Abstract
Expression of class I human leucocyte antigens (HLA) on the surface of malignant cells is critical for their recognition and destruction by cytotoxic T lymphocytes. Surface expression requires assembly and folding of HLA class I molecules in the endoplasmic reticulum with the assistance of proteins such as Transporter associated with Antigen Processing (TAP) and tapasin. Interferon-γ induces both TAP and tapasin so dissection of which protein contributes more to HLA class I expression has not been possible previously. In this study, we take advantage of a human melanoma cell line in which TAP can be induced, but tapasin cannot. Interferon-γ increases TAP protein levels dramatically but HLA class I expression at the cell surface does not increase substantially, indicating that a large increase in peptide supply is not sufficient to increase HLA class I expression. On the other hand, transfection of either allelic form of tapasin (R240 or T240) enhances HLA-B*5001 and HLA-B*5701 antigen expression considerably with only a modest increase in TAP. Together, these data indicate that in the presence of minimal TAP activity, tapasin can promote substantial HLA class I expression at the cell surface.
Keywords: antigen presentation, cancer, human leucocyte antigen, major histocompatibility complex, tapasin, transporter associated with antigen processing (TAP), tumour
Introduction
The role of human leucocyte antigen (HLA) class I antigens in interactions between target cells and cytotoxic T lymphocytes (CTL) has stimulated interest in examining their expression on malignant cells. There is convincing evidence that HLA class I antigens can be downregulated or lost on malignant cells and that these variations may be associated with a poor prognosis.1–3 Characterizing the mechanisms underlying efficient class I expression at the cell surface is expected to contribute to the design of strategies that aim to enhance class I expression at the surface for optimal recognition by CTL.
The expression of HLA class I molecules at the cell surface depends upon the appropriate assembly of three major components, a heavy chain, β2-microglobulin and peptide in the endoplasmic reticulum (ER). Peptides are generated by proteasomes4 and aminopeptidases5,6 in the cytosol followed by transport into the ER lumen by the ATP-dependent transporter associated with antigen processing (TAP).7,8 Within the ER, peptides are further trimmed by aminopeptidases.9–11 The binding of peptides by class I molecules is thought to occur within a multimolecular assemblage termed the peptide loading complex. The peptide loading complex consists of a TAP heterodimer surrounded by multiple class I molecules,12,13 tapasin,14,15 calreticulin15 and ERp57.16–18 While the chaperone calreticulin19 and the thiol-oxidoreductase ERp5720–23 promote appropriate folding and disulphide bond formation, tapasin plays multiple roles in the peptide loading complex. It promotes the stabilization of the peptide loading complex,14,24 aids in the appropriate selection of peptides,25–27 maintains appropriate HLA class I redox status21 and enhances TAP levels.24,28,29
The magnitude by which tapasin enhances TAP appears to be different in murine cells compared to the one human cell line examined to date. Murine cells deficient for tapasin show a much larger defect, (several hundred-fold) in TAP levels than the human tapasin-deficient cell line .220.B8 (two-fold) when compared to their wild-type counterparts.29,30 Furthermore, two alleles of tapasin have been identified that exist at approximately equal frequencies in the human population, yet their functional differences or similarities are unknown.31
The importance of proteins in the peptide loading complex is highlighted by impaired class I assembly and surface expression in cells that are deficient for any one of these proteins.14,19,22,30,32–35 Although each component of the peptide loading complex is required for optimal assembly and transport of HLA class I molecules to the surface, defects in TAP or tapasin show the most marked defects.14,32 To date, the relative contributions of TAP and tapasin expression to the level of HLA class I expression has been difficult to separate because these two molecules are coordinately regulated at the transcriptional level.36–39
In the present study, we have compared the effects of both tapasin alleles on TAP stabilization and HLA class I expression in a melanoma cell line. Furthermore, we have examined the relative contributions of TAP and tapasin to HLA class I expression, which addresses the functional significance of variations in TAP and tapasin expression in both normal and malignant cells.
Materials and methods
Cell lines and reagents
The melanoma cell lines M553 (HLA-A28, A2-haplotype loss40 and HLA-B*5001 and HLA-B*5701, this report) and M501 (HLA-A2, A24) have been described previously.1,40 All cell lines were grown in RPMI-1640 medium (Life Technologies, Grand Island, NY) containing 10% bovine calf serum (Hyclone Laboratories, Logan, UT) and 10 mm HEPES.
Antibodies
The monoclonal antibody (mAb) W6/32 recognizes β2-microglobulin-associated HLA-A, -B and -C molecules.41 The mAb SFR8.B6 recognizes the HLA-Bw6 epitope42 and is highly dependent upon the presence of residues Arg82 and Gly83. SFR8.B6 reactivity is abolished by the replacement of these residues with either Leu82 or Arg83.43,44 Based on this rationale, we reasoned that SFR8.B6 probably reacts with the HLA-B*5001 (Arg82, Gly83) product but not HLA-B*5701 (Leu82, Arg83R). The mAb TT4-A2045,46 recognizes the HLA-Bw4 epitope, which is highly dependent upon residues 79–83 that are present in HLA-B*5101.47 These residues 79–83 are completely conserved between HLA-B*5101 and HLA-B*5701, suggesting that the TT4-A20 recognizes HLA-B*5701 expressed on M553 cells. The rabbit antiserum R.Ring4c recognizes the C-terminal peptide of TAP1.12 The TAP1-specific antibody NOB-1 was generated by immunizing BALB/c mice with a synthetic peptide containing residues 777–794 (GGAIREGGTHQQLMEKKG).48 The mAb TO-3 specific for human tapasin was generated in BALB/c mice immunized with a synthetic peptide containing residues 29–42 (QGPGEPPPRPDLDP).48 The mAb specific for α-tubulin was purchased from Sigma-Aldrich (St Louis, MO). The mAb PaSta1 and the R.SinB rabbit polyclonal antiserum recognize lumenal epitopes within tapasin.21 The antiserum, Clyde, recognizes the C-terminal region of calnexin.24 The BAP31-specific rabbit antiserum recognizes the C-terminal peptide (CLEEHAKLQAAVDGPMDKKKAE).49
Flow cytometric analysis
Cells were washed and incubated with saturating concentrations of the HLA-specific or control antibodies for 30 min at 4°. After washing, 500 000 cells were resuspended in saturating concentrations (10 μg/ml) of fluorescein isothiocyanate-conjugated, goat anti-mouse immunoglobulin G antibodies. After washing, cells were fixed in 2% paraformaldehyde and analysed on a FACScan instrument (Becton Dickinson, San Jose, CA). Data were analysed with WinMDI software (Joe Trotter, The Scripps Research Institute, La Jolla, CA).
