Abstract
Purpose
Pancreatic cancer still has one of the worst prognoses in gastrointestinal cancers with a 5-year survival rate of 5%, making it necessary to find markers or gene sets that would further classify patients into different risk categories and thus allow more individually adapted multimodality treatment regimens. In this study, we investigated the prognostic values of HIF1a, bFGF, VEGF, and PDGFA gene expressions as well as their interrelationships.
Experimental Design
Formalin-fixed paraffin-embedded tissue samples were obtained from 41 patients with pancreatic adenocarcinoma (age, 65; range, 34–85 years). After laser capture microdissection, direct quantitative real-time reverse transcription-polymerase chain reaction assays were performed in triplicates to determine HIF1a, PDGFA, VEGF, and bFGF gene expression levels. Multivariate Cox proportional hazards regression analysis was used to assess the impact of HIF1a gene expression on prognosis.
Results
HIF1a was significantly correlated to every gene we tested: bFGF (P = .04), VEGF (P = .02), and PDGFA (P = .03). Tumor size, P = .04, and high HIF1a mRNA expression (cutoff, 75th percentile) had a significant impact on survival, P = .009 (overall model fit, P = .02). High HIF1a expression had a sensitivity of 87.1% and a specificity of 55.6% for the diagnosis short (<6 months) versus long (6–60 months) survival.
Conclusions
Measuring PDGFA, bFGF, and HIF1a expression may contribute to a better understanding of the prognosis of patients with pancreatic cancer and may even play a crucial role for the distribution of patients to multimodal therapeutic regimens. Larger studies including patients treated with actual chemotherapeutics seem to be warranted.
Introduction
Although rising for over 30 years, mortality rates of adenocarcinoma of the pancreas in Europe have leveled off in the last 10 to 15 years and have even decreased approximately 4% in the United States [1,2]. Nonetheless, pancreatic cancer still has one of the worst prognoses in gastrointestinal cancers, with a 5-year survival rate of 5%. Due to the late symptoms of pancreatic cancer and therefore often late diagnosis, only 10% to 20% of the patients are eligible for complete resection with curative intention, making it necessary to find markers or gene sets that would further classify patients into different risk categories and thus allow more individually adapted multimodality treatment regimens [3].
Several good candidate prognostic markers for pancreatic adenocarcinoma have been identified by previous work. Hypoxia-inducible factor 1 alpha (HIF1a) has been shown to correlate with an unfavorable prognosis in many cancers and is known to regulate some genes in the angiogenesis pathway [4]. Recently, Sun et al. [5] and Tao et al. [6] showed with immunohistochemical methods that HIF1a not only has a strong impact on the prognosis of patients with pancreatic ductal adenocarcinoma but is also correlated with vascular endothelial growth factor (VEGF) expression. Studies on human pancreatic cancer cells revealed that hypoxia and paracrine secretion of insulin induced HIF1a expression, which in turn led to stimulated glycolysis, cell proliferation, and VEGF secretion [7]. HIF1a binds to the promoter region of VEGF and apparently thereby increases the expression on VEGF under hypoxic conditions [8]. VEGF is not only a key factor in the angiogenesis pathway but also a molecular drug target for several U.S. Food and Drug Administration-approved chemotherapeutics [9]. Platelet-derived growth factor alpha (PDGFA) has recently been discussed as a potential drug target in pancreatic cancer [10]. It has to be further investigated in what respect HIF1a and PDGFA are coexpressed in pancreatic cancer. The expression of basic fibroblast growth factor (bFGF) is known to have a strong association with the prognosis of patients with pancreatic cancer [11]. Some study groups showed a correlation between bFGF and HIF1a expression in patients with breast and non-small cell lung cancer [12,13]. However, this association is, like that of PDGFA and HIF1a, little studied in pancreatic cancer.
In this study, we investigated the prognostic values of HIF1a, bFGF, VEGF, and PDGFA gene expressions as well as their interrelationships in pancreatic cancer. We measured the mRNA expression levels of these genes with quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR) in formalin-fixed paraffin-embedded (FFPE) tissue samples of pancreatic carcinoma. We then further analyzed the previously mentioned genes and their correlation with clinical and histopathologic variables such as tumor size (diameter/volume), primary tumor stage [pT, based on the International Union Against Cancer (UICC), 1997], regional lymph node metastasis, grading, and especially the survival time.
Materials and Methods
Study Population, Demographic Data, and Staging Procedures
Formalin-fixed paraffin-embedded samples were obtained from 41 patients with pancreatic adenocarcinoma with a median age of 65 years (range, 34–85 years) at the time of operation who were scheduled for primary surgical resection. None of the patients had received neoadjuvant or adjuvant radio-/chemotherapy. All patients were treated at the University hospital of Cologne, North Rhine-Westphalia, Germany, between December 1999 and July 2004. Demographic, clinical, and histopathologic parameters are shown in Table 1. Informed consent was obtained from each patient in accordance with the requirements of our institution's board of ethics. TNM staging was performed according to the criteria of the UICC [14].
