Abstract
Spectrin is an important structural component of the plasma membrane skeleton. Heretofore-unidentified isoforms of spectrin also associate with Golgi and other organelles. We have discovered another member of the β-spectrin gene family by homology searches of the GenBank databases and by 5′ rapid amplification of cDNA ends of human brain cDNAs. Collectively, 7,938 nucleotides of contiguous clones are predicted to encode a 271,294-Da protein, called βIII spectrin, with conserved actin-, protein 4.1-, and ankyrin-binding domains, membrane association domains 1 and 2, a spectrin dimer self-association site, and a pleckstrin-homology domain. βIII spectrin transcripts are concentrated in the brain and present in the kidneys, liver, and testes and the prostate, pituitary, adrenal, and salivary glands. All of the tested tissues contain major 9.0-kb and minor 11.3-kb transcripts. The human βIII spectrin gene (SPTBN2) maps to chromosome 11q13 and the mouse gene (Spnb3) maps to a syntenic region close to the centromere on chromosome 19. Indirect immunofluorescence studies of cultured cells using antisera specific to human βIII spectrin reveal a Golgi-associated and punctate cytoplasmic vesicle-like distribution, suggesting that βIII spectrin associates with intracellular organelles. This distribution overlaps that of several Golgi and vesicle markers, including mannosidase II, p58, trans-Golgi network (TGN)38, and β-COP and is distinct from the endoplasmic reticulum markers calnexin and Bip. Liver Golgi membranes and other vesicular compartment markers cosediment in vitro with βIII spectrin. βIII spectrin thus constitutes a major component of the Golgi and vesicular membrane skeletons.
Spectrin is an essential component of the cellular membrane skeleton. First characterized in erythrocytes (1), spectrin maintains the structural integrity of the red blood cell membrane and is defective in several inherited hemolytic anemias (2). Isoforms of spectrin probably are present in all cells, where they help to establish and maintain an organized distribution of membrane proteins (3). Recently, spectrin homologues and their binding partners have been identified in association with several intracellular organelles and have been found to participate in membrane protein sorting and organelle transport (4–11). However, the precise identity of the Golgi- and vesicle-associated spectrin (or spectrins) has remained elusive. In humans and mice, four spectrin genes, encoding the αI-, αII-, βI-, and βII-spectrin subunits, have been identified (12), yet exhaustive PCR analysis has failed to identify any βI- or βII-spectrin gene products as the Golgi-associated spectrin (unpublished observations). We therefore searched the GenBank expressed sequence tags (EST) database for cDNAs related, but not identical, to known β spectrins, and based on candidates identified by that search have completed the cloning and identification of a βIII spectrin that fulfills the criteria of a Golgi- and vesicle-associated protein.‡‡
METHODS
DNA and RNA Analyses.
GenBank database searches and nucleotide sequence analysis were performed by using the National Center for Biotechnology Information blast programs and the Genetics Computer Group (Madison, WI) Sequence Analysis Software Package (13). IMAGE consortium cDNA clones were obtained from Genome Systems (St. Louis) or the American Type Culture Collection. A clone representing EST clone KIAA0302 (GenBank accession no. AB002300; ref.14) was kindly provided by O. Ohara (Kazusa DNA Research Institute, Chiba, Japan). Nucleotide sequencing used the dideoxy terminator method with AmpliTaq FS polymerase (Applied Biosystems) and a model 373 automated DNA sequencer (Applied Biosystems) in a stretch configuration. Analysis of gene expression utilized the Human RNA Master Blot and Multiple Tissue Northern blot (CLONTECH) according to manufacturer’s instructions. Identification of the 5′ end of the βIII spectrin clone by a 5′ rapid amplification of cDNA ends (RACE) reaction followed CLONTECH’s user manual for Marathon Ready cDNA using the antisense primer (5′-GTCCCAGTCCGAGTCAGGAAG-3′), human fetal brain adapter-ligated cDNA, the Marathon Adapter primer (AP-1), and Advantage KlenTaq Polymerase mix containing polymerases suitable for long-distance PCR (CLONTECH). Further amplification used nested primer pairs with the RACE products, which were then inserted into the TA cloning vector (Invitrogen). Four such clones were isolated and sequenced. All other procedures involving DNA followed standard methods (15).
Chromosome Localization.
Chromosome localization utilized the Stanford G3 radiation hybrid panel (Research Genetics, Huntsville, AL; ref. 16). Hybrid clones were assayed for the presence of the human βIII spectrin gene by using PCR. Using the primers 5′-TGCGCTGGAGAAGCTTACTG-3′ and 5′-TGGTGTCAGAAGCTGTCTGG-3′, a 149-bp product was generated when the βIII spectrin gene was present. Data were analyzed by the Stanford Human Genome Center Radiation Hybrid (RH) Server at http://shgc-www.stanford.edu (16). The βIII gene in mice was localized using the Jackson Laboratory BSS-interspecific [(C57BL/6JEi × SPRET/Ei)F1 females × SPRET/Ei males] backcross panel (17), human clone 42742, and mouse clone 479050. These probes detect a KpnI restriction fragment-length polymorphism corresponding to a hybridizing band of 6.0 kb in C57BL/6JEi and 6.5 kb in SPRET/Ei. The segregation pattern of the restriction fragment-length polymorphism in progeny of the cross was used to determine the map location of the gene.
Antibody Preparation and Immunologic Procedures.
