Abstract
More than half of all colorectal carcinomas are known to exhibit an activated mitogen-activated protein kinase pathway. The NF1 gene, a negative regulator of KRAS, has not previously been examined in a series of colorectal cancer. In the present study, primary colorectal carcinomas stratified according to microsatellite instability status were analyzed. The whole coding region of NF1 was analyzed for mutations using denaturing high-performance liquid chromatography and sequencing, and the copy number alterations of NF1 were examined using multiple ligation-dependent probe amplification and real-time polymerase chain reaction. The mutational hot spots in KRAS and BRAF were sequenced, and promoter hypermethylation status of RASSF1A was assessed with a methylation-specific polymerase chain reaction. One sample had two missense mutations in NF1, whereas nine additional tumors had intronic mutations likely to affect exon splicing. Interestingly, 8 of these 10 tumors were microsatellite-unstable. Four other tumors showed a duplication of NF1. Mutations in KRAS and BRAF were mutually exclusive and were present at 40% and 22%, respectively. RASSF1A was hypermethylated in 31% of the samples. We show that the RAS signaling network is extensively dysregulated in colorectal carcinomas, because more than 70% of the tumors had an alteration in one or more of the four examined components.
Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer-related deaths in the western world today, and at least 50% of CRCs are thought to have a dysregulation of the RAS-RAF-MEK-ERK pathway, also known as the mitogen-activated protein kinase (MAPK) pathway [1]. When activated, this pathway leads to increased proliferation and reduced apoptosis, two of six crucial abilities of a cancer cell, as described by Hanahan and Weinberg [2]. There are several components in this pathway, which, theoretically, could be affected in cancer, and some are known mutational targets in cancer such as KRAS and BRAF. KRAS has been widely established as an important oncogene since its first mutational report in 1984 [3], and it is now known that it is mutated in 21% of all human sporadic cancers, including one third of CRCs [1].2 BRAF was shown to be a mutational target in cancer 5 years ago [4], and 20% of all human cancers harbors a mutation, including an estimated 13% of colorectal carcinomas.2 Another potential target of this pathway is the NF1 gene, which encodes neurofibromatosis type 1, a GTPase-activating protein (GAP), governing hydrolysis of KRAS-GTP to KRAS-GDP [5], thereby functioning as a negative regulator of KRAS signaling. The NF1 gene is approximately 280 kb in size and maps to chromosome 17q11.2. It contains 61 exons, with an 11- to 13-kb transcript and an open reading frame coding for 2818 amino acids. There are two catalytical domains in NF1, which are important for its function, namely, the cAMP/PKA domain comprising exons 11 to 17 and the RAS-GRD (RAS GAP-related domain) domain comprising exons 21 to 27a [6–8]. Neurofibromatosis type 1, a dominant disorder, is caused by mutations in NF1, but somatic mutations in this gene can also contribute to tumorigenesis. Since the first mutation report of the gene in 1992 showing that one colorectal tumor (of 22) was mutated in NF1 [9], it has been speculated to play a role in colorectal tumorigenesis. However, due to the large size of the gene and the fact that there are no mutational hot spots, mutation analysis of NF1 in tumors has been very scarce. RASSF1 (Ras association domain family 1) gene maps at chromosome 3p21.3, and its isoform A (RASSF1A) has been found hypermethylated in 40% of lung tumors [10] and in a large variety of human cancers, including CRC [11,12]. As implied by its designation, RASSF1A is thought to interact with KRAS through a Ras association domain that alters its effects. RASSF1A has several effects, including promotion of apoptosis, cell cycle arrest, and maintenance of genomic stability, abilities typical of tumor suppressor genes. Some of these effects refer to the negative regulation of KRAS [13]. Its association to, and its effect on, KRAS is still not solved, although increasing evidence points to a direct binding between RASSF1A and farnesylated KRAS (reviewed in the study of Donninger et al. [11]). The KRAS and BRAF mutation status together with the alteration of other upstream components affecting the RAS signaling have been reported for other cancers [14], but only two previous studies have examined alterations in KRAS, BRAF, and RASSF1A in the same series of colorectal neoplasms [15,16], and independent of cancer type, no previous study has included a detailed analyses of the NF1 gene.