Interferon-γ treatment
Cells were incubated with 100 IU/ml interferon-γ (IFN-γ), (PBL Biomedical Laboratories, New Brunswick, NJ) for 20–24 hr. This dose effectively increases HLA class I expression at the cell surface and TAP and tapasin protein levels within the cell (ref. 50 and Figs 1–3).
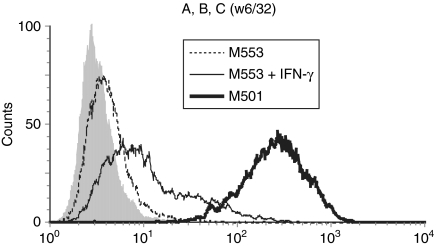
Figure 1.
Defective HLA class I expression on M553 cells is not restored by IFN-γ. The cell lines indicated were either untreated or treated with IFN-γ for 20–24 hr before analysis by flow cytometry using the HLA-A, -B, -C specific monoclonal antibody W6/32 or a control antibody (filled histogram). A representative value from at least three independent experiments is shown.
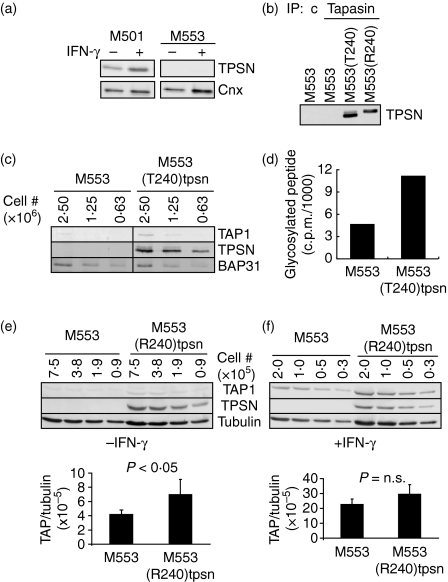
Figure 3.
Defective tapasin expression is not inducible by IFN-γ, but can be restored by transfection. (a) The cell lines indicated were either untreated (−) or treated (+) with IFN-γ for 20–24 hr before lysis and analysed as in Fig. 2a. (b) M553 cells and transfectants containing R240tpsn or T240tpsn alleles were lysed and immunoprecipitated using tapasin-specific antibody (PaSta1) or control antibody. (c). Western blots were probed for tapasin using rabbit antiserum (R.SinB). (c) Lysates from M553 and M553(T240)tpsn cells were serially diluted and after SDS–PAGE, the indicated proteins were detected by immunoblot analysis. (d) M553 and M553(T240)tpsn cells were tested for TAP function as described in the Materials and methods. (e) Serial dilutions of untreated (-IFN-γ) M553 and M553(R240)tpsn cells were immunoblotted as indicated. (f) IFN-γ-treated M553 and M553(R240)tpsn cells were analysed as in (e). Quantification of the fluorescent bands in (e) and (f) was performed with ImageQuant software. Each bar represents the average of the TAP1 : tubulin signal over all dilutions. A representative figure from at least three independent experiments is shown.
Western blot analysis
Quantitative fluorescence-based Western blotting was performed as described previously.24 Briefly, cells were lysed using 1% Triton X-100 in 10 mm Tris–HCl, 150 mm NaCl, pH7·4 containing protease inhibitors phenylmethylsulphonyl fluoride (0·5 mm) and N-ethylmaleimide (5 mm). Post-nuclear supernatants were diluted to the indicated number of cell equivalents and separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE). Protein measurements by the Bradford assay (Bio Rad, Hercules, CA) indicate that (M553 or M553tpsn or M501) lysates corresponding to 10 000 cell equivalents contain 1 μg protein. Following transfer of proteins to an Immobilon-P membrane (Millipore, Bedford, MA), proteins of interest were detected using the indicated primary antibodies followed by secondary antibodies coupled to alkaline phosphatase. After washing, VistraECF substrate (Amersham Pharmacia, Piscataway, NJ) was applied and fluorescence was measured using a STORM instrument (Molecular Dynamics, Palo Alto, CA). Fluorescence measurements were acquired and analysed using ImageQuant software. The amount of fluorescence for each band (i.e. tapasin, TAP1, tubulin) was quantified by integration of the area under the curve generated for each band. For loading controls, the same membrane was reprobed for α-tubulin and/or calnexin. The ratio of fluorescence signal of TAP1 over tubulin was utilized to compare between different membranes.
Peptide translocation assay
Peptide translocation was measured as previously described.24 Cells were permeabilized in 0·005% digitonin, 50 mm HEPES, 78 mm KCl, 4 mm MgCl2, 8·37 mm CaCl2, 10 mm ethyleneglycoltetraacetic acid, 0·4% bovine serum albumin for 5 min at 37°. The peptide termed B27#3 glycopeptide1 (RRYQNSTEL) containing an NXT glycosylation acceptor site was iodinated using Chloramine T. Permeabilized cells, supplemented with 1 mm ATP, were added to 2 294 200 counts/min of 125I-labelled B27#3 peptide at 37° for 3 min. Three minutes of peptide translocation represents an initial rate that is proportional to the number of functional heterodimers present.24 The translocation reaction was stopped by adding Triton X-100 to a final concentration of 1%. Immediately thereafter, samples were placed on ice. Glycosylated 125I-labelled peptide was recovered from post-nuclear supernatants using concanavalin A–Sepharose. After washing, concanavalin A–Sepharose-associated counts were measured in a Cobra II Auto-Gamma counter (Packard Bioscience, Meriden, CT).