An anti-peptide antiserum to βIII spectrin (Spb3C2) was produced by immunizing rabbits with a keyhole limpet hemocyanin-conjugated synthetic peptide (Research Genetics, Huntsville, AL). The peptide sequence was EGPGPGSGDEANGPRGER, corresponding to codons 2165–2182 of βIII spectrin, a region adjacent to the pleckstrin homology domain (PH) that shares little homology to other spectrins (Fig. 1B). A second rabbit antibody (PAb R-βIII7–11) was generated to a glutathione S-transferase (GST)-βIII spectrin fusion protein representing repeat units 7–11 (codons 1019–1464), prepared as before (4, 18). mAbs used were to β-COP and p58 (Sigma), TGN38 (Affinity BioReagents, Golden CO), and mannosidase II (ManII) (Babco, Richmond, CA). Polyclonal antibody (PAb) jasmin against AnkG119 has been described (4). mAb to calnexin was a gift from A. Helenius (Yale University); antibody to Bip was obtained from StressGen Biotechnologies, Victoria, Canada. HeLa, NRK, and MDCK cells were grown to subconfluent density in Eagle’s or Dulbecco’s minimal essential medium with 10% fetal calf serum (Biocell Laboratories) on Falcon tissue-culture chamber slides. Cells were washed with PBS (pH 7.4), fixed with 2–4% paraformaldehyde, permeabilized with 0.1% Triton-X100 or saponin in PBS, and incubated with specific antisera diluted 1:200 (Spb3C2) or 1:10,000 (PAb R-βIII7–11) in PBS with 2–3% BSA (fraction V; Sigma). In double-immunofluorescence studies, the incubation also included other specific antibodies diluted 1:100 or more. The cells were then washed with PBS and incubated with 1:1,000 Cy3-conjugated goat anti-rabbit antibody and 1:500 fluorescein isothiocyanate- or Cy2-conjugated goat anti-mouse antibody (Jackson ImmunoResearch). Fluorescence microscopy of the cells was done with a Nikon Microphot-FXA microscope or an Olympus IX70 dual-laser scanning confocal microscope. Western blotting of whole-cell lysates or of RIPA buffer extracts (50 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1% Nonidet P-40/0.5% deoxycholate) after SDS/PAGE was as described (4).
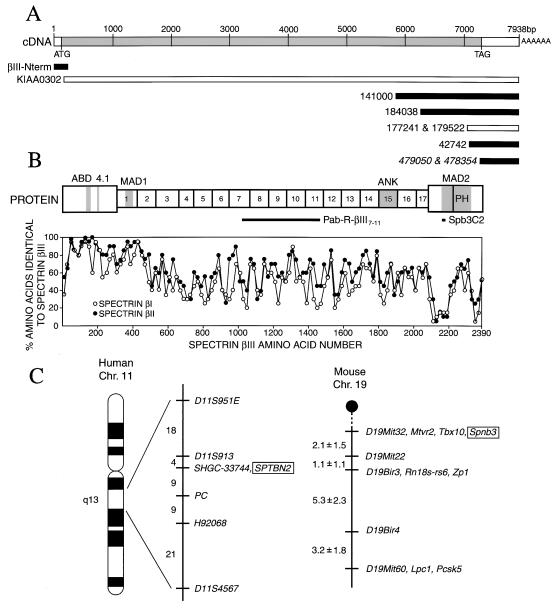
Figure 1.
Structure of βIII spectrin. (A) Relationship of the cDNA clones forming the βIII spectrin contig representing 7,938 nt. Homologous mouse clones are shown in italics. Clones fully sequenced in this work are shaded. After this work was completed, the full-length sequences of human and rat βIII spectrin were published independently in two separate reports (47, 48). (B) The ORF is predicted to encode a protein of 271,294 Da (2,391 amino acids) with potential functional domains similar to other spectrins. The actin-binding domain (ABD, shaded area) and protein 4.1-binding domain (4.1) are near the N terminus. They are followed by 17 tandem spectrin repeats. Repeats 1, 15, and 17 contain a potential membrane-association domain (MAD1), ankyrin-binding domain (ANK), and spectrin self-association site, respectively. Near the C terminus is a second membrane-association domain (MAD2, shaded) and the PH (boxed). The two peptides used to generate βIII spectrin-reactive antibodies are shown by the black bars. A comparison of the amino acid sequences of βIΣ2 and βIIΣ1 spectrins with βIII spectrin reveals extensive homology in the N-terminal domain, the repeats domain, and the PH domain, as shown by the identity-vs.-amino acid number plot. Each point represents the percent identity of 20 aligned amino acids. (C) Chromosome localization of βIII spectrin. (Left) The location of the human βIII spectrin gene (SPTBN2) on the long arm of chromosome 11 (11q13) is shown relative to other markers mapped in the panel. Gene symbols are on the right and genetic distances between markers in centirays are on the left. PC denotes the gene for pyruvate carboxylase. (Right) The location of the mouse βIII spectrin gene (Spnb3) on chromosome 19 is shown relative to other markers. Gene symbols are on the right and genetic distances between markers (centimorgans ± SE) are on the left. Gene symbols are Mtvr2, a mammary tumor virus receptor; Tbx10, a member of the Tbx1-subfamily of developmental genes; Rn18s-rs6, 18S-ribosomal RNA-related sequence 6; Zp1, zona pellucida glycoprotein 1; Lpc1, lipocortin 1; and Pcsk5, proprotein convertase subtilisin/kexin type 5. Detailed references are available at http://www.jax.org/resources/documents/cmdata/.
