dependent Fashion.
The ompA transcript is comparatively stable when cells are cultured at a slow growth rate, whereas the stability of the lpp message remains constant for a variety of growth rates (2, 3). The rate of ompA mRNA degradation parallels the rate of RNase E cleavage in its 5′ UTR (32).
To further elucidate the mechanism that regulates the growth rate-dependent turnover of the ompA transcript, we examined the level of the key enzymes of RNA decay (RNase E, PNPase, and RhlB helicase) in cells growing at different growth rates. It has been shown that the mRNA stability is related primarily to the growth rate that can be controlled by the media composition (3). E. coli MC4100 was cultured at 30°C, either in Mops-acetate medium, leading to a generation time of 150 min, or in LB, leading to a generation time of 35 min. The half-lives of the ompA mRNA were tested for two different culture conditions, and the formerly observed growth rate dependence was confirmed (ref. 2 and Table 1). Equal amounts of cells were removed from the exponential cultures and analyzed by Western blotting (Fig. 1). Cells grown in both media contained similar amounts of RNase E, PNPase, and RhlB proteins. Thus, the change in growth rate does not cause a change in the level of the major degradosome components. Based on these data we concluded that the key enzymes of RNA decay are not the targets of growth rate-dependent regulation. The notion that the stability of the majority of transcripts is controlled by RNase E (33) and that the decay of bulk mRNA is independent of the growth rate (4) supports our conclusion.
Table 1.
Half-lives of ompA and lpp mRNAs (min) in Mops/acetate and LB
| Strain | Mops/acetate
|
LB
|
Regulation
|
|||
|---|---|---|---|---|---|---|
| ompA | lpp | ompA | lpp | ompA | lpp | |
| Wild type | 7 ± 0.9 | 17 ± 2.8 | 14 ± 0.3 | 21 ± 4.1 | 2.0 | 1.2 |
| hfq1 | 43 ± 0.6 | 21 ± 3.9 | 25 ± 1.7 | 26 ± 0.6 | 0.6 | 1.2 |
| hfq1/pHFQ607 | 9 ± 0.8 | 22 ± 2.5 | 19 ± 2.1 | 28 ± 4.5 | 2.1 | 1.3 |
| hfq2 | 5 ± 0.9 | 20 ± 4.3 | 18 ± 0.1 | 26 ± 2.1 | 3.6 | 1.3 |
| rpoS | 8 ± 1.0 | 22 ± 1.7 | 24 ± 3.9 | 22 ± 1.4 | 3.0 | 1.0 |
Half-lives of the ompA and lpp mRNAs as the average of two determinations by Northern blotting and subsequent quantification by PhosphorImaging. The relative changes in half-lives are shown as fold regulation. The wild-type strain MC4100 and the mutant strains are described in Materials and Methods.
Figure 1.
Comparison of the levels of the key degradosome components in MC4100 cells grown exponentially with high (LB) or slow (Mops-acetate medium) growth rate by Western blot analysis. Equal amounts of cells were withdrawn from each medium, and cell lysates were probed with antibodies against RNase E, PNPase, or RhlB helicase as described in Materials and Methods.
Growth Rate Determines the Intracellular Level of an ompA mRNA-Specific Binding Activity.
