Abstract
Idiopathic hypogonadotropic hypogonadism (IHH) and Kallmann syndrome (KS) are clinically and genetically heterogeneous disorders caused by a deficiency of gonadotrophin-releasing hormone (GnRH). Mutations in three genes—KAL1, GNRHR and FGFR1—account for 15–20% of all causes of IHH/KS. Nearly all mutations are point mutations identified by traditional PCR-based DNA sequencing. The relatively new method of multiplex ligation-dependent probe amplification (MLPA) has been successful for detecting intragenic deletions in other genetic diseases. We hypothesized that MLPA would detect intragenic deletions in ∼15–20% of our cohort of IHH/KS patients. Fifty-four IHH/KS patients were studied for KAL1 deletions and 100 were studied for an autosomal panel of FGFR1, GNRH1, GNRHR, GPR54 and NELF gene deletions. Of all male and female subjects screened, 4/54 (7.4%) had KAL1 deletions. If only anosmic males were considered, 4/33 (12.1%) had KAL1 deletions. No deletions were identified in any of the autosomal genes in 100 IHH/KS patients. We believe this to be the first study to use MLPA to identify intragenic deletions in IHH/KS patients. Our results indicate ∼12% of KS males have KAL1 deletions, but intragenic deletions of the FGFR1, GNRH1, GNRHR, GPR54 and NELF genes are uncommon in IHH/KS.
Keywords: Kallmann syndrome, KAL1 gene, hypogonadotropic hypogonadism, idiopathic hypogonadotropic hypogonadism, MLPA
Introduction
Idiopathic hypogonadotropic hypogonadism (IHH) is comprised of absent puberty, infertility and low serum gonadotrophins in the absence of a pituitary tumor. IHH is due to impaired gonadotrophin releasing hormone (GnRH) release/action or gonadotrophin secretion. Kallmann syndrome (KS) consists of IHH with anosmia. KS appears to be the result of impairment of GnRH and olfactory neuron migration from the olfactory placode to the hypothalamus (Bhagavath and Layman, 2007).
Mutations in genes involved in GnRH neuron migration (KAL1 and FGFR1), as well as those expressed in either the hypothalamus or pituitary, including GNRHR, NROB1, GPR54, LEP, LEPR and PCSK1 account for the molecular etiology of ∼15–20% of all IHH/KS patients (Hardelin, 2001; de Roux, 2005; Pitteloud et al., 2006; Bhagavath and Layman, 2007; Seminara, 2007). Three genes—KAL1, GNRHR and FGFR1—account for the majority of causative mutations. The molecular basis for most IHH/KS cases, however, remains unknown. Digenic disease has been reported in two families with FGFR1 and either a GNRHR or a NELF mutation (Pitteloud et al., 2007).
Most mutations identified in genetic diseases, including IHH/KS, are point mutations and small deletions/duplications detected by PCR-based DNA sequencing (Kim et al., 2008). PCR based sequencing may not detect heterozygous gene deletions, however, since the remaining normal allele will be amplified. Therefore, it is possible that a substantial number of mutations remain undiagnosed.
Multiplex ligation-dependent probe amplification (MLPA) is a relatively new method for detecting copy number variations in genomic sequences (Schouten et al., 2002). MLPA involves overnight hybridization of adjacent primer pairs to the individual exons of multiple target genes simultaneously, followed by a ligation reaction of the probe pairs, which then serve as a template for multiplex PCR. Each ligated probe has a varying length of ‘stuffer’ sequences that produce different sized fragments for resolution based upon size (Schouten et al., 2002). Since all probes have identical primer binding sites, only a single primer pair is required for multiplex PCR of all genes being studied.