Isolation of Golgi Fractions from Rat Liver.
Diced fresh livers (100 g) from 16-hr-fasted adult male Sprague–Dawley rats were Polytron-homogenized at 4°C in 210 ml of homogenization buffer (10 mM Tris, pH 7.4/0.5 M sucrose/5 mM EDTA) and fractionated after low-speed centrifugation (756 × g, 10 min) by floatation through a discontinuous 1.2–0.5 M sucrose gradient, as published (19). Golgi-rich fractions were verified by the presence of ManII activity as measured by Western blotting and colorimetry at 410 nm (20).
RESULTS
Identification of βIII Spectrin.
We searched the GenBank EST database for cDNAs related to known β spectrins. Nucleotide sequences of human βIΣ1 spectrin (21), βIΣ2 spectrin (22), and βII spectrin (23) were used as query sequences. Five ESTs were initially identified that satisfied the screening criteria. They correspond to IMAGE Consortium clones 141000 (GenBank accession no. AA339388), 184038 (GenBank accession no. H30688), 177241 (GenBank accession no. H41849), 179522 (GenBank accession no. H51456), and 42742 (GenBank accession no. R61787). Nucleotide sequencing showed that the five clones overlap and encode portions of the same cDNA. All five clones contain a poly(A) tail. Clone 141000, the furthest 5′ clone, encompasses the other four clones and extends 2,099 bp to the poly(A) tail. Clones 42742, 184038, and 141000 were fully sequenced and are identical, with three exceptions: (i) a 293-bp insertion in clone 184038 after bp 530, (ii) a C → T polymorphism in clone 184038 at bp 1589, and (iii) variations in the sequence preceding the polyadenylation sequence (… TTCTC in 184038, … TTCTCTC in 141000, and … TTCTCTCAC in 42742). The insert in clone 184038 is bounded by consensus splice sequences, contains numerous stop codons, and appears to be a retained intron. Subsequent to these analyses, clone KIAA0302 (GenBank accession no. AB002300) appeared in the database. The 3′ portions of this clone overlapped our previously identified clones and extended these sequences nearly 5 kb upstream. The published sequence of clone KIAA0302, however, contains an extra G after bp 1032 (numbering according to clone 141000), which is predicted to cause a frameshift that disrupts the PH and leads to a new 44-aa C-terminal sequence. Sequencing of this clone (as obtained from O. Ohara) did not confirm the presence of the extra G, a fact verified by prokaryotic expression of this region of βIII spectrin (data not shown). Using primers derived from clone KIAA0302, 5′ RACE was carried out to complete the 5′ sequence. Four clones were generated by this reaction; all of them displayed identical sequences downstream of a putative ATG start codon. The largest (βIII-Nterm, GenBank accession no. AF079569) extended 126 bp upstream of the ATG site. Although the reading frame remains open upstream of the ATG codon in this clone, it is likely that translation of βIII spectrin is initiated at this codon because it is flanked by a perfect Kozak sequence and is homologous to the 5′ sequences of βI and βII spectrin.
A contig reveals a cDNA with 7,938 nucleotides (or slightly more if the variations in 3′ untranslated sequence are included; Fig. 1A). Its ORF predicts a 2,391-residue protein of 271,294 Da with a pI of 5.92. This protein is highly homologous to both βIΣ2 and βIIΣ1 spectrins (Fig. 1B) and preserves several functional sites characteristic of β spectrins, including the actin-binding domain (24), the protein 4.1 binding site (25), and MAD1 (18). The middle portion of the protein contains 17 spectrin repeats, the same number found in other β spectrins. The potential ankyrin-binding site in repeat 15 and the self-association site created by a partial 17th repeat (26) are also preserved. The latter feature suggests that βIII spectrin may associate with an as-yet-unidentified α-subunit. The first part of MAD2 (18) is less conserved, but the PH (18, 28) is similar to that in spectrins βIΣ2 and βIIΣ1. These homologies and distinctive features define this protein as another β spectrin. In accordance with current practice (12), we name this protein βIII spectrin (human gene symbol SPTBN2).
The nucleotide sequence of the human βIII spectrin contig also was used to search the EST database for cDNAs that may represent the mouse homologue. Two nearly identical clones were identified: clones 479050 (GenBank accession no. AA048840) and 478354 (GenBank accession no. AA049581). We completely sequenced clone 479050. It corresponds to the last 15 amino acids of human βIII spectrin (96% nucleotide identity) and the 3′ untranslated region (80% identity).
βIII Spectrin Is Widely Expressed and Is Located Near α-Actinin 3 on Chromosome 11q13.
