Abstract
Genetic aberrations are crucial in renal tumor progression. In this study, we describe loss of heterozygosity (LOH) and DNA-copy number abnormalities in clear cell renal cell carcinoma (cc-RCC) discovered by genome-wide single nucleotide polymorphism (SNP) arrays. Genomic DNA from tumor and normal tissue of 22 human cc-RCCs was analyzed on the Affymetrix GeneChip Human Mapping 10K Array. The array data were validated by quantitative polymerase chain reaction and immunohistochemistry. Reduced DNA copy numbers were detected on chromosomal arm 3p in 91%, on chromosome 9 in 32%, and on chromosomal arm 14q in 36% of the tumors. Gains were detected on chromosomal arm 5q in 45% and on chromosome 7 in 32% of the tumors. Copy number abnormalities were found not only in FHIT and VHL loci, known to be involved in renal carcinogenesis, but also in regions containing putative new tumor suppressor genes or oncogenes. In addition, microdeletions were detected on chromosomes 1 and 6 in genes with unknown impact on renal carcinogenesis. In validation experiments, abnormal protein expression of FOXP1 (on 3p) was found in 90% of tumors (concordance with SNP array data in 85%). As assessed by quantitative polymerase chain reaction, PARK2 and PACRG were down-regulated in 57% and 100%, respectively, and CSF1R was up-regulated in 69% of the cc-RCC cases (concordance with SNP array data in 57%, 33%, and 38%). Genome-wide SNP array analysis not only confirmed previously described large chromosomal aberrations but also detected novel microdeletions in genes potentially involved in tumor genesis of cc-RCC.
Introduction
Renal cell carcinoma (RCC) is the most common tumor of the adult kidney and represents approximately 2% of all adult malignancies. Histopathologically, RCC can be subdivided in clear cell carcinomas (approximately 80% of sporadic RCC) and a few other subtypes such as papillary, chromophobe, and medullary RCC. The molecular genetics of RCC have been studied intensively. More than any other group of epithelial malignancies, some subtypes of adult RCC, such as clear cell renal cell carcinoma (cc-RCC) or papillary RCC, are readily distinguishable by gene expression profiling [1,2].
Genetic events seem to play an important role in the pathogenesis of primary sporadic cc-RCC. Inactivation of tumor suppressor genes (TSGs) on chromosome 3p is the most common event in cc-RCC and loss of heterozygosity (LOH) at 3p12, 3p14.2, 3p21.3, and 3p25 was often described [3–5].
The von Hippel-Lindau (VHL) gene, located at 3p25-p26, shows intragenic mutations in approximately 50% to 57% and LOH in approximately 80% to 90% of the cases of sporadic cc-RCC [6,7]. It seems that mutations of VHL alone are not sufficient to cause malignant disease but that additional mutations of other genes are required for malignant tumor growth. Hitherto, it is unclear if cc-RCC without VHL mutations takes other routes of VHL inactivation or alternative non-VHL tumor pathways [3,4]. It was demonstrated that LOH at 3p in cc-RCC occurs in association with allelic loss on one or more chromosomal arms such as 6p, 8p, 9pq, and 14q [8].
In papillary renal tumors, additional regions of LOH were observed on chromosomes 6p, 9pq, 11q, 14q, and 21q, suggesting that TSGs localized on chromosomes 6p, 9pq, and 14q are involved in development of both papillary and nonpapillary renal cancer [8].
Reports concerning the prognostic relevance of chromosomal aberrations in RCC are conflicting. VHL status in RCC has no association with tumor grade and stage [4]. Thrash-Bingham et al. [8] found no correlation between LOH and allelic imbalances with tumor size, nodal involvement, metastasis, or nuclear grade in RCC. No correlation was found between 3p loss and tumor size, nodal involvement, tumor grade, or metastasis in RCC, but metastasizing tumors showed more aberrations than nonmetastasizing tumors [9], and patients with increased DNA losses showed a shorter time to recurrence than patients with few DNA losses [10].
Conversely, a positive correlation between allelic loss at chromosome 14q, especially 14q24-q31, 14q31-q32, and a poor prognosis were described [11–13]. Patients with 3p loss and gain at 5q22.3, due to an unbalanced translocation between these chromosomes, had a significantly better disease-specific survival [14].
It is certain that genetic aberrations play an important role in RCC, but it is still unclear which genetic events are involved in tumor initiation and progression. There are obviously many, still unknown, small mutations that could be important for carcinogenesis and progression in RCC.
Historically, two key techniques were used to detect DNA copy number variations in DNA samples: comparative genomic hybridization (CGH) and LOH analysis. The array CGH technique can use bacterial artificial chromosome, cDNA, and oligonucleotides, and it is more sensitive because its higher resolution could be studied.
Hybridization to single nucleotide polymorphism (SNP) arrays is an efficient method to simultaneously detect genome-wide LOH and DNA-copy number aberration (DNA-CNA) [15,16].
To the best of our knowledge, only one study on RCC applying high-resolution techniques on only three cases exists up to date [17].
In the present investigation, we studied a representative cohort of patients (n = 22) with cc-RCC with clinicopathological correlations to uncover regions of LOH and DNA-CNA affecting genes in the whole genome.
Materials and Methods
Patients
Fresh frozen material from 22 primary cc-RCC and matched normal kidney tissues were collected at the Department of Urology of the University of Technology, Dresden, between 1994 and 2001. All samples were obtained directly after surgery. Fresh material was snap-frozen and stored in liquid nitrogen until further processing. Each sample was evaluated microscopically to ensure a pure population of tumor and normal cells, respectively. The average of normal cell contamination in tumors was <5%. The normal tissue samples showed no tumor cell contamination. Histological typing of tumors was performed in accordance to the UICC classification (1997).