Heterozygous intragenic deletions have been found in 15–20% of patients with cystic fibrosis (Audrezet et al., 2004), deafness (Del Castillo et al., 2003) and breast/ovarian cancer (Hogervorst et al., 2003) who did not have point mutations in the corresponding genes. We hypothesized that a similar percentage of IHH/KS patients would have heterozygous deletions of autosomal genes (FGFR1, GNRH1, GNRHR, GPR54 or NELF) and/or hemizygous KAL1 gene deletions, deletions which would be missed by the traditional PCR-based sequencing methods. Additionally, since KAL1 has a pseudogene homolog on the Y chromosome, this may complicate the DNA sequencing analysis (Del Castillo et al., 1992).
Materials and Methods
Patients
One-hundred and twelve probands with IHH/KS were studied by MLPA (Table I). The diagnosis of IHH was based upon criteria published previously—absent/impaired puberty by age ≥17 in girls and age ≥18 in boys, low serum gonadotrophins, no evidence of a pituitary tumor and otherwise normal pituitary function (Crowley et al., 1985; Bhagavath et al., 2006). All males had total testosterone <100 ng/dl (normal 300–1100 ng/dl), and all females had hypoestrogenic amenorrhea. Complete IHH was defined as absent breast development in females and testicular size ≤3 ml in males (Bhagavath et al., 2006). Incomplete IHH suggested the presence of prior steroid production and was defined as breast development of Tanner ≥2 in females and testis size >3 ml in males. Patient characteristics are shown in Table I. The Medical College of Georgia Human Assurance Committee approved this study; each patient signed informed consent.
Table I.
Classification of patients by gene(s) tested.
| KAL1 Kit | KAL2 Kit | KAL1&KAL2 Kits | Total Patients Studied | ||||
|---|---|---|---|---|---|---|---|
| All subjects studied n = 54 | All subjects studied n = 100 | Studied with both kits n = 42 | Individual subjects n = 112 (154–42) | ||||
| Male | 45 | Male | 72 | Male | 38 | Male | 79 |
| Anosmic/Hypos | 33 | Anosmic/Hypos | 37 | Anosmic/Hypos | 26 | Anosmic/Hypos | 44 |
| Normosmic | 12 | Normosmic | 18 | Normosmic | 12 | Normosmic | 18 |
| Unknown | 0 | Unknown | 17 | Unknown | 0 | Unknown | 17 |
| Complete IHH | 13 | Complete IHH | 15 | Complete IHH | 10 | Complete IHH | 18 |
| Incomplete IHH | 20 | Incomplete IHH | 19 | Incomplete IHH | 18 | Incomplete IHH | 21 |
| Unknown | 12 | Unknown | 38 | Unknown | 10 | Unknown | 40 |
| Female | 9 | Female | 28 | Female | 4 | Female | 33 |
| Anosmic/Hypos | 7 | Anosmic/Hypos | 7 | Anosmic/Hypos | 3 | Anosmic/Hypos | 11 |
| Normosmic | 2 | Normosmic | 10 | Normosmic | 1 | Normosmic | 11 |
| Unknown | 0 | Unknown | 11 | Unknown | 0 | Unknown | 11 |
| Complete IHH | 3 | Complete IHH | 7 | Complete IHH | 2 | Complete IHH | 8 |
| Incomplete IHH | 2 | Incomplete IHH | 6 | Incomplete IHH | 0 | Incomplete IHH | 8 |
| Unknown | 4 | Unknown | 15 | Unknown | 2 | Unknown | 17 |
| Totals | 54 | Totals | 100 | Totals | 42 | Totals | 112 |
| Anosmic/Hypos | 40 | Anosmic/Hypos | 44 | Anosmic/Hypos | 29 | Anosmic/Hypos | 55 |
| Normosmic | 14 | Normosmic | 28 | Normosmic | 13 | Normosmic | 29 |
| Unknown | 0 | Unknown | 28 | Unknown | 0 | Unknown | 28 |
| Complete IHH | 16 | Complete IHH | 22 | Complete IHH | 12 | Complete IHH | 26 |
| Incomplete IHH | 22 | Incomplete IHH | 25 | Incomplete IHH | 18 | Incomplete IHH | 29 |
| Unknown | 16 | Unknown | 53 | Unknown | 12 | Unknown | 57 |
Fifty-four patients were studied for the presence of KAL1 deletions (KAL1 kit) and 100 IHH/KS patients were studied for deletions of an autosomal panel of FGFR1, GNRH1, GNRHR, GPR54 and NELF genes (KAL2 kit). We estimated that we needed a sample size of ∼50 for study of KAL1 gene deletions, since the prevalence of mutations is about 6% in anosmic males (Bhagavath et al., 2007). The sample size for the autosomal gene panel (KAL2 kit) was estimated to be ∼100. The prevalence of FGFR1 mutations in both normosmic IHH and KS is about 10% (Pitteloud et al., 2006), whereas GNRHR mutations occur in 3–4% of normosmic IHH (Bhagavath et al., 2005). This sample should allow detection if they occur in this frequency. The prevalence of GNRH1, NELF and GPR54 mutations in IHH and KS is currently unknown, but is probably low. By screening ∼100 patients, we could determine if the prevalence was at least 1%.