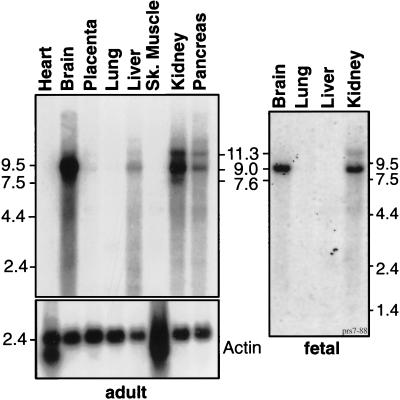
Hybridizing βIII spectrin clone 184038 to mRNAs from 50 adult and fetal human tissues spot-blotted onto a charge-modified nylon membrane reveals that βIII spectrin is widely expressed, but at different levels (Table 1). It is concentrated in the brain (the source of all of the sequenced clones) and is moderately abundant in the kidneys, liver, and testes and the prostate, pituitary, adrenal, and salivary glands. This pattern of expression is broader than βI spectrin but similar to that of βII spectrin. Northern blots of both adult and fetal tissues reveal a major 9.0-kb transcript easily detected in the brain, kidney, pancreas, and liver, with lesser amounts in placenta and lung (Fig. 2). Smaller amounts of an 11.3-kb mRNA were also present in most tissues. In kidneys, an additional 7.6-kb mRNA was noted.
Table 1.
Expression of βIII spectrin in 50 human tissues
| Tissue type | Expression level |
|---|---|
| Neurological | |
| Whole brain | 4+ |
| Cerebral cortex | 3+ |
| Frontal lobe | 3+ |
| Temporal lobe | 3+ |
| Occipital lobe | 3+ |
| Hippocampus | 3+ |
| Cerebellum | 3+ |
| Putamen | 3+ |
| Amygdala | 2+ |
| Thalamus | 2+ |
| Subthalamic nucleus | 2+ |
| Caudate nucleus | 2+ |
| Substantia nigra | 1+ |
| Medulla oblongata | 1+ |
| Spinal cord | 1+ |
| Genitourinary | |
| Kidney | 3+ |
| Prostate | 2+ |
| Bladder | tr |
| Gastrointestinal | |
| Liver | 2+ |
| Stomach | tr |
| Small intestine | tr |
| Appendix | tr |
| Colon | tr |
| Secretory | |
| Salivary gland | 3+ |
| Pituitary | 2+ |
| Adrenal | 2+ |
| Mammary gland | 1+ |
| Pancreas | 1+ |
| Thyroid | tr |
| Pulmonary | |
| Lung | 1+ |
| Trachea | 1+ |
| Muscle | |
| Heart | tr |
| Skeletal muscle | tr |
| Uterus | tr |
| Lymphoid | |
| Thymus | tr |
| Lymph node | tr |
| Spleen | tr |
| Vascular hematopoietic | |
| Aorta | tr |
| Peripheral leukocytes | tr |
| Bone Marrow | tr |
| Germ cells | |
| Testes | 3+ |
| Ovary | tr |
| Fetal/Placenta | |
| Placenta | tr |
| Fetal brain | 1+ |
| Fetal kidney | 1+ |
| Fetal heart | tr |
| Fetal liver | tr |
| Fetal lung | tr |
| Fetal thymus | tr |
| Fetal spleen | tr |
P-labeled cDNA from clone 184038 was hybridized to constant amounts of dot-blotted human poly(A)+ RNA. Hybridization signals were graded as tr, trace to 4+, strong expression. It is not clear if the weakest signals represent βIII spectrin or are due to cross-hybridization with other β spectrins.
Figure 2.
Northern blots of electrophoretically separated human poly(A)+ mRNAs (≈2 mg). (Left) Adult tissues from various sources (as indicated) hybridized to clone 184038 (Upper Left) to identify βIII spectrin transcripts or hybridized to actin (Lower Left) as a control for loading. Note that actin is overexpressed in heart and skeletal muscle and cannot be used as a control in those tissues. (Right) Similar analysis of human fetal tissues hybridized to a PCR product representing nucleotides 6,445–6,737.
Using the Stanford G3 radiation hybrid panel (16) and a primer pair located at the beginning of the low-homology region following the spectrin repeats (Fig. 1B), tight linkage was found to the marker SHGC-33744 (lod score 9.66). SHGC-33744 is flanked by D11S913 and the EST F02128 [pyruvate carboxylase (PC)] in the G3 panel database, both of which are located on chromosome 11q13 (Fig. 1C). A 3-Mb contig of this region has recently been described that includes both markers (29). The ordering of genetic markers in the contig is CEN–MLK3–FRA1–SEA–HNP36–D11S913–ACTN3–PC–GSTP1–qter. Given the size of yeast artificial chromosomes in the contig, PC and D11S913 appear less than 500 kb apart (http://www.genetics.wustl.edu/gerhard/3mb/3mb.html). Thus, α-actinin 3 (ACTN3) and βIII spectrin are near-neighbors in the chromosome. Similarly, the pattern of segregation of a KpnI restriction fragment length polymorphism in 94 progeny of the Jackson Laboratory BSS interspecific backcross panel (17) localized the mouse βIII spectrin gene (Spnb3) to chromosome 19 (Fig. 1C). Based on the consensus map in the mouse (http://www.informatics.jax.org), Spnb3 is positioned very close (0 cM) to the centromere, a region of the mouse genome that is homologous to human chromosome 11q13.
βIII Spectrin Associates with Golgi and Cytoplasmic Vesicles.