Table 1.
Patient Characteristics (N = 41).
| Parameter | n (%) |
| Median age: 65 years (range, 34–85 years) | |
| Sex | |
| Male | 23 (56.1) |
| Female | 18 (43.9) |
| Histologic diagnosis | |
| Adenocarcinoma | 41 (100) |
| pT category | |
| pT1 | 1 (2.4) |
| pT2 | 6 (14.6) |
| pT3 | 32 (78.0) |
| pT4 | 2 (4.9) |
| pN category | |
| N - | 7 (17.1) |
| N + | 33 (80.5) |
| Not evaluated | 1 (2.4) |
| c/pM category | |
| c/pM0 | 41 (100) |
| Grading | |
| G2 | 22 (53.7) |
| G3 | 19 (46.3) |
| Residual tumor category | |
| R0 | 41 (100) |
| Tumor size (diameter; cm) | |
| Minimum | 1 |
| Maximum | 7 |
| Range | 6 |
UICC 1997 Tumor-Node-Metastasis (pTNM) Pathological Classification: pT indicates primary tumor; pN, regional lymph node metastasis; c/pM, distant metastasis; G, grade of differentiation; R, residual tumor category.
Microdissection
After a review of representative hematoxylin and eosin-stained slides of the FFPE blocks by a pathologist to estimate the tumor load per sample, section slides of 10-µm thickness were obtained for lasercaptured microdissection (P.A.L.M. Microlaser Technologies AG, Munich, Germany). All tumor slides were prepared as described extensively by Vallbohmer et al. [15].
Isolation of RNA and cDNA Synthesis
The isolation of RNA from tumor tissue isolated by the microdissection was performed in accordance with a patented procedure at Response Genetics Inc (Los Angeles, CA; U.S. Patent No. 6248,535). The cDNA preparation steps were accomplished as described previously [16].
Quantitative Real-Time Polymerase Chain Reaction
To quantify HIF1a, PDGFA, VEGF, and bFGF mRNA expression levels, we used an endogenous reference gene (β-actin) and our gene set on a method based on real-time fluorescence detection of amplified cDNA (ABI PRISM 7900 Sequence Detection System [TaqMan]; Perkin-Elmer Applied Biosystems, Foster City, CA). The RT-PCR was implemented as previously described by Kuramochi et al. [17]. All genes were run on all samples in triplicates. The detection of amplified cDNA results in a cycle threshold (Ct) value that is inversely proportional to the amount of cDNA. The higher the ensuing Ct value, the more PCR cycles were necessary to attain detection limit, which means less cDNA. Colon, liver, and St. Universal Mix RNA (Stratagene, La Jolla, CA) were used as control calibrators on each plate. All primers were selected using the Gene Express software (Applied Biosystems) but were adapted to the needs of RNA/cDNA as extracted out of the paraffin-embedded tissue. All primers were validated before use analogical to the described method of Schneider et al. [18]. All results are expressed as ratios between two absolute measurements (gene of interest/endogenous reference gene) to account for loading differences. We used a log transformation before statistical analysis including a multiplier that accounts the average Ct values maintained for each gene during the validation process on the calibrators and therefore allows comparing samples that were run on different RT-PCR well plates.
Statistical Analysis
The correlation among gene expression levels, those and clinicopathologic parameters were tested with Spearman test for bivariate correlations. Each gene was tested with the Kaplan-Meier method to estimate overall survival and relapse-free survival. Differences in survival between the high- and low-expression groups were analyzed with the log rank test. To evaluate independent prognostic factors associated with survival, multivariate Cox proportional hazards regression analysis with stepwise selection was used, with the gene set, tumor stage, tumor size (diameter/volume), and histologic characteristics (grading) as covariates. A data mining technique provided by the SAS Institute was used to split gene expression in high- and low-level groups based on a platform that recursively partitions data according to a relationship between the X and Y values, creating a tree of partitions (recursive descent partition analysis). By searching all possible cuts, it finds a set of cut points of X values (Gene Expression) that best predict the Y value (Survival Time). These data splits are done, recursively forming a tree of decision rules until the desired fit is reached; the most significant split is determined by the largest likelihood ratio chi-square statistic. In either case, the split is chosen to maximize the difference in the responses between the two branches of the split. This method was previously used by Lu et al. [19]. We used receiver operating characteristic (ROC) curve analysis to test the ability of the chosen cutoffs to discriminate short survivors (<6 months) from long survivors (6 months to 5 years). The level of significance was set to P < .05. All P values reported were based on two-sided tests.