To provide further insight into the role of MAP kinase signaling in CRC, we carried out the first comprehensive mutation analyses of the NF1 gene in colorectal carcinomas in comparison with alterations of BRAF, KRAS, and RASSF1A in a sample series selected to include a comparable number of samples with and without the microsatellite instability phenotype.
Materials and Methods
Tissue Specimen
Sixty-five sporadic colorectal carcinomas from 64 patients with a mean age of 70 years (range 33–92 years), and an equal distribution of male-female were included in the present study. Twenty-nine samples displayed microsatellite instability (MSI), whereas 36 were microsatellite-stable (MSS). All tumors were nonfamilial as assessed by written questionnaires and cross check with the Norwegian Cancer Registry [17]. The colon, including the rectum, was divided into proximal and distal sections: the proximal, or right side, spans from cecum to two thirds of the way across transversum; the distal, or left side, comprises the last third of the transversum, sigmoideum, and the rectum. Of the 65 samples, 23 were located in the proximal colon and 42 were located in the distal colon. The carcinomas are from a prospective series collected from seven hospitals in the Southeast region of Norway during 1987–1989 and contain, on average, 84% tumor cells [18]. The tumors have been selected to achieve a consistently higher number of MSI-positive tumors compared to the normal distribution (15%). By stratifying the samples according to the MSI status, we ensured that any results associated with the MSI or MSS group would be detected.
NF1 Mutation Screening — DNA Amplification and Denaturing High-Performance Liquid Chromatography Analysis
Twenty-four representative CRC samples were analyzed for mutations in the NF1 gene. These samples were selected to resemble the remaining series with regards to sex, age, tumor location, MSI status, and KRAS and BRAF mutation status. The 61 NF1 gene exons were amplified in 61 polymerase chain reaction (PCR) fragments of 172 to 579 bp. The primers were generally positioned approximately 50 to 60 bp from the intron-exon boundary to allow the detection of splicing defects while minimizing intronic polymorphisms. In total, 19,843 bases were screened per sample to obtain the final mutation status. The dHPLC was carried out as previously published [19], with minor alterations in the PCR protocol and denaturing high-performance liquid chromatography (dHPLC) methods. For details concerning the dHPLC, please refer to Table W1.
In short, the initial PCR was carried out in 25 µl of reaction volumes and was cooled at room temperature for 60 minutes to yield heteroduplex formation. The identification of somatic NF1 gene mutations was carried out with dHPLC on a 3500HT WAVE DNA fragment analysis system (Transgenomic, Crewe, UK) equipped with a DNASep column (Transgenomic). Polymerase chain reaction products were examined through a 5% linear acetonitrile gradient for heteroduplexes with a separation flow rate of 1.5 ml/min. Commercially availableWAVE Optimized Buffers (A, B, and D; Transgenomic) and Syringe Solution (Transgenomic) were used to provide highly reproducible retention times with WAVE System instrumentation. Resolution temperatures and starting concentrations of buffer B for dHPLC analysis are reported in Table W1.
Sequencing
For each dHPLC abnormal elution profile, genomic DNA was reamplified with dHPLC primers and directly sequenced in both directions on a 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). Forward and reverse sequences were analyzed and compared with the mRNA reference sequence and with the chromosome 17 genomic contig reference sequence (NM_000267). The first base (position +1) of the initiator methionine is taken as the start of the cDNA. All missense and splicing mutations detected were absent on 200 control chromosomes belonging to the unaffected subjects.
KRAS and BRAF Mutation Screening
The mutational hot spots of KRAS (exons 2 and 3) and BRAF (exons 11 and 15) were directly sequenced in both directions for all samples (n = 65) on an ABI PRISM 377 DNA Sequencer (Applied Biosystems) and an ABI PRISM 3730 DNA Sequencer (Applied Biosystems). All nucleotide numbers are based on the cDNA reference sequence (BRAF, GenBank Accession No. NM_004333; KRAS, GenBank Accession No. NM_004985). For primer details please see Table W1.