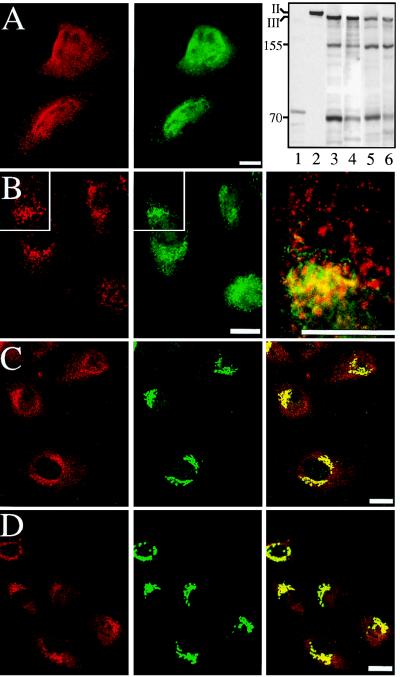
Two antibodies (Spb3C2 and PAb R-βIII7–11) were used to detect the cytoplasmic distribution of βIII spectrin in MDCK, HeLa (not shown), and NRK cells (Fig. 3). All of the cell lines and both antibodies gave comparable indirect immunofluorescence results, although only PAb R-βIII7–11 worked well in Western blots. βIII spectrin migrated at a molecular mass of ≈220 kDa despite a calculated mass of 271,294 (Fig. 3A). In all cases, it was clearly resolved from βII spectrin (≈235 kDa, calculated mass 274,658 Da), which migrated more slowly. Although PAb R-βIII7–11 did not recognize βII spectrin on Western blots, it did crossreact weakly with βI spectrin in erythrocyte ghosts (not shown). The migration of βIII spectrin in SDS/PAGE analysis of MDCK and NRK cells was indistinguishable from the band at 220 kDa previously identified as Golgi spectrin with βI spectrin antibodies (4–6, 8). Immunoreactive bands at 155 and 70 kDa were also occasionally evident in blots of MDCK and NRK cells. Their origin is undetermined, although their variable appearance suggests that they are βIII proteolytic fragments rather than alternative gene products.
Figure 3.
Cellular distribution of βIII spectrin. Indirect immunofluorescence studies of MDCK (A and B) and NRK (C and D) cells stained with (Left) anti-βIII spectrin antibodies (Spb3C2, A, C, and D; PAb R-βIII7–11, B) or with (Center) anti-β-COP (A), anti-Bip (B); anti-ManII (C), or anti-TGN38 (D). Merged images are also shown (Right). Areas marked by rectangles in B were magnified (B, Right) to show the vesicle-like structures stained with anti-βIII-spectrin antibody (in red) or anti-Bip antibody (in green). βIII spectrin is largely distinct from the distribution of Bip, a marker of the ER. In contrast, there is coincident staining of βIII spectrin with Golgi markers (Right in C and D) and variably with the punctate vesicles stained by those markers. Similar results were also observed with HeLa and COS cells (not shown). (Top, Right) Western blot analysis of MDCK cells (lanes 1–4) and NRK cells (lanes 5 and 6) using PAb R-βIII7–11 (lanes 3–6) reveals an immunoreactive band of ≈220 kDa that is distinct from βII spectrin (lane 2, stained with PAb-10D) and that is not present in pre-immune antisera for PAb R-βIII7–11 (lane 1). The βIII spectrin band was evident in both whole-cell lysates (lanes 1–3, 5) and in RIPA buffer extracts (lanes 4 and 6). Additional immunoreactive bands were inconsistently observed at ≈155 and ≈70 kDa. (Bars = 20 μm.)
The distribution of βIII spectrin by indirect immunofluorescence was striking for its punctate cytoplasmic and eccentric perinuclear distribution, a pattern reminiscent of that observed with βI spectrin-reactive antibodies (4, 6, 8). The βIII distribution overlapped partially that of β-COP (a marker of COPI transport vesicles; ref. 30), p58 (a marker of the vesiculotubular compartment and Golgi; ref. 31 and data not shown); as well as ManII and TGN38 (markers of the Golgi compartment; refs. 20, 32; Fig. 3). In preliminary experiments, we also have noted fluorescence resonance-energy transfer between some of these markers and βIII spectrin, suggesting their very close association in vivo (M.C.S. and J.S.M., unpublished data). Thus, βIII spectrin appears to be a Golgi-associated spectrin. However, its punctate pattern even when over the Golgi suggests that it may not simply form a homogeneous Golgi coat. βIII spectrin’s distribution also appears distinct from the distribution of ER markers such as Bip and calnexin, although in some areas the two are in close apposition (Fig. 3B). The nature of the punctate cytosolic structures remains uncertain. The punctate distribution of βIII spectrin extends throughout the cell, often in a curvilinear pattern (Fig. 3). Many spots, with an average diameter of about 300–350 nm (as determined from fluorescent images), appear too large to represent individual 50-nm transport vesicles (Fig. 3B). These dimensions are approximately twice the length of an extended spectrin molecule, and up to five times the nominal diameter of small transport vesicles. Thus, it is interesting to speculate that the clusters of βIII spectrin correspond to the vesiculotubular clusters of the intermediate compartment (31), to larger and more tubular TGN-derived vesicles (32), and to endolysosomes (9), all structures in which a spectrin/ankyrin-associated skeleton has been previously implicated (for review, see ref. 33).
βIII Spectrin Codistributes with Golgi Membranes in Vitro.