Polymerase chain reaction (PCR) amplification of HLA-B alleles from M553 cells
Total RNA from M553 cells was extracted using TRIzol (Invitrogen, Carlsbad, CA) and cDNA was generated using oligo-dT primers and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's recommendations. The PCR amplification of HLA-B alleles was performed using the high-fidelity enzyme Accuprime Taq (Invitrogen) and primers 5′-ATGCGGGTCACGGCGCCCCGAACC-3′ and 5′-TCAAGCTGTGAGAGACACATCAGA-3′. The amplicons were cloned into pGEM-T-EASY (Promega, Madison, WI) and single colonies were isolated and sequenced using the SP6 and T7 primers (Roswell Park Biopolymer Facility). Translated sequences were aligned to all known HLA-B alleles in the IMGT database.51–53
Restoration of tapasin expression in M553 cells
The expression construct encoding the R240 allele of tapasin was obtained by reverse transcription-PCR amplification from the melanoma cell line 1195 using the primers 1F (5′-AGCGCCATGAAGTCCCTGTCTCTGCTC-3′) and 1R (5′-GTGCCCTCACTCTGCTTTCTTCTTTGA-3′) followed by cloning into pDRIVE (Qiagen, Valencia, CA) and subcloning into a modified pCDNA3.1(-)neo (Invitrogen) in which the gene encoding puromycin resistance replaced the neomycin-resistance gene. The cloned product was verified by sequencing the DNA inserted into the pDRIVE plasmid. The T240 allele of tapasin was amplified from 721.45.1 cells and cloned into the expression construct pmcfr.puro (originally created by Tom Novak, Yale University School of Medicine). Both expression constructs [pCDNA3.1(-)puro and pmcfr.puro] contain the human cytomegalovirus (CMV) promoter to produce constitutive tapasin transcription that is unaffected by incubation with IFN-γ. Expression constructs were transfected into M553 cells using Effectene (Qiagen) according to the manufacturer's recommendations. After transfection, clones were isolated by limiting dilution in the presence of 1 μg/ml puromycin.
Results
Barely detectable HLA class I antigen expression on melanoma cells M553
Flow cytometric analysis with the HLA class I-specific mAb W6/32 showed minimal staining of M553 cells, but high class I expression on the melanoma cell line, M501 (Fig. 1). Increased class I expression on M553 cells treated with IFN-γ suggested that the structural genes (i.e. heavy chain, β2-microglobulin) were probably intact. However, the limited increase by IFN-γ was compatible with functional defects in antigen presentation components.
Defects in TAP and tapasin in M553 cells
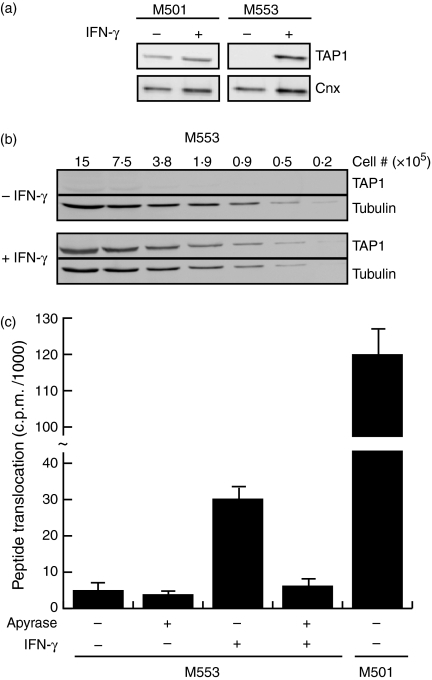
Using 50 000 cell equivalents (5 μg), TAP1 protein was undetectable in M553 cells but detectable in M501 cells (Fig. 2a). Treatment of M553 cells with IFN-γ induced TAP1 expression appreciably (Fig. 2a). To quantify the magnitude of TAP1 induction by IFN-γ, several dilutions of M553 lysates were assayed (Fig. 2b). In the absence of IFN-γ, TAP1 was only marginally detectable, even when using 1 500 000 cell equivalents (150 μg, Fig. 2b). However, TAP1 was detectable from as little as 50 000 cell equivalents (5 μg) from IFN-γ-treated M553 cells (Fig. 2b). The presence of IFN-γ strongly induced TAP1 protein levels (with at least a 10- to 30-fold increase). Given the increase in TAP1 protein levels, we expected an increase in TAP function. Indeed, M553 cells treated with IFN-γ showed a substantial increase in peptide translocation ability compared to untreated M553 cells (Fig. 2c). Addition of apyrase, which cleaves ATP, demonstrated the dependence of peptide translocation on ATP. The observation of greater peptide translocation in M501 cells compared to IFN-γ-induced M553 cells (Fig. 2c), in the face of similar or slightly lower TAP1 protein levels (Fig. 2a), suggested that peptide transport was impaired even in the IFN-γ-treated M553 cells.
Figure 2.
Defective TAP expression in M553 cells can be overcome by IFN-γ treatment. The cell lines indicated were either untreated (−) or treated (+) with IFN-γ for 20–24 hr before lysis. Recovered proteins were subject to SDS–PAGE followed by detection of the indicated proteins by immunoblot. (a) Fifty thousand cell equivalents (5 μg total protein) were applied to each lane. (b) Cell lysates were serially diluted to the indicated cell equivalents. (c) TAP function was measured as described in the Materials and methods. The amount of translocated peptide in the presence (+) or absence (−) of apyrase is plotted for each cell type and treatment indicated. A representative figure of at least three independent experiments is shown.