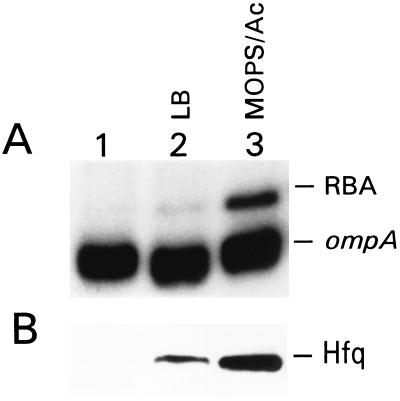
Because the 5′ UTR has been shown to determine the stability of the full-length ompA mRNA (34), we probed for binding of the regulatory factor(s) to this region. We used mobility-shift assays to identify and compare RBAs in S30 extracts obtained from the MC4100 strain cultured at different growth rates (see Materials and Methods). The 5′ end of the labeled ompA transcript was incubated with extracts to allow the formation of RNA–protein complexes and then was separated on native polyacrylamide gels (Fig. 2A). To ascertain that the up-shift of the RNA in lane 3 was caused by RNA–protein interaction, we treated the RNA samples after preincubation with extracts either with phenol/chloroform or proteinase K and loaded them on the gel alongside the untreated samples. No RNA shift was observed after such treatments (data not shown). A detectable level of RBA was seen only in cells growing in Mops-acetate medium (Fig. 2A). We found a correlation among the amount of this RBA, the generation time, and the rate of ompA mRNA degradation: an increase of RBA in slow-growing cells was accompanied by a decrease of ompA mRNA half-life and vice versa (see Table 1). We suspected this RNA-binding protein to be a regulator of mRNA decay.
Figure 2.
The level of the 5′ UTR ompA RBA in MC4100 cells grown exponentially with high (LB) or slow (Mops-acetate medium) growth rate. (A) Mobility-shift assay. S30 extracts (10 μg) were incubated with 32P-labeled ompA transcript and analyzed, as described in Materials and Methods. Lane 1, the ompA transcript alone; lanes 2 and 3, the ompA transcript incubated with extract obtained from cells grown in LB and Mops-acetate medium, respectively. (B) Western blot analysis of the same extracts as in A tested with Hfq-specific antibodies, as described in Materials and Methods.
The Growth-Regulated RBA Is Hfq.
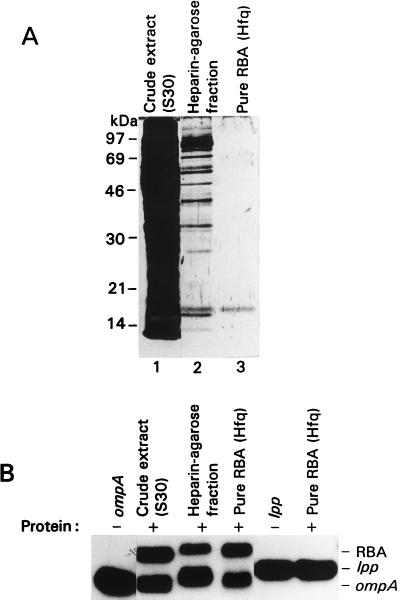
For identification of the RBA enhanced in slow-growing cells, it was purified to near homogeneity (see Materials and Methods) (Fig. 3). The predominant protein of the purest fraction was cut out from the SDS-gel and sequence-analyzed by mass spectrometry. The obtained protein sequence corresponds to the known protein, Hfq, which previously has been shown to bind Qβ and R17 phage RNAs (17–19).
Figure 3.
Analysis of the purified ompA mRNA-binding protein RBA. (A) Electrophoresis of crude and purified materials on an SDS-polyacrylamide gel visualized by silver staining. Purification steps of RBA described in Materials and Methods. Lane 1, crude extract (10 μg); lane 2, heparin-agarose fraction (0.1 μg); lane 3, finely purified material (Hfq) (0.005 μg). (B) Mobility-shift assay of crude cell extract and purified material as in A, using as an RNA substrate either ompA or lpp. The same amounts of protein as in A were incubated with 32P-labeled substrates and applied to the 4% native polyacrylamide gel.