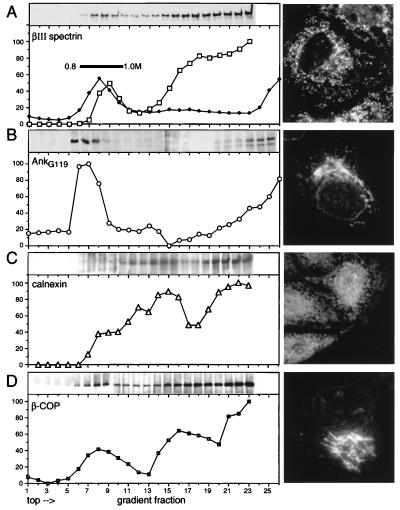
The distribution of βIII spectrin with various rat-liver fractions was evaluated by floatation of homogenized rat-liver membranes in a discontinuous sucrose gradient (Fig. 4). As in cultured cells, rat-liver βIII spectrin migrated in SDS/PAGE analysis as a ≈220-kDa band distinct from βII spectrin. In these gradients, membranes with morphological and biochemical features characteristic of Golgi float between 0.77 and 0.99 M sucrose (Fig. 4A; ref. 19). Of the total membrane-associated βIII spectrin, ≈16% migrated at this density, coincident with ManII enzymatic activity and AnkG119 (ref. 4; Fig. 4B). The remainder of the βIII spectrin was associated with denser fractions, overlapping the distribution of calnexin (a calcium-binding transmembrane chaperone protein resident in the endoplasmic reticulum; ref. 34) and β-COP (a COPI coatomer component associated with transport vesicles and cis-Golgi; ref. 35). Thus (and in accordance with the immunofluorescence studies) a substantial fraction of βIII spectrin is associated not only with the Golgi, but also with other cytoplasmic-membrane organelles.
Figure 4.
βIII spectrin cosegregates with Golgi membranes and other vesicle markers. Fresh rat-liver membranes were floated in a sucrose gradient, and fractions were analyzed for the presence of βIII spectrin and other organelle markers by Western blotting (above each gradient) and densitometry. Also shown (Right) is the intracellular distribution of each marker in MDCK cells by indirect immunofluorescence with the antibody used for the blotting the gradient fractions. (A) βIII spectrin measured with 1:5,000 PAb R-βIII7–11. The enzymatic activity of ManII in each fraction was also measured (•) as a marker of Golgi. Morphologically identifiable Golgi membranes sediment between 0.8 and 1.0 M sucrose (bar). About 16% of βIII spectrin sediments in the Golgi fraction, the rest in denser membrane fractions. (B) The distribution of ankyrin AnkG119. (C) The distribution of calnexin, an ER marker. (D) The distribution of β-COP, a marker of Golgi and COPI transport vesicles.
DISCUSSION
We have identified a β spectrin whose cDNA sequence, chromosome location, and intracellular distribution distinguish it from any other known spectrin. Human βI and βII spectrins (SPTB and SPTBN1) map to chromosomes 14q23 and 2p21, respectively, and the corresponding mouse genes (Spnb1 and Spnb2) reside on chromosomes 12 and 11 (36–39). The present study places βIII spectrin on human chromosome 11q13 (SPTBN2) and mouse chromosome 19 (Spnb3). Our mapping data show that human βIII spectrin resides next to α-actinin 3, a member of the spectrin superfamily. Previous studies have shown that α-actinin 1 and βI spectrin are also closely linked (40); together, these findings suggest that α-actinin and β spectrin share a common ancestral lineage, with each evolving separately after a gene duplication event.
A number of human diseases map to chromosome band 11q13, including type 1 Bardet–Biedl syndrome, vitelliform macular dystrophy (Best disease), and osteoporosis–pseudoglioma syndrome (http://www.ncbi.nlm.nih.gov/omim), but none seems likely to be caused by a spectrin defect. In the mouse genome, Spnb3 maps to the same position on mouse chromosome 19 as neuromuscular degeneration (nmd), an autosomal recessive disorder in the mouse that produces spinal motor neuron atrophy and progressive hind-limb weakness (41). However, markers that map near SPTBN2 on human chromosome 11 map distal to nmd on fine maps of chromosome 19 (G. Cox, personal communication), suggesting that Spnb3 is not a candidate gene for the disease. Other nearby mouse mutations include dancer (Dc), muscle deficient (mdf), osteosclerotic (oc), and osteochondrodystrophy (ocd); however, each of these maps to a position 6 cM more distal on chromosome 19 and is hence unlikely to be caused by a defect in βIII spectrin.
Recently, isoforms of spectrin and ankyrin (a major adapter protein linking spectrin to membranes) have been identified on Golgi membranes (4–8, 10), on lysosomal membranes (9), and on intracellular vesicles in cerebellar neurons (42), as well as on the plasma membrane. Spectrin also has been shown to associate with centractin (ARP-1), a subunit of the dynactin complex (5, 11) which, in association with the microtubule-based motor cytoplasmic dynein, plays a major role in the anterograde transport of vesicles in the secretory pathway from the vesiculotubular compartment to the cis-Golgi (43, 44). Disruption of the Golgi and vesicular spectrin skeleton also disrupts anterograde cargo transport at a point before the medial Golgi (5, 8). A role for spectrin in post-Golgi transport of trafficking vesicles—lysosomes and endocytic vesicles—possibly in association with kinesin-based motors also has been proposed (33, 44, 45). Even though accumulated data on Golgi and cytoplasmic spectrin is extensive, all of the existing work has been based on the crossreactivity of βI spectrin antibodies and on the ability of βI spectrin peptides to compete with the function of Golgi spectrin. For this reason, and because the precise nature of the Golgi-associated spectrin has remained elusive, these cytoplasmic spectrins have been termed βIΣ∗, reflecting the ambiguity in their identification (reviewed in refs. 33 and 46). The work presented here indicates that at least one Golgi-associated spectrin, the protein previously referred to as βIΣ∗ spectrin, is βIII spectrin.