Methylation-Specific PCR of RASSF1A
Methylation-specific PCR (MSP) of RASSF1A were performed with published primers [20]. Polymerase chain reaction conditions were as follows: denaturation and enzyme activation at 95°C for 15 minutes; 35 cycles of 30 seconds of denaturation at 95°C, 30 seconds of annealing at 62°C, and 30 seconds of elongation at 72°C; final extension at 72°C for 7 minutes.
Human placental DNA (Sigma Chemical Co., St. Louis, MO) treated in vitro with SssI methyltransferase (New England Biolabs Inc., Beverly, MA) was used as a positive control for MSP of methylated alleles, whereas DNA from normal lymphocytes was used as a control for unmethylated alleles. The PCR products were separated using a 2% agarose gel before individual visual scoring by two people. Methylated samples with intensity equal to, or higher than, the positive control were considered to be hypermethylated.
Multiple Ligation-Dependent Probe Amplification Analysis
Screening for NF1 single- and multiexon deletions was carried out in 24 of the colorectal carcinomas using the SALSA P081/082 NF1 (version 04, 05-02-2005) multiple ligation-dependent probe amplification (MLPA) assay (MRC Holland, Amsterdam, The Netherlands), as instructed by the manufacturer and previously reported [21]. In brief, two probes in each exon were hybridized to the individual tumor DNA, followed by a ligation of the nick between the probes, and PCR amplification with 6-FAM-labeled universal primers. The amplified product was analyzed on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems), and the results were exported to Coffalyzer v.5 Software (MRC Holland). As controls, and in each experiment, we used five normal blood samples taken from healthy individuals who do not show NF1 phenotypic traits as determined by clinical evaluation. Furthermore, these controls are verified to have an unaltered NF1, both at the nucleotide and at the copy number level. A ratio of ∼1 should be obtained if both alleles are present. A reduction or increase in the peak area values to <0.7 or >1.3 was considered an indication of a deletion or a gain, respectively. DNA samples showing a reduction or increase in the MLPA peak area according to the chosen threshold values were re-analyzed by MLPA, and only the samples showing consistent results between the two experiments were scored as deleted or gained.
Real-Time Polymerase Chain Reaction
The gene gains identified by MLPA were also confirmed with a TaqMan Real-Time PCR experiment using an ABI 7000 Sequence Detection System (Applied Biosystems). Two TaqMan probes mapping in NF1 exons 25 and 28, respectively, were designed by File Builder 3.1 software (Applied Biosystems). These were amplified separately together with the endogenous control (RNaseP) in 96-well fast plates following the recommended protocol (Applied Biosystems). All samples were analyzed in parallel, and the mean value was used for data analysis. In cases where N-fold was below the maximum N-fold copy number observed among the nondeleted DNA used as negative controls, it was accepted that the test sample harbored one copy of the target gene. In cases where N-fold resulted above the minimum N-fold copy number observed among the nondeleted DNA, it was accepted that the test sample harbored two or more copies of the target gene.
Statistics
For this study, 2 x 2 contingency tables were analyzed using Fisher's exact test, whereas 3 x 2 tables were analyzed by the Pearson chisquare test. An independent t test was performed when comparing continuous normally distributed data between two groups. All P values were derived from statistical tests using the SPSS Version 15.0 software (SPSS, Chicago, IL), and considered statistically significant at P ≤ .05.
Results
NF1
One of the 24 carcinomas contained two missense mutations (D1302Y and V2577G), the first located within the RAS-GRD domain.
In silico protein modeling showed that D1302Y has lost an exposed negative charge, which may be important in protein folding and in binding to other proteins. The V2577G most likely has no effect on the neurofibromin function. Additional nine tumors displayed intronic mutations in the range of 4 to 57 bases away from the intron-exon boundary (Table W2).
Using MLPA, we found that another 4 (17%) of 24 samples had a gain of parts or of the whole gene, also confirmed with real-time analysis (Table W2).
Comparison of the molecular results with clinicopathologic data showed that 8 of 10 samples with exonic or intronic alterations in NF1 occurred in MSI-positive tumors (P = .047), whereas 3 of 4 duplications occurred in MSS tumors.