Transfection of tapasin cDNA enhances TAP protein levels and peptide transport
Since tapasin is known to increase peptide transport,28 we hypothesized that a defect in tapasin could be responsible for the lower peptide translocation in M553 cells. Indeed, we found that tapasin protein was undetectable in M553 cells both under basal conditions and following incubation with IFN-γ (Fig. 3a). To test the possibility that defective tapasin expression in M553 cells is responsible for impaired peptide transport, we stably transfected M553 cells with each of the two known tapasin alleles under control of the constitutively active CMV promoter. It is noteworthy that this single amino acid polymorphism (R240 versus T240) subtly, but reproducibly, altered the mobility of tapasin as measured by SDS–PAGE (Fig. 3b, comparing the tapasin immunoprecipitate of M553(T240)tpsn with M553(R240)tpsn). The expression of the T240 tapasin allele in M553(T240)tpsn led to a two-fold enhancement of the barely detectable levels of TAP1 in M553 cells (Fig. 3c, comparing the TAP1 bands in M553 with those of M553(T240)tpsn). Support for the two-fold increase in TAP1 protein levels by tapasin was provided by a corresponding two-fold increase in the ability to translocate peptide by M553(T240)tpsn cells compared to M553 cells (Fig. 3d). Transfection of M553 cells with the R240 allele resulted in a similar two-fold increase in the faint TAP1 band in M553(R240)tpsn cells compared to M553 cells (Fig. 3e). Therefore, although tapasin increased TAP1 protein levels, this effect was modest (two-fold) compared to the IFN-γ-induced TAP1 levels (> 10-fold, see Fig. 2a). Confirmation of the idea that human tapasin had a less dramatic effect in increasing TAP levels compared to IFN-γ came from the observation that the presence of tapasin in M553(R240)tpsn cells did not further enhance M553(R240)tpsn TAP1 levels (normalized to tubulin) after IFN-γ treatment (Fig. 3f).
Enhancement of HLA-B*5001 and -B*5701 antigen expression by tapasin
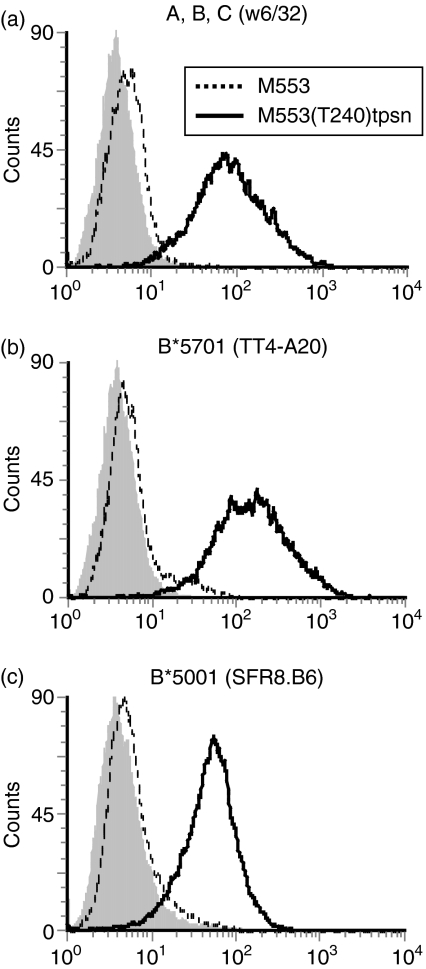
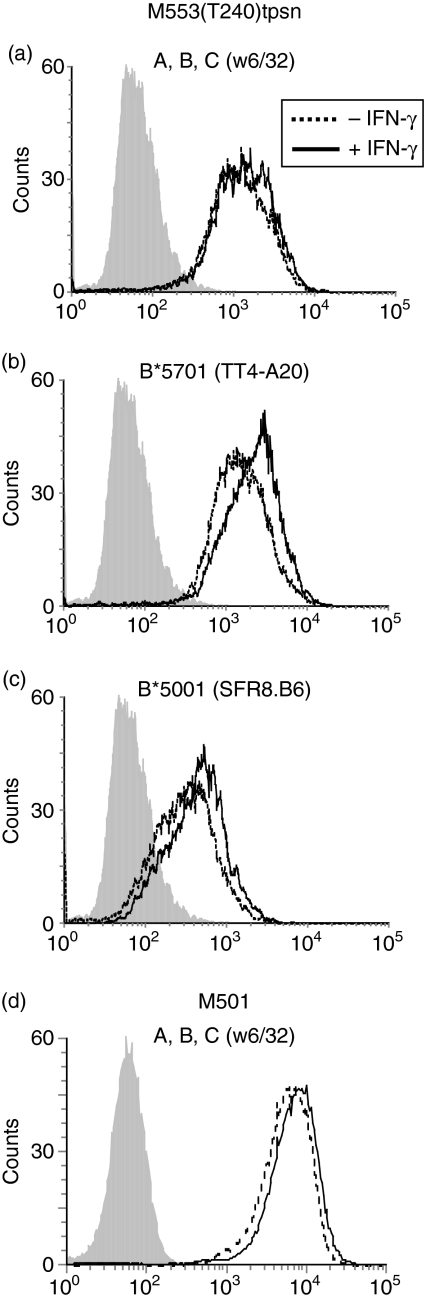
To investigate whether the lack of tapasin was responsible for the marked downregulation of class I molecules on M553 cells, we compared class I expression on M553 and M553(T240)tpsn cells by flow cytometry. Stable transfection of the T240 tapasin cDNA enhanced the levels of overall HLA-A, -B, -C antigen expression by 30-fold on M553 cells (Fig. 4a). Comparable results were obtained in M553(R240)tpsn cells (data not shown). Since tapasin affects the surface expression of different class I allelic products to varying degrees, the cDNA encoding the HLA-B alleles was amplified by PCR and sequenced to determine which alleles were expressed in M553 cells. Comparison with the IMGT database51–53 revealed that M553 cells expressed HLA-B*5001 and HLA-B*5701. The surface expression of HLA-B*5701 was enhanced 30-fold upon transfection of tapasin (Fig. 4b, compare dotted line to solid line). HLA-B*5001 was below the detection limit on M553 cells but its induction was increased 10-fold on M553(R240)tpsn cells (Fig. 4c, compare dotted line to solid line). HLA-A alleles were not detectable in the presence or absence of tapasin (data not shown). Increased HLA class I expression on tapasin-transfected M553 cells was therefore the result of the HLA-B*5701 and HLA-B*5001 alleles.