The clinicopathological characteristics and the follow up data of the patients are given in Table 1.
Table 1.
Clinicopathological Characteristics.
| Case ID | Age at Diagnosis (years) | Sex (M/F) | Grading | pT Stage | pN Stage | Metastasis | Status | |
| 4 | 51 | M | G3 | pT2 | pN0 | Pulmonary | NA | |
| 7 | 80 | F | G1 | pT3b | pN1 | No | NA | |
| 26 | 66 | F | G3 | pT2 | pN2 | Pulmonary | NA | |
| 28 | 64 | F | G2 | pT2 | pN2 | Other* | DOD | |
| 31 | 61 | F | G2 | pT3b | pN0 | No | NA | |
| 44 | 60 | F | G2 | pT3b | pN0 | Pulmonary + other | DOD | |
| 65 | 64 | M | G1 | pT1 | pN0 | Other | DOD | |
| 69 | 51 | M | G2 | pT2 | pN0 | Pulmonary + other | DOD | |
| 81 | 56 | M | G2 | pT2 | pN0 | Other | NA | |
| 88 | 39 | M | G2 | pT2 | pNx | No | NA | |
| 122 | 41 | M | G2 | pT3b | pN0 | Pulmonary + other | DOD | |
| 126 | 71 | M | G2 | pT1 | pN0 | Pulmonary + other | DOD | |
| 132 | 66 | M | G3 | pT3b | pN1 | Pulmonary + other | DOD | |
| 135 | 61 | M | G2 | pT1 | pN1 | Other | DOD | |
| 139 | 57 | F | G2 | pT1 | pNx | No | ANED | |
| 154 | 52 | F | G2 | pT1 | pN0 | Other | DOD | |
| 163 | 62 | M | G2 | pT1 | pNx | No | DAD | |
| 167 | 35 | F | G2 | pT4 | pN0 | Pulmonary | ANED | |
| 178 | 63 | F | G2 | pT1 | pNx | No | NA | |
| 182 | 77 | M | G3 | pT2 | pNx | Other | DOD | |
| 195 | 70 | M | G2 | pT1 | pN0 | Other | DOD | |
| 201 | 51 | M | G3 | pT1 | pN2 | Other | DOD |
ANED indicates alive no evidence of disease; DAD, dead of another disease; DOD, dead of disease; NA, not available.
Other: cerebral, mediastinal, pancreas, adrenal glands, liver, bones, or glandula parotis metastasis.
Signed informed consent was obtained from all patients in this study. All procedures were reviewed by the ethical committee of the University of Technology, Dresden (no. 195092004).
DNA Preparation
Genomic DNA of both tumor and normal tissue was extracted from 80 to 100 frozen sections of 10-µm thickness (corresponding to 30–40 mg of tissue) with the Invisorb Spin Tissue Kit (Invitek GmbH, Berlin, Germany) according to the manufacturer's instructions.
GeneChip Human Mapping 10K Array
The tumor and normal tissue samples were analyzed on the Affymetrix GeneChip Human Mapping 10K Array Xba 131 (Affymetrix, Santa Clara, CA). The DNA for hybridization was prepared according to the Affymetrix Mapping Assay protocol. Briefly, 250 ng of DNA was digested with the restriction enzyme XbaI; fragments were ligated to an adaptor and amplified using a universal polymerase chain reaction (PCR) primer in six PCR reactions in parallel. Fragment sizes were confirmed by electrophoresis on 2% agarose gel, and the PCR products were pooled and purified using the MinElute 96 UF PCR Purification Plate (Qiagen, Valencia, CA).
The fragmented and labeled DNA was hybridized to the GeneChip Human Mapping 10K Array Xba 131 overnight at 48°C. The arrays were washed, stained, and scanned according to Affymetrix standard protocols using the Affymetrix GeneChip Fluidic Station and the Agilent GeneArray 2500 Scanner (Agilent Technologies, Palo Alto, CA).
Array Scan and Data Collection
Image analysis was performed using the Affymetrix GeneChip Analysis Software MAS 5.0, and results were assessed using GeneChip Genotyping Analysis Software Version 4.0 (Affymetrix). The LOH regions were detected and demarcated with the Chromosome Copy Number Analysis Tool software version 3.0 (CNAT 3.0) from Affymetrix. The concordance check was performed between tumor and normal tissue of the same specimen. The overall SNP call rate was determined by the software as the number of SNPs assigned AA, BB, or AB divided by the total number of SNPs on the microarray. Results containing information about the identity of each SNP and its chromosomal location were exported to Excel worksheets for further analysis.
Data Analysis
Raw intensity data were retrieved in CEL files and analyzed using self-developed analysis algorithm as described previously [18]. In brief, for copy number analysis, individual probe intensities were locally intensity corrected and merged in intensity values for each probe set. Tumor and normal tissue values were combined to ratios, and the log2 of these ratios were exported for further analysis. For the determination of the allelic configuration of each individual SNP, the intensity distribution between the two alleles was compared. A biallelic SNP, which is characterized by the same intensity for both alleles, is expressed in (single allele) scores of 0.0. LOH and therefore SNPs, which feature only a single allele with noteworthy intensity, result in single allele scores (SAS) of 1.0. LOH was assumed when the same normal tissue biallelic marker was found to be monoallelic in the tumor tissue. In three cases, we were not able to generate analyzable DNA profiles of normal tissues (#4, #7, and #139) due to lack or low amount of starting material. For these cases, we looked at the SAS values only for the tumor DNA and stated LOH by judging the local absence of biallelic markers. For the generation of copy number profiles for these cases, an average of all normal tissuederived intensities was used as a reference.
For the detection of microdeletions, we considered three affected SNP in a row as the threshold.