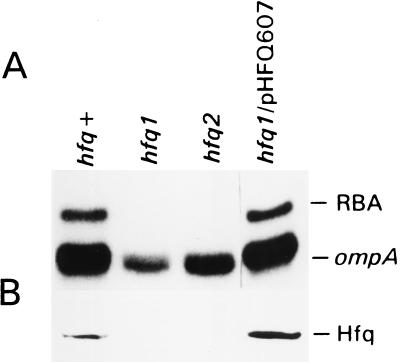
To confirm that Hfq can bind the 5′ UTR of the ompA mRNA we tested the effect of mutations in the hfq gene on the formation of the RNA-binding complex. For this purpose we used the congenic strains AM111 (hfq1) and AM112 (hfq2) with insertions of an Ω (Kan) cassette in the hfq gene (22). The hfq1 mutation is a disruption approximately in the middle of the gene, whereas the hfq2 insertion is located close to the end of the hfq coding region. Both mutations are equally polar on the expression of the hflA region located downstream of the hfq gene on the chromosome. Mobility-shift assays showed a total loss of the RBA in both mutant cells (Fig. 4). To exclude a possible polarity effect of the hfq mutation on the RBA we determined whether the provision of the hfq gene in trans would restore this RBA. Providing the hfq1 mutant cells with pHFQ607 plasmid, which contains the hfq gene (28), reconstituted the RNA-binding complex as well as Hfq synthesis (Fig. 4). When the hfq gene under control of the T7 RNA-polymerase promoter was overexpressed in the E. coli BL21(DE3) strain producing T7 RNA-polymerase, the level of the RNA–protein complex did not increase to the same extent as the level of Hfq protein (data not shown).
Figure 4.
The hfq gene is responsible for RBA. (A) Mobility-shift assay of the ompA mRNA-binding activity of MC4100 strain (hfq+), its derivatives AM111 (hfq1) and AM112 (hfq2), and hfq1 strain provided with the plasmid pHFQ607, containing the hfq gene. Cells were grown in Mops-acetate medium, and 10 μg of S30 extracts were used for the RNA–protein complex formation. (B) Western blot analysis of the same extracts as in A tested with Hfq-specific antibodies, as described in Materials and Methods.
The purified RBA also was probed with GroEL-specific antibodies and no detectable amount of GroEL was found (data not shown). Equal amounts of extracts, as analyzed for RNA binding (Fig. 2A), were submitted to Western blot using Hfq-specific antibodies. The amount of Hfq was much higher in slow-growing cells than in fast-growing cells (Fig. 2B).
Taken together, our data show that Hfq itself is involved in the complex formation with the ompA 5′ UTR.
Hfq Facilitates ompA mRNA Decay.
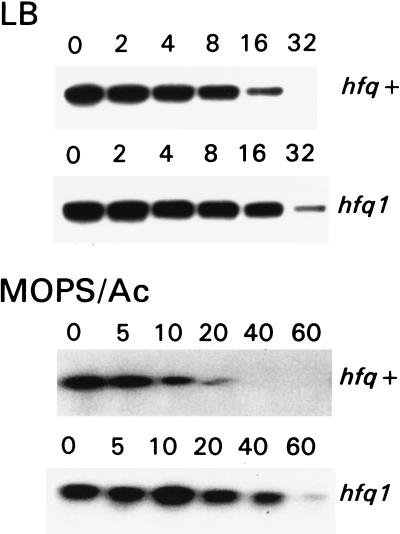
To examine whether Hfq plays a role in mRNA decay, we determined the ompA mRNA half-life in the hfq mutant strains. Mutant and parent cells were cultured in either Mops-acetate medium or LB to the middle of the exponential phase. Rifampicin was added to block transcription, and aliquots were withdrawn in time intervals thereafter. Total cellular RNA was isolated, and degradation of the ompA transcript was analyzed by Northern blotting (Fig. 5, Table 1). The half-life of the ompA mRNA was found to increase two times in LB and six times in Mops-acetate medium, if the hfq1 mutant is compared with the parent strain. This stabilization occurred even though the mutant cells grew slower in both media. Thus, we demonstrated the participation of the hfq gene in ompA mRNA degradation. Moreover, in the hfq1 mutant the stability of the ompA transcript was higher in cells grown in Mops-acetate medium than in cells grown in LB. This result shows that the hfq1 mutation eliminates the growth rate-dependent regulation of the ompA mRNA stability. In the hfq2 background no aberration of the ompA regulation was observed, though the RNA binding was still impaired in the in vitro assay (see above). Indeed, it previously has been reported that the hfq2 mutant has a similar phenotype as the parent strain and served as a control for polarity effect of downstream genes (21–23). To verify that Hfq plays a role in growth rate-regulated degradation of mRNA we analyzed the stability of the lpp transcript, which previously was shown to be independent of bacterial growth rate (2) and served as a control. The half-life of the lpp mRNA was similar in hfq1 and parent strains, irrespective of the growth rate. We tested the binding of purified Hfq to the lpp 5′ UTR and found no specific complex formation with this mRNA (Fig. 3). Furthermore, no hfq-dependent mRNA-binding activity of cells was found to bind to the lpp transcript (data not shown). These data support the conclusion that Hfq facilitates specifically the degradation of the ompA mRNA by direct binding to the transcript.