Figure 4.
Enhancement of HLA-B*5001 and HLA-B*5701 expression by tapasin. M553 cells (dotted lines) and M553(T240)tpsn cells (solid lines) were stained using the mAbs (a) w6/32, (b) TT4-A20, (c) SFR8.B6 which detect the indicated allelic products on these cells (open histograms). Staining with a control antibody is shown in the filled histograms.
Lack of detectable effects of IFN-γ on HLA class I antigen expression on M553tpsn cells
Since the enhancement of TAP1 by IFN-γ was greater than that observed by tapasin transfection, we tested whether IFN-γ induction of TAP in the presence of high tapasin expression (i.e. M553(T240)tpsn cells) could enhance class I levels further. Given that the transfected tapasin gene is under the control of the CMV promoter, which is not induced by IFN-γ, we reasoned that the effects of IFN-γ on M553tpsn cells would only reflect the enhancement of TAP levels, but not of tapasin levels. Although overall HLA-A, -B, -C expression was not enhanced by IFN-γ (Fig. 5a, mean fluorescence intensity for untreated 107 versus 132 with IFN-γ), the staining of HLA-B*5001 (mean fluorescence intensity for untreated 47 versus 74 with IFN-γ, Fig. 5b) and HLA-B*5701 (mean fluorescence intensity for untreated 140 versus 209 with IFN-γ, Fig. 5c) both appeared to be modestly enhanced by IFN-γ. The enhancement of Bw4 and Bw6 staining by IFN-γ may be the result of the influence of a different, or more optimal, peptide repertoire because peptides can affect staining of these epitopes by some antibodies.54 These data further indicate that low versus high quantitative variations in TAP protein levels in M553tpsn cells did not alter the overall levels of surface class I expression. In contrast, high levels of tapasin expression maximized class I expression at the cell surface in M553tpsn cells. This finding is reproducible in the M501 cell line, which contains high tapasin levels (Figs 3a and 5d) but does not show increased class I expression on the surface upon IFN-γ treatment (Fig. 5d).
Figure 5.
Inability of IFN-γ to increase class I expression on cells containing high levels of tapasin. (a–c) Untreated M553(T240)tpsn cells (dotted lines) and IFN-γ-treated M553(T240)tpsn cells (solid lines) were stained using the mAbs described in Fig. 4. (d) Untreated or IFN-γ-treated M501 cells were stained using the mAb W6/32.
Discussion
In the present report we utilize a tapasin-deficient, but TAP-inducible, cell line to reveal the relative contributions of TAP and tapasin to HLA class I expression at the cell surface. The phenotype of negligible class I expression on the melanoma cell line M553 could have been explained by low TAP level or lack of tapasin protein. We find that although IFN-γ effectively induces TAP expression and activity, class I expression does not increase appreciably. These data suggest that, for HLA-B*5001 and HLA-B*5701, low versus high TAP levels do not alter overall class I status. On the other hand, tapasin transfection increases class I expression at the cell surface 30-fold. Combined, these observations imply that in a fully competent antigen-presenting cell, although IFN-γ induces both TAP and tapasin, it is tapasin that is the major contributor to expression of these alleles at the cell surface.
Further support for the idea that the level of TAP is not a major factor in expression of HLA class I at the cell surface comes from the observation that TAP induction (by IFN-γ) does not increase class I levels in tapasin-transfected M553 cells. Additionally, the fact that tapasin increases TAP by only two-fold and enhances HLA class I expression 30-fold substantiates the idea that the presence of TAP—although not necessarily its abundance—is important for efficient expression of these alleles. Although we suggest that TAP levels are not important for HLA class I expression, elegant studies using cell lines in which the TAP genes are deleted demonstrate that TAP is required for class I expression at the surface.32 These results can be reconciled with ours by acknowledging that a minimum basal level of TAP is required to support HLA class I transport to the cell surface. However, we suggest that further increases in TAP above the minimum level do not alter the expression of tapasin-dependent alleles such as HLA-B*5001 and HLA-B*5701.
Our findings that tapasin controls HLA class I expression are consistent with previous evidence indicating that tapasin is limiting and recycles to assemble class I molecules sequentially.55 By extension, one may predict a linear relationship between tapasin expression levels and class I surface expression. Indeed, regardless of the HLA haplotype, tapasin appears to be the only antigen presentation component that directly correlates with surface HLA class I expression in a panel of twenty-five melanoma cell lines (T. Ogino, S. Ferrone unpublished observations). Furthermore, high tapasin levels (in M553tpsn cells or M501 cells) appear to maximize cell surface class I expression such that even IFN-γ treatment fails to upregulate class I expression further. It is however possible that, in other cell types, other components of the peptide loading complex may be responsible for low surface expression of HLA class I molecules.56 Consistent with this idea, BAP31 has been shown to be limiting for HLA class I expression on the surface of HeLa cells.57
Before the discovery of tapasin, M553 cells were reported as having a single genomic haplotype loss in which both HLA-A2 and HLA-B40 had been deleted.40 We confirm that the HLA-A2 antigen is not detectable by flow cytometry (data not shown). However, HLA-B (HLA-B*5001 and HLA-B*5701) expression at the cell surface is detectable when M553 cells are transfected with tapasin. Tapasin expression is, therefore, an important parameter to consider when examining the HLA class I phenotype of tumour cells by flow cytometry. Although recent studies have begun to examine tapasin expression in cell lines and in malignant lesions,38,58,59 to our knowledge, the present report represents the first demonstration of the causal effects of tapasin on HLA class I antigen expression and TAP activity in a tumour cell line.