Quantitative PCR
To validate the microarray data by quantitative PCR (qPCR), 16 of 22 tumor samples and matched normal tissue were available.
Total RNA was isolated from tissue samples according to the manufacturer's instructions (Invisorb Spin Cell/Tissue RNA Mini Kit; Invitek, Berlin, Germany) and was transcribed into cDNA using random hexamer primers (Amersham, Freiburg, Germany) and the SuperScript II Reverse Transcriptase (Invitrogen, Karlsruhe, Germany). Transcript levels of three selected genes (CSF1R, PACRG, and PARK2) and the reference gene PPIA (peptidyl-prolyl cis-trans isomerase A) were determined by qPCR on the LightCycler 480 instrument (Roche, Mannheim, Germany) using commercially available assays on demand (HS_00234622-m1, HS_00247755-m1, and HS_00330069-m1) and the TaqMan Gene Expression Master Mix (all from Applied Biosystems, Darmstadt, Germany).
Using the ΔΔCT method [19], expression levels of the genes of interest were normalized to those of the internal reference gene PPIA, which was detected in all samples (crossing point, <36). As threshold for altered expression in tumor samples, we considered a twofold upor down-regulation compared with the expression in matched normal tissues. Absence of expression of target gene in tumor tissue (crossing point, ≥36) combined with gene expression in matched normal tissue was considered as a complete loss. In contrast, absence of expression of target gene in normal tissues accompanied by detectable target gene expression in the matched tumor sample was considered as a tumor-associated up-regulation. If the target gene expression was under the detection limit (crossing point, <36) in both, tumor and nonmalignant tissue, the sample pair was excluded from analysis for the corresponding target gene.
Immunohistochemistry
Immunohistochemical staining for FOXP1 was performed using tissue microarrays (TMAs) from formalin-fixed, paraffin-embedded archival material. The TMA contains one to five cores each of tumor and normal tissue from 20 of the patients analyzed by SNP microarrays. The sections were deparaffinized, and the antigen retrieval was done at 95°C with sodium citrate buffer (pH = 6.0). The sections were immersed in 0.3% hydrogen peroxide for 10 minutes to block endogenous peroxidase activity. Thereafter, the slides were incubated overnight at 4°C with a polyclonal antibody (1:250) against FOXP1 (Clone ab16645; Abcam, Cambridge, UK), followed by incubation with the EnVision Plus detection system (DAKO, Copenhagen, Denmark). Staining was visualized with diaminobenzidine solution followed by counterstaining with hematoxylin.
The normal kidney tissue was first examined to establish the normal FOXP1 staining in renal tissue. Then, tumor cores were examined, and nuclear, membranous, and cytoplasmic expression was evaluated. The FOXP1 expression was scored using the following system: 0% to 10% positive cells = 0; >10% to 30% positive cells = 1; >30% to 70% positive cells = 2; >70% positive cells = 3. Membranous expression of FOXP1 or concomitant nuclear and cytoplasmic expression of FOXP1 in tumor cells was considered as aberrant.
Results
A total of 22 cc-RCC and paired normal tissues were screened by high-resolution SNP-Chip analysis.
The median age at diagnosis was 61 years. Two patients showed metastasis at the time of diagnosis (#25 and #135). Follow-up data were available from 15 patients. The median time to tumor progression was approximately 12 months. Case 167 presented one solitary pulmonary metastasis that was removed operatively; thereafter, no further metastases were detected (Table 1).
The most common type of alteration we observed in all patients was the loss of one allele, which led to LOH (68 events). The gain of chromosomal regions was noted less frequently (25 events). All detected larger genomic imbalances are summarized graphically in Figure 1. Sporadically, we detected LOH that was not accompanied by a copy number loss (three events: on chromosomes 3, 6, and 7).
Figure 1.
Graphical representation of chromosomal imbalances detected in genomes of patients with cc-RCC. Losses associated with LOH are indicated as red lines and green lines denote regions of copy number gains. Orange line points to a single occasion where copy number loss was not associated with LOH and blue lines indicate regions with LOH without copy number loss. The most frequent alterations are LOH due to hemizygous deletion on chromosomes/arms 3p, 9, and 14 and gains on 5q and 7. Patient IDs from left to right: 122, 126, 132, 135, 139, 154, 163, 167, 178, 182, 195, 201, 26, 28, 31, 44, 4, 65, 69, 7, 81, and 88.
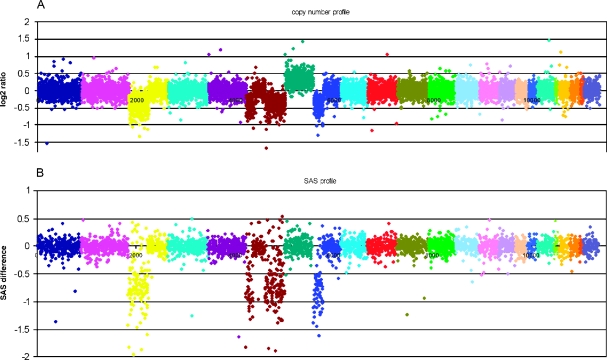
A representative example of DNA-CNA and LOH profile is shown for case 154 in Figure 2, revealing LOH on chromosomes 3, 6, and 8, each accompanied by a copy number loss and a gain of copy of chromosome 7, which did not change the allelic balance.
Figure 2.