Acknowledgments
We thank Dr. Osamu Ohara for useful discussions, for providing us with clone KIAA0302, and for generous prepublication communication; Dr. Greg Cox for sharing his unpublished work on nmd and on the arrangement of loci on mouse chromosome 19; and Dr. Anna Godi for assistance with Golgi membrane preparation. This work was supported by National Institutes of Health grants to S.E.L., J.S.M., and L.L.P., and by a grant from the March of Dimes to L.L.P.
ABBREVIATIONS
- Bip
Ig heavy chain-binding protein
- ER
endoplasmic reticulum
- EST
expressed sequence tag
- GST
glutathione S-transferase
- MAD1
MAD2, membrane association domains of spectrin
- ManII
mannosidase II
- MDCK
Madin Darby canine kidney cells
- NRK
newborn rat kidney cells
- PAb
polyclonal antibody
- PH
pleckstrin homology domain
- TGN
trans-Golgi network
- TGN38
transmembrane glycoprotein found in the trans-Golgi network
Footnotes
Data deposition: The nucleotide sequences of βIII spectrin clones 141000, 184038, 479050, and βIII-Nterm have been deposited in GenBank as AF026487, AF026488, AF026489, and AF079569, respectively. Mapping data have been deposited in the Mouse Genome Database as MGD-JNUM-42524.
Nomenclature: βIΣ1 spectrin is the erythroid isoform (Σ1) of erythroid (βI) spectrin. It is also called spectrinR. The gene name is SPTB (human) or Spnb1 (mouse). βIΣ2 spectrin is the muscle isoform (Σ2) of erythroid β spectrin. It contains a different C-terminal sequence. βII spectrin is “nonerythroid” β spectrin. It has also been called fodrin, brain spectrin, spectrinG, or spectrin beta, nonerythroid type 1. The gene name is SPTBN1 (human) or Spnb2 (mouse).
References
- 1.Marchesi V T, Steers E. Science. 1968;159:203–204. doi: 10.1126/science.159.3811.203. [DOI] [PubMed] [Google Scholar]
- 2.Lux S E, Palek J. In: Blood: Principles and Practice of Hematology. Handin R I, Lux S E, Stossel T P, editors. Philadelphia: Lippincott; 1995. pp. 1701–1818. [Google Scholar]
- 3.Morrow J S, Rimm D L, Kennedy S P, Cianci C D, Sinard J H, Weed S A. In: Handbook of Physiology. Hoffman J, Jamieson J, editors. London: Oxford Univ. Press; 1997. pp. 485–540. [Google Scholar]
- 4.Devarajan P, Stabach P R, Mann A S, Ardito T, Kashgarian M, Morrow J S. J Cell Biol. 1996;133:819–830. doi: 10.1083/jcb.133.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devarajan P, Stabach P R, De Matteis M A, Morrow J S. Proc Natl Acad Sci USA. 1997;94:10711–10716. doi: 10.1073/pnas.94.20.10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck K A, Buchanan J A, Malhotra V, Nelson W J. J Cell Biol. 1994;127:707–723. doi: 10.1083/jcb.127.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck K A, Buchanan J A, Nelson W J. J Cell Sci. 1997;110:1239–1249. doi: 10.1242/jcs.110.10.1239. [DOI] [PubMed] [Google Scholar]
- 8.Godi A, Santone I, Pertile P, Devarajan P, Stabach P R, Morrow J S, Di Tullio G, Polishuck R, Petrucci T C, Luini A, et al. Proc Natl Acad Sci USA. 1998;95:8607–8612. doi: 10.1073/pnas.95.15.8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoock T C, Peters L L, Lux S E. J Cell Biol. 1997;136:1059–1070. doi: 10.1083/jcb.136.5.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fath K R, Trimbur G M, Burgess D R. J Cell Biol. 1997;139:1169–1181. doi: 10.1083/jcb.139.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holleran E A, Tokito M K, Karki S, Holzbaur E L. J Cell Biol. 1996;135:1815–1829. doi: 10.1083/jcb.135.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winkelmann J C, Forget B G. Blood. 1993;81:3173–3185. [PubMed] [Google Scholar]
- 13.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagase T, Ishikawa K, Nakajima D, Ohira M, Seki N, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. DNA Res. 1997;4:141–150. doi: 10.1093/dnares/4.2.141. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Stewart E A, McKusick K B, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, et al. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- 17.Rowe L B, Nadeau J H, Turner R, Frankel W N, Letts V A, Eppig J T, Ko M S, Thurston S J, Birkenmeier E H. Mamm Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- 18.Lombardo C R, Weed S A, Kennedy S P, Forget B G, Morrow J S. J Biol Chem. 1994;269:29212–29219. [PubMed] [Google Scholar]