Tapasin is reported to enhance TAP by two- to four-fold when transfected into the only human cell line, .220 (a B lymphocyte cell line), known to be deficient for tapasin.24,28 However, the stabilizing effect of tapasin on TAP levels in murine fibroblasts or splenocytes has been reported to be in the order of several hundred-fold.29,30 Given the consistent results between human .220 B cells and M553 melanoma cells, a species difference (rather than differences between cell types) probably accounts for the differential effects of mouse versus human tapasin on TAP protein levels. Furthermore, the expression of human tapasin in insect cells does not alter (human) TAP activity, suggesting that human tapasin does not have a large effect on TAP activity.60
Overall, we have demonstrated that tapasin plays a dominant role in dictating surface HLA-B*5001 and HLA-B*5701 in the presence of minimal TAP activity. In the endogenous setting, we suggest that IFN-γ contributes to antigen presentation via two major mechanisms. First, IFN-γ induces TAP protein levels and activity to increase peptide supply into the ER, which may increase the opportunity to bind viral peptides for example, but this induction does not appear to increase HLA-B*5001 or HLA-B*5701 expression substantially. Second, IFN-γ induces tapasin, which promotes the expression of HLA class I molecules at the cell surface. The increase in TAP levels caused by tapasin is modest compared to that induced by IFN-γ directly. Together, these findings suggest that modulation of tapasin in tumour cells may have a profound effect on HLA class I expression and their potential to be recognized by CTLs.
Acknowledgments
We thank Dilek Oktay Oflazoglu, Nancianne Maynard, Brenda Pritchett, Violet Tredo and Jessica Eygnor for technical assistance and Julia Jane Hart-Dewey for assistance with manuscript preparation. We appreciate the antibody gifts from Dr Frank Momburg (German Cancer Research Centre, Heidelberg, TT4-A20), Dr Ari Helenius (Swiss Federal Institute of Technology, Zurich, Switzerland, calnexin-specific antiserum) and Dr Michael Edidin (Johns Hopkins University, Baltimore MD, BAP31-specific antiserum). We appreciate the technical assistance of Michelle Detwiler and Christian A. Borrelli at the Roswell Park Biopolymer Facility. We are grateful to the two anonymous reviewers who greatly improved the quality of the manuscript. This work was supported in part by Investigator Start-Up funds (N.B.), NIAID PHS grant AI063341 (N.B.), NCI PHS grants CA67108 (S.F.), CA105500 (S.F.), CA55791 (S.G.), CA98156 (S.G.) and by NCI Cancer Center Support Grant (CA016156) to Roswell Park Cancer Institute.
Glossary
Abbreviations
- BAP31
B-cell receptor associated protein 31 kiloDaltons
- HLA
human leucocyte antigen
- IFN-γ
interferon-γ
- TAP
transporter associated with antigen processing
- Tpsn
tapasin
References
- 1.Cormier JN, Panelli MC, Hackett JA, et al. Natural variation of the expression of HLA and endogenous antigen modulates CTL recognition in an in vitro melanoma model. Int J Cancer. 1999;80:781–90. doi: 10.1002/(sici)1097-0215(19990301)80:5<781::aid-ijc24>3.0.co;2-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrone S, Marincola FM. Loss of HLA class I antigens by melanoma cells: molecular mechanisms, functional significance and clinical relevance. Immunol Today. 1995;16:487–94. doi: 10.1016/0167-5699(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 3.Kageshita T, Hirai S, Ono T, Hicklin DJ, Ferrone S. Down-regulation of HLA class I antigen-processing molecules in malignant melanoma: association with disease progression. Am J Pathol. 1999;154:745–54. doi: 10.1016/S0002-9440(10)65321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–79. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 5.Stoltze L, Schirle M, Schwarz G, et al. Two new proteases in the MHC class I processing pathway. Nat Immunol. 2000;1:413–8. doi: 10.1038/80852. [DOI] [PubMed] [Google Scholar]
- 6.Seifert U, Maranon C, Shmueli A, et al. An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nat Immunol. 2003;4:375–9. doi: 10.1038/ni905. [DOI] [PubMed] [Google Scholar]
- 7.Shepherd JC, Schumacher TN, Ashton-Rickardt PG, et al. TAP1-dependent peptide translocation in vitro is ATP dependent and peptide selective. Cell. 1993;74:577–84. doi: 10.1016/0092-8674(93)80058-m. [DOI] [PubMed] [Google Scholar]
- 8.Androlewicz MJ, Anderson KS, Cresswell P. Evidence that transporters associated with antigen processing translocate a major histocompatibility complex class I-binding peptide into the endoplasmic reticulum in an ATP-dependent manner. Proc Natl Acad Sci USA. 1993;90:9130–4. doi: 10.1073/pnas.90.19.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–3. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 10.York IA, Chang SC, Saric T, et al. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8–9 residues. Nat Immunol. 2002;3:1177–84. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 11.Saric T, Chang SC, Hattori A, et al. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat Immunol. 2002;3:1169–76. doi: 10.1038/ni859. [DOI] [PubMed] [Google Scholar]
- 12.Ortmann B, Androlewicz MJ, Cresswell P. MHC class I/beta 2-microglobulin complexes associate with TAP transporters before peptide binding. Nature. 1994;368:864–7. doi: 10.1038/368864a0. [DOI] [PubMed] [Google Scholar]
- 13.Suh WK, Cohen-Doyle MF, Fruh K, Wang K, Peterson PA, Williams DB. Interaction of MHC class I molecules with the transporter associated with antigen processing. Science. 1994;264:1322–6. doi: 10.1126/science.8191286. [DOI] [PubMed] [Google Scholar]