KAL1 gene deletions were studied in 45 males and nine females. Since KAL1 mutations have been exclusively identified in anosmic/hyposmic patients, the majority of patients studied (40/54) were anosmic/hyposmic. All patients were randomly selected from available DNA samples without regard to prior mutation analysis from our large IHH/KS cohort.
Molecular analysis/MLPA
DNA was extracted from white blood cells in all IHH/KS using standard methods. MLPA was performed in patients according to Schouten (Schouten et al., 2002; Hogervorst et al., 2003). A total of 125 ng of DNA was hybridized overnight to the probe sets of the commercially available kits. The P132 KAL1 kit (MRC Holland, Amsterdam, Netherlands) contains 34 probe pairs for all 14 KAL1 exons and control sequences for other regions on Xp, Xq and Yq11. The P133 KAL2 kit contains 39 probe pairs covering the following exons of autosomal genes: GNRH1 (exons 1–3 of 4 exons), GNRHR (exons 1–3 of 3 exons), GPR54 (exons 1, 4 and 5 of 5 exons), FGFR1 (1–3, 5, 6, 8,10,13,14 and 18 of 18 exons) and NELF (exons 5, 11 and 15 of 16 exons). These probes also include control sequences on chromosomes 1–3, 6, 11, 15–17, 19, 20 and Y.
Following hybridization, a ligation reaction was performed which served as a template for 35 cycles of multiplex PCR using universal primers. PCR products were analyzed on an ABI 310 autoanalyzer using the Gene Scan program and data were evaluated using Genotyper 2.0 (both from Applied Biosystems, Foster City, CA, USA). Peak areas for each exon were converted into an Excel file and the relative copy number of each fragment was compared to the same fragments from 2 to 3 controls. MLPA was repeated at least three times for patients with putative deletions. Deletions were confirmed by PCR and DNA sequencing (Bhagavath et al., 2007).
Results
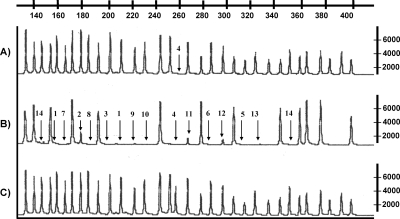
Using MLPA for the KAL1 gene, deletions were identified in 4/54 (7.4%) patients and in 4/40 (10%) of anosmic/hyposmic patients (Table II). When only anosmic males were considered, KAL1 deletions were present in 4/33 (12.1%). Three whole gene deletions were detected, as indicated by absence of peaks for each exon (Fig. 1). All three whole gene deletions were confirmed by the absence of bands by PCR. In one of these patients, deletion of exons 1–13 had previously been identified (Bhagavath et al., 2007); however, DNA sequencing of exon 14 was consistent with pseudogene sequence, thereby confirming the whole gene deletion by MLPA. One patient had an exon 4 deletion (Fig. 1) which, upon DNA sequencing, revealed a 3 bp deletion (TGT) of codon 164, deleting a Cys (Cys164del) rather than the deletion of the entire exon. No heterozygous/homozygous deletions were identified in the FGFR1, GNRH1, GNRHR, GPR54 or NELF genes in 100 IHH/KS patients.