KRAS
Direct sequencing of exons 2 and 3 of KRAS revealed that 26 (40%) of 65 tumors harbored a mutation (Table W2). All but two mutations were missense mutations and occurred in codon 12, 13, or 61. These two were a 3-bp insertion (TGG) in exon 2 (c.49insTGG) and a 3-bp deletion in exon 3, codons 62 to 63 (c.184_189delGAG; Table W2). Furthermore, two of the tumors had two KRAS mutations each. One displayed both G12A and V14I mutations, and the second had both G12D and G13D.
Mutations in KRAS were seemingly more often present in MSS tumors than in MSI tumors, 69% versus 46% (P = .08).
BRAF
Mutational analysis of BRAF gene revealed that 14 (22%) of 64 samples harbored a mutation. Eleven of these were the typical V600E mutation; the remaining three were D594G, L597Q, and G1406C (Table W2).
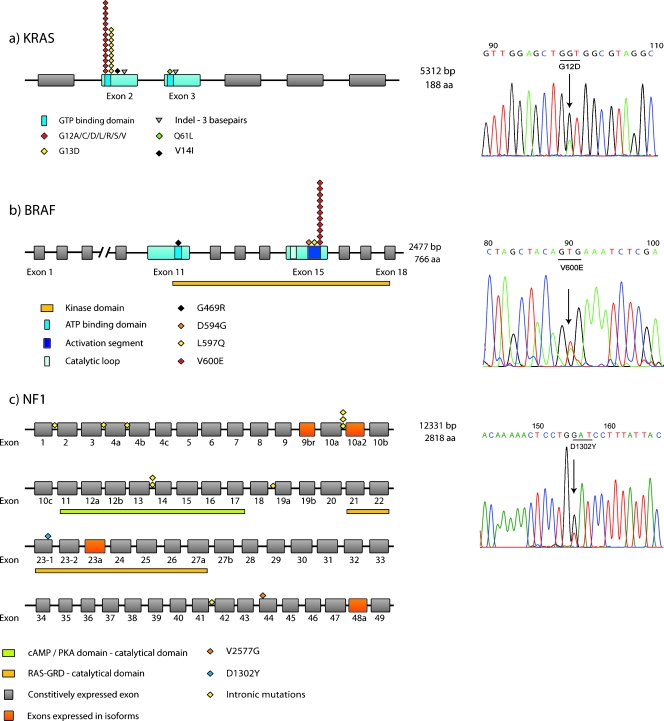
Mutations in BRAF were strongly associated to MSI, female gender, and proximal location (P = .006, P = .015, and P = .025, respectively). Figure 1 illustrates the individual localization of each mutation in KRAS, BRAF, and NF1.
Figure 1.
Site distribution of mutations within each gene. The mutations for the respective gene are placed according to their sequence position. In (a) and (b), only the exons in blue have been analyzed. In (c), all exons are analyzed, and the exons in orange indicate those that are only expressed in isoforms. To the right, representative sequencing results of mutant samples are presented.
RASSF1A
By MSP analysis, we found that 18 (31%) of 59 samples were hypermethylated in the promoter of RASSF1A. Methylation of the gene was more frequent in the distally located tumors (P = .041), but was not overlapping with the MSS phenotype. In eight tumors, hypermethylation of RASSF1A was the only observed alteration among the four genes analyzed here. We found no covariance between RASSF1A methylation and mutation status of either of the analyzed genes.
Dysregulation of RAS Signaling
When looking at concurrent mutations in individual tumors, we found that KRAS and BRAF were mutually exclusive because all BRAF mutations occurred in wild type KRAS tumors and vice versa (P < .0001). The sample with NF1 missense mutations was MSI-positive, proximally located, and harbored a BRAF mutation. When including the intronic mutations in the number of NF1 mutations, six of eight BRAF mutations occurred in NF1-mutated samples (P = .03), overlapping with the MSI. Three of the four duplications found in the NF1 locus occurred in tumors with wild type BRAF and KRAS. The remaining tumor had both a KRAS mutation and duplication.