- 14.Ortmann B, Copeman J, Lehner PJ, et al. A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science. 1997;277:1306–9. doi: 10.1126/science.277.5330.1306. [DOI] [PubMed] [Google Scholar]
- 15.Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996;5:103–14. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 16.Hughes EA, Cresswell P. The thiol oxidoreductase ERp57 is a component of the MHC class I peptide-loading complex. Curr Biol. 1998;8:709–12. doi: 10.1016/s0960-9822(98)70278-7. [DOI] [PubMed] [Google Scholar]
- 17.Morrice NA, Powis SJ. A role for the thiol-dependent reductase ERp57 in the assembly of MHC class I molecules. Curr Biol. 1998;8:713–6. doi: 10.1016/s0960-9822(98)70279-9. [DOI] [PubMed] [Google Scholar]
- 18.Lindquist JA, Jensen ON, Mann M, Hammerling GJ. ER-60, a chaperone with thiol-dependent reductase activity involved in MHC class I assembly. EMBO J. 1998;17:2186–95. doi: 10.1093/emboj/17.8.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao B, Adhikari R, Howarth M, et al. Assembly and antigen-presenting function of MHC class I molecules in cells lacking the ER chaperone calreticulin. Immunity. 2002;16:99–109. doi: 10.1016/s1074-7613(01)00260-6. [DOI] [PubMed] [Google Scholar]
- 20.Farmery MR, Allen S, Allen AJ, Bulleid NJ. The role of ERp57 in disulfide bond formation during the assembly of major histocompatibility complex class I in a synchronized semipermeabilized cell translation system. J Biol Chem. 2000;275:14933–8. doi: 10.1074/jbc.275.20.14933. [DOI] [PubMed] [Google Scholar]
- 21.Dick TP, Bangia N, Peaper DR, Cresswell P. Disulfide bond isomerization and the assembly of MHC class I–peptide complexes. Immunity. 2002;16:87–98. doi: 10.1016/s1074-7613(02)00263-7. [DOI] [PubMed] [Google Scholar]
- 22.Garbi N, Tanaka S, Momburg F, Hammerling GJ. Impaired assembly of the major histocompatibility complex class I peptide-loading complex in mice deficient in the oxidoreductase ERp57. Nat Immunol. 2006;7:93–102. doi: 10.1038/ni1288. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Baig E, Williams DB. Functions of ERp57 in the folding and assembly of major histocompatibility complex class I molecules. J Biol Chem. 2006;281:14622–31. doi: 10.1074/jbc.M512073200. [DOI] [PubMed] [Google Scholar]
- 24.Bangia N, Lehner PJ, Hughes EA, Surman M, Cresswell P. The N-terminal region of tapasin is required to stabilize the MHC class I loading complex. Eur J Immunol. 1999;29:1858–70. doi: 10.1002/(SICI)1521-4141(199906)29:06<1858::AID-IMMU1858>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 25.Williams AP, Peh CA, Purcell AW, McCluskey J, Elliott T. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. 2002;16:509–20. doi: 10.1016/s1074-7613(02)00304-7. [DOI] [PubMed] [Google Scholar]
- 26.Zarling AL, Luckey CJ, Marto JA, et al. Tapasin is a facilitator, not an editor, of class I MHC peptide binding. J Immunol. 2003;171:5287–95. doi: 10.4049/jimmunol.171.10.5287. [DOI] [PubMed] [Google Scholar]
- 27.Howarth M, Williams A, Tolstrup AB, Elliott T. Tapasin enhances MHC class I peptide presentation according to peptide half-life. Proc Natl Acad Sci U S A. 2004;101:11737–42. doi: 10.1073/pnas.0306294101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehner PJ, Surman MJ, Cresswell P. Soluble tapasin restores MHC class I expression and function in the tapasin-negative cell line .220. Immunity. 1998;8:221–31. doi: 10.1016/s1074-7613(00)80474-4. [DOI] [PubMed] [Google Scholar]
- 29.Garbi N, Tiwari N, Momburg F, Hammerling GJ. A major role for tapasin as a stabilizer of the TAP peptide transporter and consequences for MHC class I expression. Eur J Immunol. 2003;33:264–73. doi: 10.1002/immu.200390029. [DOI] [PubMed] [Google Scholar]
- 30.Grandea AG, III, Golovina TN, Hamilton SE, et al. Impaired assembly yet normal trafficking of MHC class I molecules in tapasin mutant mice. Immunity. 2000;13:213–22. doi: 10.1016/s1074-7613(00)00021-2. [DOI] [PubMed] [Google Scholar]
- 31.Copeman J, Bangia N, Cross JC, Cresswell P. Elucidation of the genetic basis of the antigen presentation defects in the mutant cell line .220 reveals polymorphism and alternative splicing of the tapasin gene. Eur J Immunol. 1998;28:3783–91. doi: 10.1002/(SICI)1521-4141(199811)28:11<3783::AID-IMMU3783>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 32.Spies T, DeMars R. Restored expression of major histocompatibility class I molecules by gene transfer of a putative peptide transporter. Nature. 1991;351:323–4. doi: 10.1038/351323a0. [DOI] [PubMed] [Google Scholar]
- 33.Powis SJ, Townsend AR, Deverson EV, Bastin J, Butcher GW, Howard JC. Restoration of antigen presentation to the mutant cell line RMA-S by an MHC-linked transporter. Nature. 1991;354:528–31. doi: 10.1038/354528a0. [DOI] [PubMed] [Google Scholar]
- 34.Van Kaer L, Ashton-Rickardt PG, Ploegh HL, Tonegawa S. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4– 8+ T cells. Cell. 1992;71:1205–14. doi: 10.1016/s0092-8674(05)80068-6. [DOI] [PubMed] [Google Scholar]
- 35.Garbi N, Tan P, Diehl AD, et al. Impaired immune responses and altered peptide repertoire in tapasin-deficient mice. Nat Immunol. 2000;1:234–8. doi: 10.1038/79775. [DOI] [PubMed] [Google Scholar]
- 36.Abarca-Heidemann K, Friederichs S, Klamp T, Boehm U, Guethlein LA, Ortmann B. Regulation of the expression of mouse TAP-associated glycoprotein (tapasin) by cytokines. Immunol Lett. 2002;83:197–207. doi: 10.1016/s0165-2478(02)00104-9. [DOI] [PubMed] [Google Scholar]