Copy number and LOH profile of patient 154 in detail. (A) Copy number ratios in log2 scale between the tumor DNA and unaffected DNA from the same patient. Each spot indicates a unique SNP locus corresponding to its genomic position aligned from 1pter (left) to Xqter (right). Each individual chromosome is coded in a different color. (B) SAS (=LOH) values for those SNP loci that were informative (i.e., biallelic) in the control DNA. Markers that were not informative (monoallelic) in the healthy control DNA are not shown, reducing the amount of data points by roughly two-thirds. In this case, losses in the copy number profile (on chromosomes 3p, 6p, and 8p) are always associated with LOH, so that a hypodiploid karyotype has to be assumed. Interestingly, the gain on chromosome 7 is not accompanied by a shift in the allele ratio. We interpret this constellation as a sign of a mixed population of cells having either two paternal and one maternal chromosome 7 and cells with two maternal and only one paternal chromosome 7.
Frequent Large Aberrations
In 20 (91%) of 22 cases, LOH due to the loss of one copy on 3p harboring the VHL gene was found. The two cases without alterations n 3p had very few copy number alterations in general. Ten tumors (45%) showed a gain on the chromosomal arm 5q. The corresponding minimal overlapping region comprised 5q23.3.-5qter. Other frequent chromosomal alterations were as follows: 6q- (27%), 9- (32%), 7q+ (32%), and 14q- (36%). A complete list of large aberrations is provided in Table 2.
Table 2.
Frequent Large Copy Number Aberrations.
| Aberration | No. Events | % | Minimal Overlapping Region | Candidate Genes* |
| 3p- | 20 | 91 | 3pter-3p21.31 | FOXP1, FHIT, TUSC2, MHL1, VHL |
| 5q+ | 10 | 45 | 5q23.3-5qter | CSF1R |
| 14q- | 8 | 36 | 14q23.3-14q.32.31 | MLH3, FOXN3 |
| 9- | 7 | 32 | 9pter-9qter | CDKN2B, GAS1, DBC1 |
| 7q+ | 7 | 32 | 7q11.21-7qter | FZD9, MET, EPHA1 |
| 6q- | 6 | 27 | 6q23.2-6q26 | PLAGL1, LATS1, PACRG, PARK2 |
For each aberration found in more than 10% of the cases, number of events, percentage of events, and the minimal overlapping region are indicated.
Candidate genes were located in the minimal overlapping region and with an established/inferred function as a TSG (case of losses) or oncogene (case of gains).
No relation was found between the common occurrence of 3p loss and gains on chromosome 5 concerning a better disease-specific survival of the patients. In addition, no correlation between individual chromosomal aberrations and the survival of the patients was found (data not shown).
Microdeletions/Microamplifications
Due to the high variance of individual SNP intensities, aberrations as small as one to three consecutive SNPs are easy to miss. The high resolution of the SNP arrays, the small sample size, and the paired sample approach enabled us to screen for small copy number changes that have emerged in the tumor DNA. Therefore, we looked first for monoallelic deleted SNP genome-wide. It is known that the remaining allele is often inactivated by mutation or methylation. Only a very small number of microdeletions could be retrieved using a threshold of three consecutively deleted SNP loci detected at least in one single case. No small amplifications could be detected. The genomic regions that are affected by a monoallelic loss are listed in Table 3.
Table 3.
Recurrent Small Regions of Monoallelic Loss.
| Genomic Localization (I) | Patients with More Than One SNP Deleted (II) | Genes Disrupted (III) | Patients with a Single SNP Deleted at the Genomic Locus (IV) | Genes Potentially Disrupted (V) |
| Chr 1: 239.498.674–240.013.390 | 18% | FH, KMO, OPN3, CHML | 32% | RGS7, WDR64 |
| Chr 4: 155.360.637–155.520.387 | 9% | 5% | DCHS2 | |
| Chr 6: 157.688.569–157.890.930 | 27% | 14% | ZDHHC14 | |
| Chr 6: 162.491.313–163.442.156 | 5% | PARK2 | 5% | PACRG |
| Chr 8: 13.155.237–13.746.870 | 5% | 14% | DLC1 | |
| Chr 9: 8.149.670–8.258.198 | 5% | 5% | PTPRD |
The table compiles a number of recurrent small deletions that lead to a loss of a gene or parts of it.
Localizations (I) denote the mapping according to the 2007 freeze of the human genome (NCBI build 36.2). Column II indicates patients where, in at least three consecutive SNPs, a loss (ratios <1.0 in the log2 scale) could be stated. These index deletions cover at least three SNPs but could be longer. Column III specifies genes that are affected by the index deletion. Column IV denotes patients with deletion for at least one SNP in the region of the index deletion. Column V specifies genes that are located between the index deletion and the next SNP enclosing it and are potentially disrupted. The deletion on chromosome 6 covers genes (PARK2 and PACRG) with inferred function as tumor suppressor, which are also covered by a minimal overlapping region listed in Table 2.
The deletion on chromosome 6 affects genes (PARK2 and PACRG) that were also covered by a minimal overlapping region (Table 2) and have an inferred function as TSGs.
Furthermore, the data set was screened for biallelic microdeletions. We only found such completely deleted loci to harbor a single SNP and to be located in exon-poor regions (gene deserts or very large first introns, e.g., SNP_A-1513517, SNP_A-1509211, SNP_A-1507787, SNP_A-1514639, SNP_A-1517606).
Validation of Gene Expression by qPCR
For validation of the microarray results, the mRNA expression of the genes CSF1R (located on 5q), PARK2, and PACRG (both located on 6q) was analyzed by qPCR. cDNA samples from 16 tumors and matched normal tissues were available for this analysis.
As expected from the microarray data, the CSF1R relative transcript levels were up-regulated in 11 (69%) of 16 tumors (Figure 3). The PACRG gene was completely lost in all of the 15 evaluable cases. The PARK2 gene was down-regulated in 8 (57%) of 14 evaluable cases, whereby 5 of the 8 tumors showed a complete loss of PARK2 expression.
Figure 3.