The occurrence of RASSF1A hypermethylation in the presence of other mutations did not show any trends toward coexistence or mutually exclusive nature.
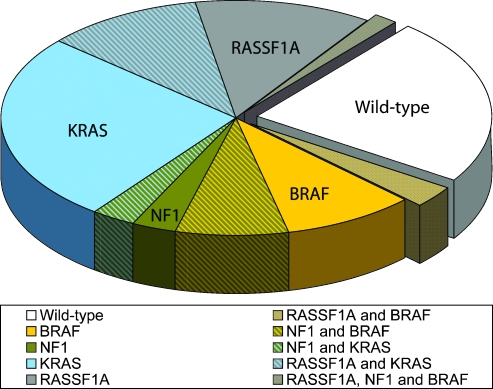
Taken together, we found that 74% (48/65) of the tumors had an overactive RAS signaling pathway due to change of at least one of the four analyzed components (one alteration in 37/48, two alterations in 10/48, and three alterations in 1/48). For the 24 samples submitted for complete analyses, the number of samples with at least one alteration was 19 (79%): one alteration was seen in 14 samples, two alterations were seen in 4 samples, and three alterations were seen in 1 sample. All samples and alterations are summarized in Figure 2 and Table W2.
Figure 2.
Alterations across the sample series. The pie chart indicates the four analyzed components and the percentage of tumors which showed alterations among these. Clockwise from the wild type pie, we see alterations in RASSF1A and BRAF; BRAF; BRAF and NF1; NF1; NF1 and KRAS; KRAS; KRAS and RASSF1A; RASSF1A; RASSF1A, NF1 and BRAF.
Discussion
This is the first report with an extensive analysis of the role of NF1 mutations in colorectal tumorigenesis. Previous mutation studies have only looked at a small number of samples, usually in a limited part of the gene, in the RAS-GAP domain. The initial mutational report on NF1 showed that 1 of 22 colorectal adenocarcinomas harbored a mutation in the RAS-GAP domain using single-strand conformation polymorphism [9]. Another study of 10 colorectal cell lines and 4 sporadic tumors using protein truncation test disclosed mutations in the NF1 coding region in four MSI cell lines (40%) and one MSI tumor (25%). Two of the cell lines had in fact two mutations each [22]. A recent study examined five hereditary non-polyposis CRC patients for mutations in five exons and found a mutation in one (20%) of the patients who had a homozygous germline mutation of MLH1 [23]. Moreover, loss of heterozygosity (LOH) at loci within the NF1 gene have been shown in primary colorectal tumors (range, 14–57%) [24,25]. One of these studies also used realtime expression studies of NF1 in 55 of the carcinomas and found an increased expression among tumors compared with normal tissue. In the COSMIC database [26], 79 carcinomas of the colorectum were apparently included among the NF1 data, yielding a mutation frequency of 11%. However, seven of nine mutations reported were from one study including seven cell lines, leaving only two of the mutations occurring in sporadic primary tumors.
In this study, we found the NF1 mutation profile to be in contrast to published germline mutation profile of NF1 patients3 as well as to the somatic mutation profiles of malignant peripheral nerve sheath tumor taken from patients with and without the NF1 disease [27,28] (Bottillo I et al., unpublished observations). Furthermore, the median age of the patients included in the present CRC series is old, suggesting that potential NF1 carriers among them should have shown a debut of an NF1-associated cancer type. As no typical NF1-associated tumors are recorded, based on written questionnaires and confirmation of cancer diagnoses from the Norwegian Cancer Registry [17], it further support that the reported mutations are somatic. The observed intronic mutations prevailing among the colorectal tumors could be involved in alternative splicing but this remains to be elucidated. Four of the nine intronic mutations were indels of one or two bases in microsatellites and reflect replication slippage (which often occurs in such repetitive stretches of bases) left unrepaired by the defective DNA mismatch repair system [29]. No such indels were found in KRAS or BRAF.