- 19.Leelavathi D E, Estes L W, Feingold D S, Lombardi B. Biochim Biophys Acta. 1970;211:124–138. [Google Scholar]
- 20.Tabas I, Kornfeld S. J Biol Chem. 1979;254:11655–11663. [PubMed] [Google Scholar]
- 21.Winkelmann J C, Chang J G, Tse W T, Scarpa A L, Marchesi V T, Forget B G. J Biol Chem. 1990;265:11827–11832. [PubMed] [Google Scholar]
- 22.Winkelmann J C, Costa F F, Linzie B L, Forget B G. J Biol Chem. 1990;265:20449–20454. [PubMed] [Google Scholar]
- 23.Hu R-J, Watanabe M, Bennett V. J Biol Chem. 1992;267:18715–18722. [PubMed] [Google Scholar]
- 24.Karinch A M, Zimmer W E, Goodman S R. J Biol Chem. 1990;265:11833–11840. [PubMed] [Google Scholar]
- 25.Becker P S, Tse W T, Lux S E, Forget B G. J Clin Invest. 1993;92:612–616. doi: 10.1172/JCI116628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy S P, Weed S A, Forget B G, Morrow J S. J Biol Chem. 1994;269:11400–11408. [PubMed] [Google Scholar]
- 27.Kennedy S P, Warren S L, Forget B G, Morrow J S. J Cell Biol. 1991;115:267–277. doi: 10.1083/jcb.115.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macias M J, Musacchio A, Ponstingl H, Nilges M, Saraste M, Oschkinat H. Nature (London) 1994;369:675–677. doi: 10.1038/369675a0. [DOI] [PubMed] [Google Scholar]
- 29.Smith C M, Ma N S, Nowak N J, Shows T B, Gerhard D S. Genome Res. 1997;7:835–842. doi: 10.1101/gr.7.8.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schekman R, Mellman I. Cell. 1997;90:197–200. doi: 10.1016/s0092-8674(00)80326-8. [DOI] [PubMed] [Google Scholar]
- 31.Tisdale E J, Plutner H, Matteson J, Balch W E. J Cell Biol. 1997;137:581–593. doi: 10.1083/jcb.137.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kreis T E, Pepperkok R. Curr Opin Cell Biol. 1994;6:533–537. doi: 10.1016/0955-0674(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 33.deMatteis M A, Morrow J S. Curr Opin Cell Biol. 1998;10:542–549. doi: 10.1016/s0955-0674(98)80071-9. [DOI] [PubMed] [Google Scholar]
- 34.Tatu U, Helenius A. J Cell Biol. 1997;136:555–565. doi: 10.1083/jcb.136.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pepperkok R, Scheel J, Horstmann H, Hauri H P, Griffiths G, Kreis T E. Cell. 1993;74:71–82. doi: 10.1016/0092-8674(93)90295-2. [DOI] [PubMed] [Google Scholar]
- 36.Fukushima Y, Byers M G, Watkins P C, Winkelmann J C, Forget B G, Shows T B. Cytogenet Cell Genet. 1990;53:232–233. doi: 10.1159/000132939. [DOI] [PubMed] [Google Scholar]
- 37.Chang J G, Scarpa A, Eddy R L, Byers M G, Harris A S, Morrow J S, Watkins P, Shows T B, Forget B G. Genome. 1993;17:287–293. doi: 10.1006/geno.1993.1323. [DOI] [PubMed] [Google Scholar]
- 38.Bloom M L, Lee B K, Birkenmeier C S, Ma Y, Zimmer W E, Goodman S R, Eicher E M, Barker J E. Mamm Genome. 1992;3:293–295. doi: 10.1007/BF00292159. [DOI] [PubMed] [Google Scholar]
- 39.Laurila P, Cioe L, Kozak C A, Curtis P J. Somat Cell Mol Genet. 1987;13:93–97. doi: 10.1007/BF02422304. [DOI] [PubMed] [Google Scholar]
- 40.Youssoufian H, McAfee M, Kwiatkowski D J. Am J Hematol. 1990;47:62–71. [PMC free article] [PubMed] [Google Scholar]
- 41.Cook S A, Johnson K R, Bronson R T, Davisson M T. Mamm Genome. 1995;6:187–191. doi: 10.1007/BF00293010. [DOI] [PubMed] [Google Scholar]
- 42.Malchiodi-Albedi F, Ceccarini M, Winkelmann J C, Morrow J S, Petrucci T C. J Cell Sci. 1993;106:67–78. doi: 10.1242/jcs.106.1.67. [DOI] [PubMed] [Google Scholar]
- 43.Presley J F, Cole N B, Schroer T A, Hirschberg K, Zaal K J, Lippincott-Schwartz J. Nature (London) 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 44.Lippincott-Schwartz J. Curr Opin Cell Biol. 1998;10:52–59. doi: 10.1016/s0955-0674(98)80086-0. [DOI] [PubMed] [Google Scholar]
- 45.Hirokawa N. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 46.Holleran E A, Karki S, Holzbaur E L. Int Rev Cytol. 1998;182:69–109. doi: 10.1016/s0074-7696(08)62168-3. [DOI] [PubMed] [Google Scholar]
- 47.Ohara O, Ohara R, Yamakawa H, Nakajima D, Nakayama M. Brain Res Mol Brain Res. 1998;57:181–192. doi: 10.1016/s0169-328x(98)00068-0. [DOI] [PubMed] [Google Scholar]
- 48.Sakaguchi G, Orita S, Naito A, Maeda M, Igarashi H, Sasaki T, Takai Y. Biochem Biophys Res Commun. 1998;248:846–851. doi: 10.1006/bbrc.1998.9067. [DOI] [PubMed] [Google Scholar]