Graphical representation of qPCR results. The mRNA expression of CSF1R was analyzed by qPCR in 16 matched pairs of malignant and nonmalignant tissues. A twofold up-regulation (solid line) was considered as threshold for altered expression in tumor samples. For one case (#154), no n-fold change could be calculated because the CSF1R expression level in the nonmalignant sample was below the detection limit. This sample pair belongs to the group with altered expression.
The best concordance of qPCR and microarray data was found for the PARK2 gene (50%, 7/14 cases) followed by CSF1R (38%, 6/16 cases) and PACRG (33%, 5/15 cases).
Validation of FOXP1 Expression by Immunohistochemistry
Large aberrations on 3p containing FOXP1 locus were found by SNP array analysis in 91% of tumors. To validate this result, immunohistochemical staining for FOXP1 was performed. In the normal renal tissue, no concomitant nuclear and cytoplasmic FOXP1 expression was noticed. Proximal tubuli showed an intense cytoplasmic FOXP1 expression, and distal tubuli showed an intense nuclear expression. No or faint FOXP1 expression was noticed in glomeruli and Henle loops.
In the analyzed tumors, an abnormal FOXP1 expression was found in 18 (90%) of 20 cases: no expression in 3 cases (15%), a weak expression in 4 cases (20%), and an aberrant expression in 11 cases (55%). Two cases showed an intense cytoplasmic FOXP1 expression (Figure 4).
Figure 4.
Expression of FOXP1 by immunohistochemistry. FOXP1 nuclear and cytoplasmic expression in normal renal tissue (A; original magnification, x20); nuclear FOXP1 expression in cc-RCC (B; original magnification, x20); lack of FOXP1 expression in cc-RCC (C; original magnification, x10); and abnormal concomitant membranous and nuclear FOXP1 expression in cc-RCC (D; original magnification, x20).
In summary, our results for FOXP1 expression compared with those of microarray analysis show a concordance of 85%.
Discussion
This study represents the first application of high-density Affymetrix 10K SNP genome-wide mapping arrays in simultaneous screening for CNA and LOH in a larger cohort of patients with cc-RCC. Previous studies showed that LOH and CNA analysis by high-density SNP arrays is an efficient and rapidly available method to detect changes in the DNA in different solid malignant neoplasms, revealing candidate genes and loci possibly involved in tumor progression and inherited diseases [20–22].
Our analysis included tumor and matching normal DNA from 22 patients with cc-RCC with known clinicopathological and survival data. The samples showed very high genotyping efficiency with SNP call rates up to 93%.
To analyze the arrays, we used a self-developed analysis algorithm [18] with a higher sensitivity for the detection of microdeletions than the Affymetrix standard software (CNAT 3.0). CNAT 3.0 assumes a normal two-allele configuration, but tumor genomes often feature a gain of a single copy leading to a 2:1 situation, which would render biallelic (i.e., “AB”) calls difficult to interpret. The algorithm is designed to reduce signal variation and therefore enabled us to detect small, locally confined aberrations. In summary, the comparison of the copy number profile obtained with our analysis approach with the results derived from CNAT 3.0 showed good concordance in general, but our generic approach yielded more stable profiles.
Frequent losses on chromosomes 3, 6q, 8p, 13q, 14q, and Xq and gains on chromosomes 5q and 7 [10,23] were previously detected by both CGH and array CGH. The most frequent large chromosomal aberration found in our study was LOH due to a loss of one copy on the short arm of chromosome 3, followed by a gain of 5q, and loss of 14q and chromosome 9.
Loss of heterozygosity without copy number aberration was noticed on chromosomes 3, 6, and 7 (Figure 1, blue lines). Therefore, there are two two-step possibilities: the first one is the doubling of one allele AB to AAB followed by loss of one allele AAB to AA. Another possibility is the loss of one allele AB in A, followed by doubling of allele A in AA. A uniparental disomy for those cases is excluded because such an aberration should also be present in normal tissue and we did not observe any case of uniparental disomy in the nonmalignant tissues.
DNA copy number loss on 3p located in a minimal overlapping region comprising 3pter-3p21.31 was found in 91% of our cases of metastasized and nonmetastasized RCC. Using CGH, Moch et al. [10] detected losses on chromosome 3 in only 56% of nonmetastasized cc-RCC. Because losses on 3p are known to be an early aberration in renal carcinogenesis [4], this discrepancy not only could be unlikely due to the fact that, in our study, metastasized tumors were also included but also more likely due to the higher resolution of the SNP array.
Chromosome 3p harbors several other TSGs such as FOXP1, FHIT, TUSC2, and MLH1 (Table 2) in addition to the well-described VHL gene. The FOXP1 (forkhead box P1) gene at 3p14.1 is a member of winged helix family of transcription factors that play an important role in cellular transformation, differentiation, and proliferation. FOXP1 may act as a TSG and is lost in several tumor types such as endometrial or breast cancer [24,25]. In some types of lymphoma, such as MALT lymphoma, FOXP1 is targeted by recurrent chromosome translocations, and its overexpression is associated with poor prognosis [26].We first studied the FOXP1 expression in normal renal tissue and compared it with the FOXP1 expression in cc-RCC. In normal tissue, only nuclear or only cytoplasmic FOXP1 expression was noticed; never both (Figure 4). In cc-RCC, an abnormal immunohistochemical FOXP1 expression (concomitant nuclear and cytoplasmic expression) or loss of FOXP1 expression (Figure 4) was found in 90% of tumors.
Gains on 5q33-q35 were detected in 45% of our cases by SNP microarray analysis. Gains of the CSF1R (colony-stimulating factor 1 receptor) are the most frequent changes in this region, and mutations of the CSF1R gene are supposedly involved in the progression of cc-RCC [27].