Figure 5.
Influence of the hfq1 mutation on the ompA mRNA stability analyzed by Northern blotting. Mutant and parent cells were cultured in either LB or Mops-acetate medium. In the middle of exponential phase, transcription was blocked by rifampicin and aliquots were taken out at the indicated (in min) time points. Total RNAs were isolated by the method of hot phenol extraction, separated on 1% agarose gels, transferred to nitrocellulose filter, and hybridized with the 32P-labeled DNA probe, complemented to the coding region of the ompA gene.
The observation that the hfq1 mutation affects neither the amounts of the key enzymes of RNA degradation [RNase E (21), PNPase, and RhlB helicase] (data not shown) nor the decay of bulk mRNA (21) agrees with our conclusion.
DISCUSSION
The rate of OmpA protein synthesis is regulated by the rate of bacterial growth and drops as the doubling time increases (35). This regulation has been reported (35) to be achieved by the changes in the degradation rate of the ompA mRNA, whose half-life can decrease by as much as a factor of four in slowly growing cells (2). The comparatively rapid decay of the ompA transcript at slow bacterial growth rates has been attributed to an increased rate of RNase E cleavage in the 5′ UTR (32). In spite of extensive research on the structure and function of RNase E and its cognate ompA substrate, no factor has been identified that could account for the growth rate-dependent RNase E cleavage that makes the ompA mRNA comparatively more vulnerable to degradation at slower growth rates.
In this report we provide evidence that Hfq plays a crucial role as a negative regulator of ompA mRNA stability, linking RNase E-controlled degradation to bacterial growth rate. The amount of Hfq was observed to be larger in cells grown at a slow rate than in cells grown more rapidly. In hfq1 cells lacking Hfq the pattern of growth rate-dependent regulation of the ompA mRNA is entirely lost. Furthermore, knockout of the hfq gene leads to a prolonged half-life of the ompA mRNA even during fast growth. We show that Hfq binds to certain mRNAs: Hfq binds to the 5′ UTR of the ompA transcript, but has no significant affinity for the lpp transcript, whose stability has been shown to be unaffected by bacterial growth rate (2). Because the RNA-binding capacity of Hfq for the 5′ UTR of the ompA mRNA parallels the rate of the full-length transcript degradation in cells, it is safe to suggest that Hfq regulates the growth rate-dependent RNase E cleavage in the 5′ end of the ompA message.
During the last few years the role of Hfq as a regulator of gene expression in E. coli has been increasingly appreciated (21–23, 36). In the case of synthesis of the stationary phase sigma factor, σs, Hfq has been suggested to act as a positive regulator by binding to the 5′ UTR of the rpoS transcript near the Shine-Dalgarno sequence, and thus stimulating ribosome binding (23, 37, 38). Recently, a negative role of Hfq in the synthesis of the MutS protein has been reported (21). Tsui et al. discovered an involvement of the hfq gene in the degradation of mutS, miaA, and hfq gene transcripts, and attributed this effect to a Hfq function as an RNA chaperone. However, none of these studies have confirmed a direct binding of Hfq to the analyzed mRNA species.