Table II.
Summary of KAL1 exon/gene deletions.
| Deletion type | Sex | Smell | IHH severity | Family history/comments |
|---|---|---|---|---|
| Whole Gene | Male | Anosmic | Incomplete | None |
| Whole Gene | Male | Hyposmic | Complete | Visual Field abnormality |
| Whole Gene | Male | Anosmic | Complete | Two first cousins with KS |
| Exon 4 | Male | Anosmic | Incomplete | Renal Agenesis |
Figure 1:
MLPA results demonstrating.
(A) The KAL1 exon 4 deletion; (B) whole KAL1 gene deletion; (C) normal control. Arrows indicate the deleted exons. The horizontal axis (top) shows the size of the fragment in base pairs. A difference in relative copy peak height or peak area indicates a copy number change of the probe target sequence.
Discussion
The genetic basis of IHH/KS has been identified in 15–20% of patients, most commonly in KAL1, GNRHR or FGFR1 genes (Bhagavath and Layman, 2007). KAL1 mutations have been identified in anosmic males—approximately 5% of KS males without a family history and 30–70% of those with clear X-linked recessive inheritance (Bhagavath et al., 2007). GNRHR mutations cause autosomal recessive IHH in ∼3–5% of IHH patients, all of whom are normosmic (Bhagavath et al., 2005). Interestingly, FGFR1 mutations occur in ∼10% of anosmic or normosmic patients (Pitteloud et al., 2006).
Nearly all mutations in these genes are point mutations or small deletions/insertions. In contrast, only a very few gross deletions been identified in IHH/KS candidate genes. Available prevalence studies likely underestimate IHH/KS gene deletions as traditional PCR-based DNA sequencing is unable to detect heterozygous gene deletions. Therefore, it is possible that a substantial number of mutations remain undiagnosed. Using MLPA, heterozygous intragenic deletions have been found in 15–20% of patients with cystic fibrosis (Audrezet et al., 2004), deafness (Del Castillo et al., 2003) and breast/ovarian cancer (Hogervorst et al., 2003) who did not have point mutations.
MLPA is an effective technique used to detect genomic deletions and duplications (Schouten et al., 2002). If probe pairs for individual exons are utilized, MLPA can successfully determine the relative copy number of all exons within gene/genes simultaneously. MLPA has gained acceptance in genetic diagnostic laboratories due to its simplicity, relatively low cost and capacity for reasonably high throughput analysis. MLPA has a variety of applications in addition to detection of deletions/duplications. Other important uses include aneuploidy detection (Gerdes et al., 2005), analysis of DNA methylation (Procter et al., 2006), relative mRNA quantification (Wehner et al., 2005), chromosomal characterization of cell lines and tissue samples (Wilting et al., 2006) and the detection of polymorphisms (Volikos et al., 2006).
The frequency of gene deletions in IHH/KS candidate genes has not been previously reported. We hypothesized that MLPA would identify intragenic deletions in ∼15–20% of IHH/KS patients. Using MLPA, we discovered that 7.4% of 54 patients had KAL1 deletions. If only anosmic males were considered, 4/33 (12.1%) had KAL1 deletions. Three patients had whole gene deletions—one of which was previously described by PCR to be deleted of exons 1–13 (Bhagavath et al., 2007). PCR and DNA sequencing subsequently confirmed a whole gene deletion in this patient. MLPA also revealed one subject who appeared to have a deletion of exon 4. In this case, DNA sequencing revealed a 3 bp deletion rather than an entire exon deletion. It is likely that the probe pair did not specifically anneal to the template of exon 4, therefore, no ligation and subsequent PCR could be performed (Schouten et al., 2002). Our findings indicate that putative deletions indicate identified by MLPA requires confirmation by another technique.