Multiple ligation-dependent probe amplification results showed that 17% of the analyzed samples had gained parts of or whole of the NF1 gene. This is not in accordance with the expectations of a tumor suppressor gene involved in tumorigenesis. A duplication of NF1 could lead to a stronger negative regulation of KRAS, with subsequent stronger control of proliferation and differentiation. The duplications may arise as a consequence of the chromosomal instability present in three of the four tumors, which yield a wide range of gains and losses of whole or parts of chromosomes. As reported by ĈaĈev et al. [30], colorectal tumors show a significant increase of NF1 mRNA expression compared with corresponding normal tissue. They also showed that the expression of NF1 isoform I (lacking exon 23a, located in the middle of the RAS-GRD domain) was significantly higher in tumor compared with normal tissue [30]. As of this, the present findings are in agreement with those of the study by ĈaĈev et al. [30].
We also found a 40% mutation frequency of KRAS, which is within the expected range [26]. A mutated KRAS (in codons 12, 13, and 61) hinders the hydrolysis of GTP to GDP, and will keep KRAS in a constitutively active state, leading to phosphorylation of downstream effectors such as BRAF [31]. BRAF mutations are known to be strongly associated with MSI and CpG island methylator phenotype [32,33] and are found very often mutated in sessile-serrated adenomas, lesions often considered as a precursor of MSI-H tumors [34–38]. We found BRAF mutation in 22% of the samples, a higher frequency than in the mutation databases [26]. This reflects a bias due to the enrichment of MSI tumors in the present series. In one study, 71% of the MSI tumors had a V600E mutation in BRAF, as opposed to 7% in the chromosomal-unstable tumors [39], a figure comparable with the present series, as 18 (62%) of 29 of MSI tumors had BRAF mutations. The most common BRAF mutation, V600E, just as the common KRAS mutations, will lead to a constitutively active protein, as the activation loop of the protein is changed [31].
Some studies indicate an indirect interaction between RASSF1A and KRAS through RASSF5 (previously annotated NORE1A) [12], whereas others argue for a direct binding between RASSF1A and activated, farnesylated, KRAS [11]. Previous studies have also included RASSF1A when analyzing the impact of KRAS and BRAF mutations in colorectal tumorigenesis [13,15,16,40], and none of them found any co-occurrence between RASSF1A methylation and BRAF or KRAS mutation, in line with the present finding.
When adding the data of the fourth component, NF1, of the RAS signaling pathway, we found that more than 70% of the samples had a hyperactive RAS signaling. As the effect of RASSF1A on RAS signaling is still unclear, the eight samples with the sole alteration being hypermethylation may not be important for an overactive RAS signaling pathway. When we exclude the RASSF1A data from the combined analysis, 62% (40/65) of the samples had an overactive RAS signaling network, all due to KRAS or BRAF mutations, as the sample with the NF1 missense mutations overlapped with BRAF mutation. If we include the NF1 changes potentially affecting the splicing, 77% of the tumors have a dysregulation of the RAS signaling pathway.
In conclusion, we show that the RAS signaling network is extensively dysregulated in colorectal carcinomas as more than 70% of the tumors have an alteration in one or more of the four components.
Supplementary Material
Abbreviations
- CRC
colorectal cancer
- MAPK
mitogen-activated protein kinase
- MSI
microsatellite instability
- MSS
microsatellite stable
- dHPLC
denaturing high-performance liquid chromatography
- MSP
methylation-specific PCR
- MLPA
multiple ligation-dependent probe amplification
Footnotes
This article refers to supplementary materials, which are designated by Tables W1 and W2 and are available online at www.neoplasia.com.
Sanger Institute—Catalogue of Somatic Mutations in Cancer (COSMIC) Web site.
NFI International Mutation Database (http://www.nfmutation.org).