In our study we demonstrate the specific binding of Hfq to the stability determinant of the ompA mRNA, encompassing the RNase E cleavage sites. The exact binding site of Hfq remains to be elucidated. However, early studies addressing the role of Hfq in bacteriophage Qβ RNA replication suggest that Hfq binds preferentially to RNA at single-stranded and AU-rich regions (18, 19), somehow resembling the feature of RNase E cleavage sites. Recent results suggest a function of Hfq in recognition and melting of highly structured RNA, thereby making RNA templates more susceptible for RNA replicase, as in the case of Qβ phage RNA (39), or for ribosomes, as in the case of the rpoS transcript (36, 38). Thus, it is appealing to propose that Hfq assists RNase E in finding the cleavage targets of the ompA message, which are perturbed by secondary structures (5, 6) and thereby stimulates mRNA degradation. Such a model is supported by reports showing that secondary structure in the vicinity of cleavage sites impedes the action of RNase E (40).
We observed that the level of Hfq and its RBA is substantially higher in cells cultured at a slow growth rate than in those growing at a fast rate. In contrast, a concomitant change of cell growth rate and Hfq content has been reported (28). This conclusion is, however, based solely on the complicated method for protein determination that includes radioactive labeling and immunoprecipitation. It is noteworthy that the rpoS gene, specifically activated during stationary phase, does not affect the synthesis of Hfq (21). In agreement, the rpoS mutation did not influence the binding activity of Hfq, as assessed by ompA mRNA-binding assays (data not shown), nor the growth rate-related regulation of ompA mRNA half-life (Table 1). Furthermore, Hfq synthesis does not seem to depend on the growth rate-specific global regulator, ppGpp (guanosine tetraphosphate), as revealed by analysis of the relA spoT strain lacking ppGpp production (27) (data not shown). Further investigations are needed to reveal how Hfq expression is coupled to growth rate.
We tested the influence of two hfq mutations on the stability of the ompA message. One, hfq1, that is reported to disrupt protein function completely (22, 23) led to the stabilization of the ompA mRNA. The other one, hfq2, mapping to the carboxyl-terminal region of the Hfq protein, which served as a control for polarity of downstream genes, had no influence on the degradation of this transcript. Our data in addition to a previous study (21) show that the hfq1, but not hfq2, mutation affects mRNA stability. Thus, the carboxyl-terminal truncation of Hfq protein derived from the hfq2 mutation remains without impact on mRNA degradation, though an in vitro effect on RNA binding is shown by our study. The discrepancy between in vivo and in vitro results could be, at least partially, explained by the stability of the RNA–protein complex in mobility-shift assay, which in the case of the truncated form of protein might be disturbed.
The pioneering work of Maaløe (4) regarding the growth rate-related physiological adjustments of macromolecular synthesis in bacteria has suggested that the degradation of bulk mRNA, i.e. most mRNA species, is not affected by the bacterial doubling time, predicting that the core of the mRNA decay machinery is not controlled by growth rate. We confirm and explain this classical view by showing that the amount of RNase E (and the major degradosome components) is insensitive to growth rate. Thus, growth rate-dependent stability of mRNA is restricted to a subset of transcripts, e.g. ompA (2), whose degradation relies on auxiliary factors, such as Hfq. In agreement with this conclusion, we found that the degradation of the lpp transcript, which is not regulated by bacterial growth rate, does not seem to involve Hfq. Furthermore, no change in bulk mRNA degradation has been observed in hfq mutants (21).