- 37.Herberg JA, Sgouros J, Jones T, et al. Genomic analysis of the Tapasin gene, located close to the TAP loci in the MHC. Eur J Immunol. 1998;28:459–67. doi: 10.1002/(SICI)1521-4141(199802)28:02<459::AID-IMMU459>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 38.Seliger B, Schreiber K, Delp K, et al. Downregulation of the constitutive tapasin expression in human tumor cells of distinct origin and its transcriptional upregulation by cytokines. Tissue Antigens. 2001;57:39–45. doi: 10.1034/j.1399-0039.2001.057001039.x. [DOI] [PubMed] [Google Scholar]
- 39.Seliger B, Hammers S, Hohne A, et al. IFN-gamma-mediated coordinated transcriptional regulation of the human TAP-1 and LMP-2 genes in human renal cell carcinoma. Clin Cancer Res. 1997;3:573–8. [PubMed] [Google Scholar]
- 40.Marincola FM, Shamamian P, Alexander RB, et al. Loss of HLA haplotype and B locus down-regulation in melanoma cell lines. J Immunol. 1994;153:1225–37. [PubMed] [Google Scholar]
- 41.Parham P, Barnstable CJ, Bodmer WF. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C, antigens. J Immunol. 1979;123:342–9. [PubMed] [Google Scholar]
- 42.Radka SF, Kostyu DD, Amos DB. A monoclonal antibody directed against the HLA-Bw6 epitope. J Immunol. 1982;128:2804–6. [PubMed] [Google Scholar]
- 43.Toubert A, Raffoux C, Boretto J, et al. Epitope mapping of HLA-B27 and HLA-B7 antigens by using intradomain recombinants. J Immunol. 1988;141:2503–9. [PubMed] [Google Scholar]
- 44.Lutz CT, Smith KD, Greazel NS, et al. Bw4-reactive and Bw6-reactive antibodies recognize multiple distinct HLA structures that partially overlap in the alpha-1 helix. J Immunol. 1994;153:4099–110. [PubMed] [Google Scholar]
- 45.Tahara T, Yang SY, Khan R, Abish S, Hammerling GJ, Hammerling U. HLA antibody responses in HLA class I transgenic mice. Immunogenetics. 1990;32:351–60. doi: 10.1007/BF00211650. [DOI] [PubMed] [Google Scholar]
- 46.Tan P, Kropshofer H, Mandelboim O, Bulbuc N, Hammerling GJ, Momburg F. Recruitment of MHC class I molecules by tapasin into the transporter associated with antigen processing-associated complex is essential for optimal peptide loading. J Immunol. 2002;168:1950–60. doi: 10.4049/jimmunol.168.4.1950. [DOI] [PubMed] [Google Scholar]
- 47.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–44. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Campoli M, Cho HS, et al. A method to generate antigen-specific mAb capable of staining formalin-fixed, paraffin-embedded tissue sections. J Immunol Methods. 2005;299:139–51. doi: 10.1016/j.jim.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 49.Spiliotis ET, Manley H, Osorio M, Zuniga MC, Edidin M. Selective export of MHC class I molecules from the ER after their dissociation from TAP. Immunity. 2000;13:841–51. doi: 10.1016/s1074-7613(00)00081-9. [DOI] [PubMed] [Google Scholar]
- 50.Diedrich G, Bangia N, Pan M, Cresswell P. A role for calnexin in the assembly of the MHC class I loading complex in the endoplasmic reticulum. J Immunol. 2001;166:1703–9. doi: 10.4049/jimmunol.166.3.1703. [DOI] [PubMed] [Google Scholar]
- 51.Robinson J, Waller MJ, Parham P, et al. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–4. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson J, Malik A, Parham P, Bodmer JG, Marsh SG. IMGT/HLA database—a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55:280–7. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- 53.Robinson J, Waller MJ, Parham P, Bodmer JG, Marsh SG. IMGT/HLA Database—a sequence database for the human major histocompatibility complex. Nucleic Acids Res. 2001;29:210–3. doi: 10.1093/nar/29.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takamiya Y, Sakaguchi T, Miwa K, Takiguchi M. Role of HLA-B*5101 binding nonamer peptides in formation of the HLA-Bw4 public epitope. Int Immunol. 1996;8:1027–34. doi: 10.1093/intimm/8.7.1027. [DOI] [PubMed] [Google Scholar]
- 55.Bangia N, Cresswell P. Stoichiometric tapasin interactions in the catalysis of major histocompatibility complex class I molecule assembly. Immunology. 2005;114:346–53. doi: 10.1111/j.1365-2567.2005.02103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson DR, Mook-Kanamori B. Dependence of elevated human leukocyte antigen class I molecule expression on increased heavy chain, light chain (beta 2-microglobulin), transporter associated with antigen processing, tapasin, and peptide. J Biol Chem. 2000;275:16643–9. doi: 10.1074/jbc.M910035199. [DOI] [PubMed] [Google Scholar]
- 57.Ladasky JJ, Boyle S, Seth M, et al. Bap31 enhances the endoplasmic reticulum export and quality control of human class I MHC molecules. J Immunol. 2006;177:6172–81. doi: 10.4049/jimmunol.177.9.6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cabrera CM, Lopez-Nevot MA, Jimenez P, Garrido F. Involvement of the chaperone tapasin in HLA-B44 allelic losses in colorectal tumors. Int J Cancer. 2005;113:611–8. doi: 10.1002/ijc.20526. [DOI] [PubMed] [Google Scholar]
- 59.Dissemond J, Kothen T, Mors J, et al. Downregulation of tapasin expression in progressive human malignant melanoma. Arch Dermatol Res. 2003;295:43–9. doi: 10.1007/s00403-003-0393-8. [DOI] [PubMed] [Google Scholar]
- 60.Raghuraman G, Lapinski PE, Raghavan M. Tapasin interacts with the membrane-spanning domains of both TAP subunits and enhances the structural stability of TAP1 × TAP2 complexes. J Biol Chem. 2002;277:41786–94. doi: 10.1074/jbc.M207128200. [DOI] [PubMed] [Google Scholar]