References
- 1.Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322–327. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Santos E, Martin-Zanca D, Reddy EP, Pierotti MA, Della PG, Barbacid M. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science. 1984;223:661–664. doi: 10.1126/science.6695174. [DOI] [PubMed] [Google Scholar]
- 4.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 5.Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104:593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 6.Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, Conroy L, Clark R, O'Connell P, Cawthon RM. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- 7.Xu GF, Lin B, Tanaka K, Dunn D, Wood D, Gesteland R, White R, Weiss R, Tamanoi F. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990;63:835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- 8.Fahsold R, Hoffmeyer S, Mischung C, Gille C, Ehlers C, Kucukceylan N, Abdel-Nour M, Gewies A, Peters H, Kaufmann D, et al. Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am J Hum Genet. 2000;66:790–818. doi: 10.1086/302809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Bollag G, Clark R, Stevens J, Conroy L, Fults D, Ward K, Friedman E, Samowitz W, Robertson M. Somatic mutations in the neurofibromatosis 1 gene in human tumors. Cell. 1992;69:275–281. doi: 10.1016/0092-8674(92)90408-5. [DOI] [PubMed] [Google Scholar]
- 10.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 11.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120:3163–3172. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 12.Hesson LB, Cooper WN, Latif F. The role of RASSF1A methylation in cancer. Dis Markers. 2007;23:73–87. doi: 10.1155/2007/291538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harada K, Hiraoka S, Kato J, Horii J, Fujita H, Sakaguchi K, Shiratori Y. Genetic and epigenetic alterations of Ras signalling pathway in colorectal neoplasia: analysis based on tumour clinicopathological features. Br J Cancer. 2007;97:1425–1431. doi: 10.1038/sj.bjc.6604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McIntyre A, Summersgill B, Spendlove HE, Huddart R, Houlston R, Shipley J. Activating mutations and/or expression levels of tyrosine kinase receptors GRB7, RAS, and BRAF in testicular germ cell tumors. Neoplasia. 2005;7:1047–1052. doi: 10.1593/neo.05514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliveira C, Velho S, Domingo E, Preto A, Hofstra RM, Hamelin R, Yamamoto H, Seruca R, Schwartz S., Jr Concomitant RASSF1A hypermethylation and KRAS/BRAF mutations occur preferentially in MSI sporadic colorectal cancer. Oncogene. 2005;24:7630–7634. doi: 10.1038/sj.onc.1208906. [DOI] [PubMed] [Google Scholar]
- 16.Miranda E, Destro A, Malesci A, Balladore E, Bianchi P, Baryshnikova E, Franchi G, Morenghi E, Laghi L, Gennari L, et al. Genetic and epigenetic changes in primary metastatic and nonmetastatic colorectal cancer. Br J Cancer. 2006;95:1101–1107. doi: 10.1038/sj.bjc.6603337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lothe RA, Peltomäki P, Meling GI, Aaltonen LA, Nystrom-Lahti M, Pylkkanen L, Heimdal K, Andersen TI, Moller P, Rognum TO. Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res. 1993;53:5849–5852. [PubMed] [Google Scholar]
- 18.Meling GI, Lothe RA, Borresen AL, Hauge S, Graue C, Clausen OP, Rognum TO. Genetic alterations within the retinoblastoma locus in colorectal carcinomas. Relation to DNA ploidy pattern studied by flow cytometric analysis. Br J Cancer. 1991;64:475–480. doi: 10.1038/bjc.1991.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Luca A, Schirinzi A, Buccino A, Bottillo I, Sinibaldi L, Torrente I, Ciavarella A, Dottorini T, Porciello R, Giustini S, et al. Novel and recurrent mutations in the NF1 gene in Italian patients with neurofibromatosis type 1. Hum Mutat. 2004;23:629. doi: 10.1002/humu.9245. [DOI] [PubMed] [Google Scholar]