Indirect evidence (such as migration in the mobility-shift assay, affinity to AU-rich RNA-region, and others) suggests that the Hfq-specific RBA is identical with the previously identified RBA1 (15). RBA1 was not purified to homogeneity, but it was suggested to contain GroEL. Because we definitively do not find GroEL to be a part of the Hfq-containing activity, our former conclusion of GroEL presence in RBA1 is not confirmed by the present study. RBA1 was found to increase in cells exposed to nutritional stress, i.e., slow growth rate plus anaerobiosis, when a retarded degradation of ompA mRNA as well as bulk mRNA was observed (16). On the basis of this correlation RBA1 was proposed to be involved in the repression of RNA decay. However, other factors that could influence the stability of bulk mRNA, for instance nuclease activities, were not characterized. In the present study we definitively show that disruption of the hfq gene (hfq1) leads to an increase of the ompA mRNA half-life but does not influence the level of degradosomal components. This result strongly suggests the destabilizing role of Hfq in the ompA mRNA turnover and supports the similar finding of other authors (21).
In vitro studies with the purified Hfq and isolated assembly of the RNase E degradosome components will allow comparison of lpp and ompA RNA templates as cleavage substrates and investigation of how the RBA of Hfq supports the mechanism of ompA mRNA degradation.
Acknowledgments
The skillful technical assistance of Mrs. L. Bayr is gratefully acknowledged. We are indebted to A. Hirsch for critically reading this manuscript. We also thank Dr. A. Ishihama (Department of Molecular Genetics, National Institute of Genetics, Mishima, Shizuoka, Japan) for the gift of the anti-Hfq antiserum and plasmid pHFQ607; Dr. A. J. Carpousis (Laboratoire de Microbiologie et Génétique Moléculaire du Centre National de la Recherche Scientifique, Toulouse, France) for antisera against the degradosome components; and Dr. R. Hengge-Aronis (Department of Biology, University of Konstanz, Konstanz, Germany) for providing the hfq mutants. This work was supported by Grant P10766-MED (to M.B.) and P11841-Mob (to A.v.G.) of the Austrian Research Fund.
ABBREVIATIONS
- UTR
untranslated region
- PNPase
polynucleotide phosphorylase
- RBA
RNA-binding activity
- LB
Luria–Bertani medium
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Lundberg U, Kaberdin V, von Gabain A. In: Manual of Industrial Microbiology and Biotechnology. Demain A L, Davies R M, Cohen G, Hershberg C L, Sherman D H, Willson R C, Wu J-H D, editors. Washington, DC: Am. Soc. Microbiol.; 1999. , in press. [Google Scholar]
- 2.Nilsson G, Belasco J G, Cohen S N, von Gabain A. Nature (London) 1984;312:75–77. doi: 10.1038/312075a0. [DOI] [PubMed] [Google Scholar]
- 3.Georgellis D, Arvidson S, von Gabain A. J Bacteriol. 1992;174:5382–5390. doi: 10.1128/jb.174.16.5382-5390.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maaløe O. In: Biological Regulation and Development. Goldberger R F, editor. New York: Plenum; 1979. pp. 487–542. [Google Scholar]
- 5.Chen L-H, Emory S A, Bricker A L, Bouvet P, Belasco J G. J Bacteriol. 1991;173:4578–4586. doi: 10.1128/jb.173.15.4578-4586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenbaum V, Klahn T, Lundberg U, Holmgren E, von Gabain A, Riesner D. J Mol Biol. 1993;229:656–670. doi: 10.1006/jmbi.1993.1070. [DOI] [PubMed] [Google Scholar]
- 7.Emory S A, Bouvet P, Belasco J G. Genes Dev. 1992;6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- 8.Mudd E A, Krisch H M, Higgins C F. Mol Microbiol. 1990;4:2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 9.Nilsson M, Lundberg U, von Gabain A. EMBO J. 1988;7:2269–2275. doi: 10.1002/j.1460-2075.1988.tb03067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghora B K, Apirion D. Cell. 1978;15:1055–1066. doi: 10.1016/0092-8674(78)90289-1. [DOI] [PubMed] [Google Scholar]