We also used MLPA to screen for heterozygous intragenic deletions of five autosomal genes—FGFR1, GNRH1, GNRHR, GPR54 and NELF. Surprisingly, no deletions were identified in 100 IHH/KS patients. Probe pairs were dispersed across the coding regions of these genes, affording the opportunity to detect intragenic deletions. Only three exons were studied for the NELF gene, but at the time that this kit was designed, no NELF mutations had been described. We cannot exclude that intragenic deletions of exons not included in the kits occur, which would underestimate the prevalence of intragenic gene deletions. Nevertheless, no deletions of any of the five autosomal genes were identified in 100 IHH/KS patients.
Although the prevalence of deletions in IHH/KS in autosomal genes was much less than that expected, the prevalence of KAL1 deletions approximated 10–15% that we hypothesized. The findings from this pilot study demonstrate the feasibility of MLPA to detect deletions in IHH/KS and suggest the benefits of future studies with an increased sample size of IHH/KS.
Funding
This work was supported by NIH grants HD33004 and HD040287 (L.C.L).
Acknowledgements
We appreciate the help of Melissa Prince and John A. Phillips, III, Medical Genetics, Department of Pediatrics, Vanderbilt University, Nashville, TN.
References
- Audrezet MP, Chen JM, Raguenes O, Chuzhanova N, Giteau K, Le Marechal C, Quere I, Cooper DN, Ferec C. Genomic rearrangements in the CFTR gene: extensive allelic heterogeneity and diverse mutational mechanisms. Hum Mutat. 2004;23:343–357. doi: 10.1002/humu.20009. [DOI] [PubMed] [Google Scholar]
- Bhagavath B, Layman LC. The genetics of hypogonadotropic hypogonadism. Semin Reprod Med. 2007;25:272–286. doi: 10.1055/s-2007-980221. [DOI] [PubMed] [Google Scholar]
- Bhagavath B, Ozata M, Ozdemir IC, Bolu E, Bick DP, Sherins RJ, Layman LC. The prevalence of gonadotropin-releasing hormone receptor mutations in a large cohort of patients with hypogonadotropic hypogonadism. Fertil Steril. 2005;84:951–957. doi: 10.1016/j.fertnstert.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Bhagavath B, Podolsky RH, Ozata M, Bolu E, Bick DP, Kulharya A, Sherins RJ, Layman LC. Clinical and molecular characterization of a large sample of patients with hypogonadotropic hypogonadism. Fertil Steril. 2006;85:706–713. doi: 10.1016/j.fertnstert.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Bhagavath B, Xu N, Ozata M, Rosenfield RL, Bick DP, Sherins RJ, Layman LC. KAL1 mutations are not a common cause of idiopathic hypogonadotrophic hypogonadism in humans. Mol Hum Reprod. 2007;13:25–30. doi: 10.1093/molehr/gal108. [DOI] [PubMed] [Google Scholar]
- Crowley WF, Jr, Filicori M, Spratt DI, Santoro NF. The physiology of gonadotropin-releasing hormone (GnRH) secretion in men and women. Recent Prog Horm Res. 1985;41:473–531. doi: 10.1016/b978-0-12-571141-8.50015-9. [DOI] [PubMed] [Google Scholar]
- de Roux N. Isolated gonadotropic deficiency with and without anosmia: a developmental defect or a neuroendocrine regulation abnormality of the gonadotropic axis. Horm Res. 2005;64(Suppl 2):48–55. doi: 10.1159/000087754. [DOI] [PubMed] [Google Scholar]