An overexpression of CSF1R was detected by qPCR in 11 (69%) of 16 tumors (Figure 3). The concordance between SNP microarray data and qPCR was 38% (Table 4), suggesting that DNA-CNA is only one pathway inducing an overexpression of CSF1R in cc-RCC.
Table 4.
Gains and Losses Detected by SNP Array and qPCR
| Patient ID | Gains on Chromosome 5 | Losses on Chromosome 6 | ||||
| Detected by | Detected by | |||||
| SNP Array | qPCR CSF1R | SNP Array* | qPCR PACRG | qPCR PARK2 | ||
| 4 | ■ | NA | □ | NA | NA | |
| 7 | ■ | NA | □ | NA | NA | |
| 26 | ■ | □ | □ | ■ | ■ | |
| 28 | □ | ■ | □ | ■ | □ | |
| 31 | □ | □ | ■ | ■ | ■ | |
| 44 | □ | ■ | □ | ■ | □ | |
| 65 | ■ | NA | □ | NA | NA | |
| 69 | ■ | ■ | □ | ■ | ■ | |
| 81 | ■ | NA | □ | NA | NA | |
| 88 | □ | □ | □ | ■ | NE | |
| 122 | □ | □ | ■ | ■ | ■ | |
| 126 | □ | ■ | □ | ■ | ■ | |
| 132 | □ | ■ | □ | ■ | ■ | |
| 135 | ■ | □ | □ | ■ | □ | |
| 139 | ■ | ■ | □ | ■ | ■ | |
| 154 | □ | ■ | ■ | NE | NE | |
| 163 | □ | ■ | ■ | ■ | □ | |
| 167 | □ | ■ | ■ | ■ | □ | |
| 178 | □ | NA | □ | NA | NA | |
| 182 | ■ | NA | ■ | NA | NA | |
| 195 | ■ | ■ | □ | ■ | □ | |
| 201 | □ | ■ | ■ | ■ | ■ | |
■ indicates detectable aberration (gain or loss); □, no detectable aberration (gain or loss); NA, cDNA not available; NE, not evaluable because the target gene in both tumor and nonmalignant tissues was under the detection limit.
Includes large aberrations and microdeletions.
In addition, gains in the region 5q22.1-23.2 were found in 27% of our cases (Figure 1). A gain in this region could be the result of an unbalanced translocation 3:5, which leads to a loss of 3p and duplication of 5q22∼qter sequences in RCC. Nagao et al. [14] found that patients with 3p loss and gain at 5q22.3-23.3 had a significantly better disease-free survival than those who had a 3p loss without such a gain. We could not detect a survival benefit for the patients with only a 3p loss in our study (data not shown).
It has been shown that cc-RCC with LOH on chromosome 9 have a higher risk of recurrence and a shorter tumor-specific survival [28,29]. In the present study, aberrations on chromosome 9 were found in 7 (32%) of 22 tumors. Five of these seven patients are dead, demonstrating the poor prognosis of cc-RCC with aberrations on chromosome 9 (Table 1 and Figure 1).
As expected, the analysis with genome-wide SNP arrays revealed additional small changes that were not yet detected by CGH or array CGH. We used this high-resolution technology as a method to detect loci crucial for tumor development in cc-RCC. Microamplifications were not found, but microdeletions leading to a monoallelic loss of a gene or gene parts were occasionally encountered (Table 3).
On chromosome 1, we found microdeletions in 55% of the tumors. An association between allelic imbalances on chromosome 1 and cc-RCC is known; Zhao et al. [30] described LOH of HRPT2, located at 1q25-q32 in various types of RCC including cc-RCC. In the present study, a monoallelic deletion in a region comprising FH, KMO, OPN3, and CHML on chromosome 1 was detected in four tumors (Table 3). There are very small genes (7.07–63.5 kbp), and no SNP was located directly in the genes. The deleted SNP are located at the beginning and at the end of this small region containing these four genes. There is currently very little information available linking these genes to tumor formation or progression. Germline FH mutations are associated with hereditary leiomyomatosis and renal cell cancer [31].
Microdeletions on chromosome 6 affecting PARK2 and PACRG were found in 5% of the tumors. Mutations in both genes are known to cause autosomal recessive juvenile parkinsonism [32,33]. Agirre et al. [34] described that abnormal methylation and regulation of PARK2 and PACRG may be involved in the pathogenesis of acute lymphoblastic and chronic myeloid leukemia. We could show by qPCR that PARK2 and PACRC are down-regulated in 57% (8/14) and 100% (15/15) of cc-RCC, respectively, compared with the normal renal tissue (Figure 3), but the relevance of these genes in renal carcinogenesis is still unknown. We also found by qPCR more tumors showing down-regulation of PACRG and PARK2 as expected after SNP microarray analysis, suggesting additional mechanisms, such as methylation, involved in switching off both genes.
Microdeletions with potential gene disruption within or close to the DLC1 (deleted in liver cancer 1) gene mapped to 8p22-p21.3 were found in three cases (Table 3). This gene is deleted in primary hepatocellular carcinoma, and studies done by Seng et al. [35] suggested that this gene is a candidate TSG for human liver cancer as well as for prostate, lung, colorectal, and breast cancers. Its role in renal carcinogenesis has still to be determined.
In conclusion, SNP arrays are an excellent tool not only to confirm the previously described large chromosomal aberrations but also to detect novel microdeletions in genes potentially involved in tumorigenesis and progression of cc-RCC. Our results will need to be validated on a larger patient cohort with known clinical outcome. The significance of the described microdeletions in tumorigenesis and progression of RCC remains to be proven.
Acknowledgments
The authors thank Annelie Zuerich, Andrea Lohse-Fischer, and Antje Zobjack for their excellent technical assistance.