All statistical tests were performed using the Statistical Package for the Social Sciences for Windows (Version 16.0; Chicago, IL) and the JMP 7.0 Software (SAS, Cary, NC).
Results
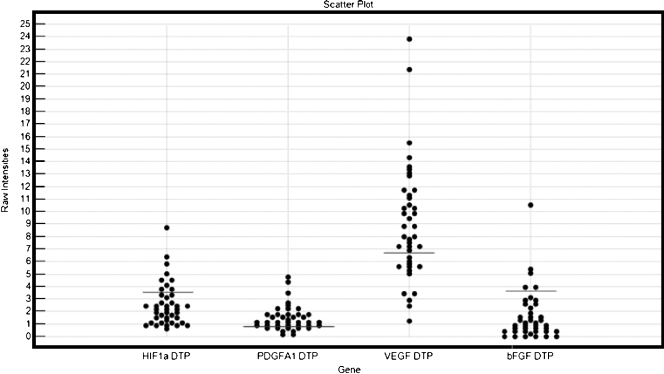
The distribution of the log-transformed ΔCt values is shown in Figure 1.
Figure 1.
Scatterplot of the log-transformed ΔCt values for the studied genes. The lines represent the cutoff values based on recursive descent partition analysis.
Spearman Test for Bivariate Correlations
Spearman test on the log-transformed ΔCt values showed significant correlations between some of the gene expressions. HIF1a was correlated to every gene we tested. HIF1a was significantly correlated to bFGF at P = .04 (P < .05), it was associated to the VEGF gene expression with a significance of P = .02 (P < .05) and to PDGFA at a level of significance of P = .03 (P < .05). VEGF gene expression and PDGFA expression levels were associated with a significance of P = .02 (P < .05). We performed partition analysis of VEGF mRNA expression based on the existence of lymph node metastasis, thereby obtaining two groups with high versus low expression profiles. This grouping showed a significant correlation to lymph node metastasis with P = .04 (P < .05), thus implicating that a high expression of VEGF is linked to a higher likelihood of lymph node metastasis. Using the same method for the other used genes, no significant correlation appeared. With a significance level of P = .01 (P < .05), a high VEGF mRNA level was also connected to the size (in diameter) of the resected pancreatic adenocarcinoma.
Partition Tree Analysis of Genes Based on Survival Time
Using the survival (in days) as the factor to perform partition analysis on the chosen gene set, bFGF showed up as the most significant divisor (with the highest log rank). The next in line were first HIF1a, then PDGFA, and then VEGF. The cutoff value for HIF1a was 3.9 (the 75th percentile) leaving the high-expression group of 11 patients with a mean of 4.8 (range, 3.9–8.7) and the low-expression group consisting of 30 patients with a mean of 3.2 (range, 0.6–3.8). The groupings, respectively, the cutoffs for the other genes are indicated in Figure 1.
Survival Analysis Using the Kaplan-Meier Method
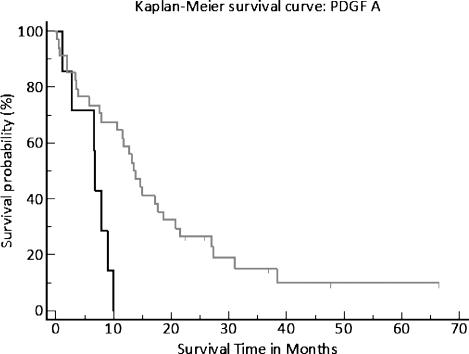
The groupings obtained by the recursive descent partition analysis were used for each gene to assign survival with Kaplan-Meier log rank analysis. The level of significance for bFGF was P = .01 (Figure 2) and for PDGFA, P = .003 (Figure 3). The P value of HIF1a only showed a trend to significance.
Figure 2.
Kaplan-Meier plot, estimating overall survival and relapsefree survival. Differences in survival between the high and the low bFGF expression groups were analyzed with the log rank test.
Figure 3.
Kaplan-Meier plot, estimating overall survival and relapse-free survival. Differences in survival between the high and the low PDGFA expression groups were analyzed with the log rank test.