- Del Castillo I, Cohen-Salmon M, Blanchard S, Lutfalla G, Petit C. Structure of the X-linked Kallmann syndrome gene and its homologous pseudogene on the Y chromosome. Nat Genet. 1992;2:305–310. doi: 10.1038/ng1292-305. [DOI] [PubMed] [Google Scholar]
- Del Castillo I, Moreno-Pelayo MA, Del Castillo FJ, Brownstein Z, Marlin S, Adina Q, Cockburn DJ, Pandya A, Siemering KR, Chamberlin GP, et al. Prevalence and evolutionary origins of the del(GJB6-D13S1830) mutation in the DFNB1 locus in hearing-impaired subjects: a multicenter study. Am J Hum Genet. 2003;73:1452–1458. doi: 10.1086/380205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes T, Kirchhoff M, Lind AM, Larsen GV, Schwartz M, Lundsteen C. Computer-assisted prenatal aneuploidy screening for chromosome 13, 18, 21, X and Y based on multiplex ligation-dependent probe amplification (MLPA) Eur J Hum Genet. 2005;13:171–175. doi: 10.1038/sj.ejhg.5201307. [DOI] [PubMed] [Google Scholar]
- Hardelin JP. Kallmann syndrome: towards molecular pathogenesis. Mol Cell Endocrinol. 2001;179:75–81. doi: 10.1016/s0303-7207(01)00462-2. [DOI] [PubMed] [Google Scholar]
- Hogervorst FB, Nederlof PM, Gille JJ, McElgunn CJ, Grippeling M, Pruntel R, Regnerus R, van Welsem T, van Spaendonk R, Menko FH, et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003;63:1449–1453. [PubMed] [Google Scholar]
- Kim HG, Bhagavath B, Layman LC. Clinical Manifestations of Impaired GnRH Neuron Development and Function. Neurosignals. 2008;16:165–182. doi: 10.1159/000111561. [DOI] [PubMed] [Google Scholar]
- Pitteloud N, Acierno JS, Jr, Meysing A, Eliseenkova AV, Ma J, Ibrahimi OA, Metzger DL, Hayes FJ, Dwyer AA, Hughes VA, et al. Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2006;103:6281–6286. doi: 10.1073/pnas.0600962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng YZ, et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procter M, Chou LS, Tang W, Jama M, Mao R. Molecular diagnosis of Prader-Willi and Angelman syndromes by methylation-specific melting analysis and methylation-specific multiplex ligation-dependent probe amplification. Clin Chem. 2006;52:1276–1283. doi: 10.1373/clinchem.2006.067603. [DOI] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seminara SB. Kisspeptin in reproduction. Semin Reprod Med. 2007;25:337–343. doi: 10.1055/s-2007-984739. [DOI] [PubMed] [Google Scholar]
- Volikos E, Robinson J, Aittomaki K, Mecklin JP, Jarvinen H, Westerman AM, de Rooji FW, Vogel T, Moeslein G, Launonen V, et al. LKB1 exonic and whole gene deletions are a common cause of Peutz-Jeghers syndrome. J Med Genet. 2006;43:e18. doi: 10.1136/jmg.2005.039875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner M, Mangold E, Sengteller M, Friedrichs N, Aretz S, Friedl W, Propping P, Pagenstecher C. Hereditary nonpolyposis colorectal cancer: pitfalls in deletion screening in MSH2 and MLH1 genes. Eur J Hum Genet. 2005;13:983–986. doi: 10.1038/sj.ejhg.5201421. [DOI] [PubMed] [Google Scholar]
- Wilting SM, Snijders PJ, Meijer GA, Ylstra B, van den Ijssel PR, Snijders AM, Albertson DG, Coffa J, Schouten JP, van de Wiel MA, et al. Increased gene copy numbers at chromosome 20q are frequent in both squamous cell carcinomas and adenocarcinomas of the cervix. J Pathol. 2006;209:220–230. doi: 10.1002/path.1966. [DOI] [PubMed] [Google Scholar]