Abbreviations
- cc-RCC
clear cell renal cell carcinoma
- DNA-CNA
DNA-copy number aberration/abnormality
- LOH
loss of heterozygosity
- SAS
single allele score
- SNP
single nucleotide polymorphism
- TSG
tumor suppressor gene
Footnotes
This study was partially supported by Deutsche Krebshilfe e. V. (70-2923).
References
- 1.Higgins JP, Shinghal R, Gill H, Reese JH, Terris M, Cohen RJ, Fero M, Pollack JR, van de Rijn M, Brooks JD. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol. 2003;162:925–932. doi: 10.1016/S0002-9440(10)63887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young AN, Amin MB, Moreno CS, Lim SD, Cohen C, Petros JA, Marshall FF, Neish AS. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. Am J Pathol. 2001;158:1639–1651. doi: 10.1016/S0002-9440(10)64120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clifford SC, Prowse AH, Affara NA, Buys CH, Maher ER. Inactivation of the von Hippel-Lindau (VHL) tumor suppressor gene and allelic losses at chromosome arm 3p in primary renal cell carcinoma: evidence for a VHL-independent pathway in clear tumourigenesis. Genes Chromosomes Cancer. 1998;22:200–209. doi: 10.1002/(sici)1098-2264(199807)22:3<200::aid-gcc5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 4.Martinez A, Fullwood P, Kondo K, Kishida T, Yao M, Maher ER, Latif F. Role of chromosome 3p12-p21 tumour suppressor genes in clear cell renal cell carcinoma: analysis of VHL dependent and VHL independent pathways of tumorigenesis. Mol Pathol. 2000;53:137–144. doi: 10.1136/mp.53.3.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higgins JP. Gene array studies in renal neoplasia. ScientificWorldJournal. 2006;6:502–511. doi: 10.1100/tsw.2006.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kondo K, Yao M, Yoshida M, Kishida T, Shuin T, Miura T, Moriyama M, Kobayashi K, Sakai N, Kaneko S, et al. Comprehensive mutational analysis of the VHL gene in sporadic renal cell carcinoma: relationship to clinicopathological parameters. Genes Chromosomes Cancer. 2002;34:58–68. doi: 10.1002/gcc.10054. [DOI] [PubMed] [Google Scholar]
- 7.Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, Latif F, Liu S, Chen F, Duh FM, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 8.Thrash-Bingham CA, Salazar H, Freed JJ, Greenberg RE, Tartof KD. Genomic alterations and instabilities in renal cell carcinomas and their relationship to tumor pathology. Cancer Res. 1995;55:6189–6195. [PubMed] [Google Scholar]
- 9.Kardas I, Mrozek K, Babinska M, Krajka K, Hadaczek P, Lubinski J, Roskiewicz A, Kuziemska E, Limon J. Cytogenetic and molecular findings in 75 clear cell renal cell carcinomas. Oncol Rep. 2005;13:949–956. [PubMed] [Google Scholar]
- 10.Moch H, Presti JC, Sauter G, Buchholz N, Jordan P, Mihatsch MJ, Waldman FM. Genetic aberrations detected by comparative genomic hybridization are associated with clinical outcome in renal cell carcinoma. Cancer Res. 1996;56:27–30. [PubMed] [Google Scholar]
- 11.Mitsumori K, Kittleson JM, Itoh N, Delahunt B, Heathcott RW, Stewart JH, McCredie MR, Reeve AE. Chromosome 14q LOH in localized clear cell renal cell carcinoma. J Pathol. 2002;198:110–114. doi: 10.1002/path.1165. [DOI] [PubMed] [Google Scholar]
- 12.Kaku H, Ito S, Ebara S, Ouchida M, Nasu Y, Tsushima T, Kumon H, Shimizu K. Positive correlation between allelic loss at chromosome 14q24-31 and poor prognosis of patients with renal cell carcinoma. Urology. 2004;64:176–181. doi: 10.1016/j.urology.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 13.Alimov A, Sundelin B, Wang N, Larsson C, Bergerheim U. Loss of 14q31-q32.2 in renal cell carcinoma is associated with high malignancy grade and poor survival. Int J Oncol. 2004;25:179–185. [PubMed] [Google Scholar]
- 14.Nagao K, Yamaguchi S, Matsuyama H, Korenaga Y, Hirata H, Yoshihiro S, Fukunaga K, Oba K, Naito K. Allelic loss of 3p25 associated with alterations of 5q22.3 approximately q23.2 may affect the prognosis of conventional renal cell carcinoma. Cancer Genet Cytogenet. 2005;160:43–48. doi: 10.1016/j.cancergencyto.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Janne PA, Li C, Zhao X, Girard L, Chen TH, Minna J, Christiani DC, Johnson BE, Meyerson M. High-resolution single-nucleotide polymorphism array and clustering analysis of loss of heterozygosity in human lung cancer cell lines. Oncogene. 2004;23:2716–2726. doi: 10.1038/sj.onc.1207329. [DOI] [PubMed] [Google Scholar]