- 11.McDowall K J, Lin-Chao S, Cohen S N. J Biol Chem. 1994;269:10790–10796. [PubMed] [Google Scholar]
- 12.Miczak A, Kaberdin V, Wei C-L, Lin-Chao S. Proc Natl Acad Sci USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Py B, Higgins C F, Krisch H M, Carpousis A J. Nature (London) 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 14.Blum E, Py B, Carposis A J, Higgins C F. Mol Microbiol. 1997;26:387–398. doi: 10.1046/j.1365-2958.1997.5901947.x. [DOI] [PubMed] [Google Scholar]
- 15.Georgellis D, Sohlberg B, Hartl F U, von Gabain A. Mol Microbiol. 1995;16:1259–1268. doi: 10.1111/j.1365-2958.1995.tb02347.x. [DOI] [PubMed] [Google Scholar]
- 16.Georgellis D, Barlow T, Arvidson S, von Gabain A. Mol Microbiol. 1993;9:375–381. doi: 10.1111/j.1365-2958.1993.tb01698.x. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez F M, Eoyang L, August J T. Nature (London) 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez F M, Hayward W S, August J T. J Biol Chem. 1972;247:824–831. [PubMed] [Google Scholar]
- 19.Senear A W, Steitz J A. J Biol Chem. 1976;251:1902–1912. [PubMed] [Google Scholar]
- 20.Kajitani M, Ishihama A. Nucleic Acids Res. 1991;19:1063–1066. doi: 10.1093/nar/19.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsui H-C T, Feng G, Winkler M E. J Bacteriol. 1997;179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsui H-C T, Leung H-C E, Winkler M E. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 23.Muffler A, Fisher D, Hengge-Aronis R. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 24.Casadaban M J. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 25.Studier F W, Moffatt B A. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 26.Touati E, Dassa E, Dassa J, Boquet P L. Res Microbiol. 1991;142:29–36. doi: 10.1016/0923-2508(91)90094-q. [DOI] [PubMed] [Google Scholar]
- 27.Bremer H, Ehrenberg M. Biochem Biophy Acta. 1995;1262:15–36. doi: 10.1016/0167-4781(95)00042-f. [DOI] [PubMed] [Google Scholar]
- 28.Kajitani M, Kato A, Wada A, Inokuchi Y, Ishihama A. J Bacteriol. 1994;176:531–534. doi: 10.1128/jb.176.2.531-534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilm M, Shevchenko A, Houthave T, Breit S, Schweigerer L, Fotsis T, Mann M. Nature (London) 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 30.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 31.Wilm M, Mann M. Anal Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 32.Melefors Ö, von Gabain A. Cell. 1988;52:893–901. doi: 10.1016/0092-8674(88)90431-x. [DOI] [PubMed] [Google Scholar]
- 33.Kuwano M, Ono H, Hori K, Hirota Y, Ohnishi Y. Mol Gen Genet. 1977;154:279–285. doi: 10.1007/BF00571283. [DOI] [PubMed] [Google Scholar]
- 34.Lundberg U, Nilsson G, von Gabain A. Gene. 1988;72:141–149. doi: 10.1016/0378-1119(88)90136-9. [DOI] [PubMed] [Google Scholar]
- 35.Belasco J G, Nilsson G, von Gabain A, Cohen S N. Cell. 1986;46:245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- 36.Brown L, Elliott T. J Bacteriol. 1996;178:3763–3770. doi: 10.1128/jb.178.13.3763-3770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muffler A, Traulsen D D, Fischer D, Lange R, Hengge-Aronis R. J Bacteriol. 1997;179:297–300. doi: 10.1128/jb.179.1.297-300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown L, Elliott T. J Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuppli D, Miranda G, Tsui H-C T, Winkler M E, Sogo J M, Weber H. Proc Natl Acad USA. 1997;94:10239–10242. doi: 10.1073/pnas.94.19.10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDowall K J, Kaberdin V R, Wu S-W, Cohen S N, Lin-Chao S. Nature (London) 1995;374:287–290. doi: 10.1038/374287a0. [DOI] [PubMed] [Google Scholar]