- 20.Koul S, Houldsworth J, Mansukhani MM, Donadio A, McKiernan JM, Reuter VE, Bosl GJ, Chaganti RS, Murty VV. Characteristic promoter hypermethylation signatures in male germ cell tumors. Mol Cancer. 2002;1:8. doi: 10.1186/1476-4598-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Luca A, Bottillo I, Dasdia MC, Morella A, Lanari V, Bernardini L, Divona L, Giustini S, Sinibaldi L, Novelli A, et al. Deletions of NF1 gene and exons detected by multiplex ligation-dependent probe amplification. J Med Genet. 2007;44:800–808. doi: 10.1136/jmg.2007.053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Montmain G, Ruano E, Upadhyaya M, Dudley S, Liskay RM, Thibodeau SN, Puisieux A. Neurofibromatosis type 1 gene as a mutational target in a mismatch repair-deficient cell type. Hum Genet. 2003;112:117–123. doi: 10.1007/s00439-002-0858-4. [DOI] [PubMed] [Google Scholar]
- 23.Alotaibi H, Ricciardone MD, Ozturk M. Homozygosity at variant MLH1 can lead to secondary mutation in NF1, neurofibromatosis type I and early onset leukemia. Mutat Res. 2008;637:209–214. doi: 10.1016/j.mrfmmm.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Cawkwell L, Lewis FA, Quirke P. Frequency of allele loss of DCC, p53, RBI, WT1, NF1, NM23 and APC/MCC in colorectal cancer assayed by fluorescent multiplex polymerase chain reaction. Br J Cancer. 1994;70:813–818. doi: 10.1038/bjc.1994.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leggett B, Young J, Buttenshaw R, Thomas L, Young B, Chenevix-Trench G, Searle J, Ward M. Colorectal carcinomas show frequent allelic loss on the long arm of chromosome 17 with evidence for a specific target region. Br J Cancer. 1995;71:1070–1073. doi: 10.1038/bjc.1995.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forbes S, Clements J, Dawson E, Bamford S, Webb T, Dogan A, Flanagan A, Teague J, Wooster R, Futreal PA, et al. COSMIC 2005. Br J Cancer. 2006;94:318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Luca A, Buccino A, Gianni D, Mangino M, Giustini S, Richetta A, Divona L, Calvieri S, Mingarelli R, Dallapiccola B. NF1 gene analysis based on DHPLC. Hum Mutat. 2003;21:171–172. doi: 10.1002/humu.9111. [DOI] [PubMed] [Google Scholar]
- 28.Upadhyaya M, Han S, Consoli C, Majounie E, Horan M, Thomas NS, Potts C, Griffiths S, Ruggieri M, von Deimling A, et al. Characterization of the somatic mutational spectrum of the neurofibromatosis type 1 (NF1) gene in neurofibromatosis patients with benign and malignant tumors. Hum Mutat. 2004;23:134–146. doi: 10.1002/humu.10305. [DOI] [PubMed] [Google Scholar]
- 29.Perucho M. Cancer of the microsatellite mutator phenotype. Biol Chem. 1996;377:675–684. [PubMed] [Google Scholar]
- 30.Ĉaĉev T, Radosevic S, Spaventi R, Pavelic K, Kapitanovic S. NF1 gene loss of heterozygosity and expression analysis in sporadic colon cancer. Gut. 2005;54:1129–1135. doi: 10.1136/gut.2004.053348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 32.Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS, Ogino S. IGFBP3 promoter methylation in colorectal cancer: relationship with microsatellite instability, CpG island methylator phenotype, and p53. Neoplasia. 2007;9:1091–1098. doi: 10.1593/neo.07760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, Hernandez NS, Chen X, Ahmed S, Konishi K, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci USA. 2007;104:18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higuchi T, Jass JR. My approach to serrated polyps of the colorectum. J Clin Pathol. 2004;57:682–686. doi: 10.1136/jcp.2003.015230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jass JR. Serrated adenoma of the colorectum and the DNA-methylator phenotype. Nat Clin Pract Oncol. 2005;2:398–405. doi: 10.1038/ncponc0248. [DOI] [PubMed] [Google Scholar]
- 36.Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makinen MJ. Colorectal serrated adenocarcinoma. Histopathology. 2007;50:131–150. doi: 10.1111/j.1365-2559.2006.02548.x. [DOI] [PubMed] [Google Scholar]
- 38.Wynter CV, Walsh MD, Higuchi T, Leggett BA, Young J, Jass JR. Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut. 2004;53:573–580. doi: 10.1136/gut.2003.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 40.Derks S, Postma C, Moerkerk PT, van den Bosch SM, Carvalho B, Hermsen MA, Giaretti W, Herman JG, Weijenberg MP, de Bruine AP, et al. Promoter methylation precedes chromosomal alterations in colorectal cancer development. Cell Oncol. 2006;28:247–257. doi: 10.1155/2006/846251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.