- 16.Zheng HT, Peng ZH, Li S, He L. Loss of heterozygosity analyzed by single nucleotide polymorphism array in cancer. World J Gastroenterol. 2005;11:6740–6744. doi: 10.3748/wjg.v11.i43.6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam CW, To KF, Tong SF. Genome-wide detection of allelic imbalance in renal cell carcinoma using high-density single-nucleotide polymorphism microarrays. Clin Biochem. 2006;39:187–190. doi: 10.1016/j.clinbiochem.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Herr A, Grutzmann R, Matthaei A, Artelt J, Schrock E, Rump A, Pilarsky C. High-resolution analysis of chromosomal imbalances using the Affymetrix 10K SNP genotyping chip. Genomics. 2005;85:392–400. doi: 10.1016/j.ygeno.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindblad-Toh K, Tanenbaum DM, Daly MJ, Winchester E, Lui WO, Villapakkam A, Stanton SE, Larsson C, Hudson TJ, Johnson BE, et al. Loss-of-heterozygosity analysis of small-cell lung carcinoma using single-nucleotide polymorphism arrays. Nat Biotechnol. 2000;18:1001–1005. doi: 10.1038/79269. [DOI] [PubMed] [Google Scholar]
- 21.Hoque MO, Lee CC, Cairns P, Schoenberg M, Sidransky D. Genome-wide genetic characterization of bladder cancer: a comparison of high-density single-nucleotide polymorphism arrays and PCR-based microsatellite analysis. Cancer Res. 2003;63:2216–2222. [PubMed] [Google Scholar]
- 22.Primdahl H, Wikman FP, von der Maase H, Zhou XG, Wolf H, Orntoft TF. Allelic imbalances in human bladder cancer: genome-wide detection with high-density single-nucleotide polymorphism arrays. J Natl Cancer Inst. 2002;94:216–223. doi: 10.1093/jnci/94.3.216. [DOI] [PubMed] [Google Scholar]
- 23.Wilhelm M, Veltman JA, Olshen AB, Jain AN, Moore DH, Presti JC, Kovacs G, Waldman FM. Array-based comparative genomic hybridization for the differential diagnosis of renal cell cancer. Cancer Res. 2002;62:957–960. [PubMed] [Google Scholar]
- 24.Giatromanolaki A, Koukourakis MI, Sivridis E, Gatter KC, Harris AL, Banham AH. Loss of expression and nuclear/cytoplasmic localization of the FOXP1 forkhead transcription factor are common events in early endometrial cancer: relationship with estrogen receptors and HIF-1α expression. Mod Pathol. 2006;19:9–16. doi: 10.1038/modpathol.3800494. [DOI] [PubMed] [Google Scholar]
- 25.Fox SB, Brown P, Han C, Ashe S, Leek RD, Harris AL, Banham AH. Expression of the forkhead transcription factor FOXP1 is associated with estrogen receptor alpha and improved survival in primary human breast carcinomas. Clin Cancer Res. 2004;10:3521–3527. doi: 10.1158/1078-0432.CCR-03-0461. [DOI] [PubMed] [Google Scholar]
- 26.Banham AH, Beasley N, Campo E, Fernandez PL, Fidler C, Gatter K, Jones M, Mason DY, Prime JE, Trougouboff P, et al. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res. 2001;61:8820–8829. [PubMed] [Google Scholar]
- 27.Strefford JC, Stasevich I, Lane TM, Lu YJ, Oliver T, Young BD. A combination of molecular cytogenetic analyses reveals complex genetic alterations in conventional renal cell carcinoma. Cancer Genet Cytogenet. 2005;159:1–9. doi: 10.1016/j.cancergencyto.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 28.Presti JC, Wilhelm M, Reuter V, Russo P, Motzer R, Waldman F. Allelic loss on chromosome 8 and 9 correlates with clinical outcome in locally advanced clear cell carcinoma of the kidney. J Urol. 2002;167:1464–1468. [PubMed] [Google Scholar]
- 29.Schraml P, Struckmann K, Bednar R, Fu W, Gasser T, Wilber K, Kononen J, Sauter G, Mihatsch MJ, Moch H. CDKN2A mutation analysis, protein expression, and deletion mapping of chromosome 9p in conventional clear-cell renal carcinomas. Am J Pathol. 2001;158:593–601. doi: 10.1016/s0002-9440(10)64001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, Yart A, Frigerio S, Perren A, Schraml P, Weisstanner C, Stallmach T, Krek W, Moch H. Sporadic human renal tumors display frequent allelic imbalances and novel mutations of the HRPT2 gene. Oncogene. 2007;26:3440–3449. doi: 10.1038/sj.onc.1210131. [DOI] [PubMed] [Google Scholar]
- 31.Sudarshan S, Pinto PA, Neckers L, Linehan WM. Mechanisms of disease: hereditary leiomyomatosis and renal cell cancer—a distinct form of hereditary kidney cancer. Nat Clin Pract Urol. 2007;4:104–110. doi: 10.1038/ncpuro0711. [DOI] [PubMed] [Google Scholar]
- 32.Lucking CB, Durr A, Bonifati V, Vaughan J, De Michele G, Gasser T, Harhangi BS, Meco G, Denefle P, Wood NW, et al. Association between early-onset Parkinson's disease and mutations in the parkin gene. N Engl J Med. 2000;341:1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 33.West AB, Lockhart PJ, O'Farell C, Farrer MJ. Identification of a novel gene linked to parkin via a bi-directional promoter. J Mol Biol. 2003;326:11–19. doi: 10.1016/s0022-2836(02)01376-1. [DOI] [PubMed] [Google Scholar]
- 34.Agirre X, Roman-Gomez J, Vazquez I, Jimenez-Velasco A, Garate L, Montiel-Duarte C, Artieda P, Cordeu L, Lahortiga I, Calasanz MJ, et al. Abnormal methylation of the common PARK2 and PACRG promoter is associated with downregulation of gene expression in acute lymphoblastic leukemia and chronic myeloid leukemia. Int J Cancer. 2006;118:1945–1953. doi: 10.1002/ijc.21584. [DOI] [PubMed] [Google Scholar]
- 35.Seng TJ, Low JS, Li H, Cui Y, Goh HK, Wong ML, Srivastava G, Sidransky D, Califano J, Steenbergen RD, et al. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26:934–944. doi: 10.1038/sj.onc.1209839. [DOI] [PubMed] [Google Scholar]