Multivariate Cox Proportional Hazards Regression Analysis
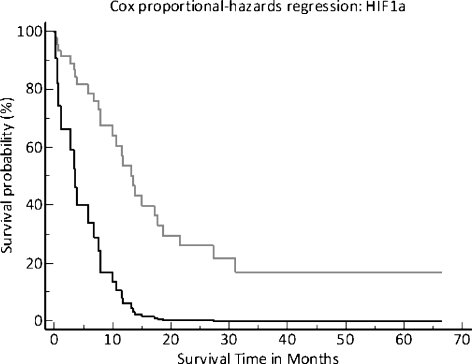
We put all clinical and histopathologic parameters along with the measured gene expressions in a stepwise multivariate Cox proportional hazards regression model. The overall model fit had a significance level of P = .02. There were two factors that had a significant impact on the survival time in this patient cohort, namely, tumor size, P = .04, and a high HIF1a mRNA expression (75th percentile cutoff ), with a significance level of P = .009 (Figure 4).
Figure 4.
Survival plot from multivariate Cox regression analysis estimating overall survival and relapse-free survival. The upper light gray line represents the patients with a HIF1a expression of lower than the 75th percentile.
Receiver Operating Characteristic
The 75th percentile cutoff of the HIF1a mRNA expression showed a sensitivity (true-positive rate) of 87.1% and a specificity (true-negative rate) of 55.6% for the diagnosis short (<6 months) versus long (6 months to 5 years) survival. The area under the curve was 0.713 (confidence interval, 0.549–0.845) with a significance level of P = .043. The positive likelihood ratio (true-positive rate/false-positive rate) was 1.96, and the negative likelihood ratio (falsenegative rate/true-negative rate) was 0.23.
Discussion
In this study, we determined the gene expressions of HIF1a, VEGF, bFGF, and PDGFA in the FFPE samples of pancreatic cancer patients who did not receive any chemotherapy. By using laser capture microdissection to isolate tumor tissue from the clinical specimens along with quantitative RT-PCR, we hoped to achieve a more precise characterization of the associations of these gene expressions with each other and with patients' prognosis than was previously available. By multivariate Cox regression analysis, HIF1a gene expression showed the most significant impact on prognosis in our patient cohort (P = .009).
It has to be mentioned that HIF1a is known to undergo a rapid posttranscriptional degradation under normoxic conditions by the ubiquitin-proteasome system, which is restricted by an oxygen-dependent degradation domain within HIF-1, but under hypoxic conditions, the accumulation of HIF1a involves stabilization of the protein [20]. For several genes, such as thymidylate synthase and dihydropyrimidine dehydrogenase, there have been studies that provided information on a generally close linear correlation between the expression of mRNA and the protein expression [21,22]. Although the regulation of HIF1a RNA expression seems to respond rapidly to the oxygen concentrations in the cell [23], meaning that the mechanisms of HIF1a regulation are transcriptional as well as posttranscriptional, it is controversially discussed whether this mechanisms correlate well to each other [24]. In this study, we show that the determination of quantitative levels of gene expressions may be valuable and important in itself, regardless of whether it matches the protein or not. Although there is posttranscriptional regulation of the HIF1a subunit, an approach to use mRNA expression for assessing the prognosis of patients with pancreatic ductal adenocarcinoma seems to be feasible.
The results of this pilot study also show a strong correlation of HIF1a to VEGF and underline the meaning of HIF1a for the angiogenesis and its most prominent marker VEGF on pancreatic cancer. Although we could not verify the results of Sun et al. [5] regarding the meaning of VEGF for the survival of the patients, we were also able to show that a high VEGF expression significantly correlated to lymph node metastasis (P = .04) and tumor size (P = .01).
The heparin-binding bFGF is one of the more frequently described genes in pancreatic cancer. The association of HIF1a and bFGF has recently been shown in some cancers. Bos et al. [12] investigated 45 samples of invasive breast cancer with immunohistochemistry. They were able to show a significant association between HIF1a and bFGF but could not show this for VEGF and HIF1a. Also, a different member of the PDGF family than we studied had a significant connection to HIF1a expression. Giatromanolaki et al. [13] examined 120 samples of patients with non-small cell lung cancer with monoclonal antibodies on FFPE samples, assessing the relationship between HIF1a, bFGF, VEGF, and PD-ECGF. In their patient cohort, the association between the expression of these proteins was significant. A literature search disclosed no studies focusing on the relationship between bFGF and HIF1a in pancreatic cancer. However, in our patient group, the association between these two genes was significant, suggesting that the bFGF-HIF1a relationship also has a role in pancreatic cancer, especially because of the impact on prognosis both factors had. The survival probability was significantly higher with P = .013 of patients with lower bFGF expression. As previously mentioned, the impact of HIF1a expression on prognosis was the strongest genetic factor on our patient cohort and even stronger than clinicopathologic parameters.
Although members of the PDGF family and their receptors have recently been discussed as potential drug targets in cancer [10], little is known about their role specifically in pancreatic cancer. As of today, especially the connection between PDGFA and HIF1a, although described in renal cell carcinoma, seems to be unevaluated in pancreatic cancer [25]. So far, the correlated mRNA expression of PDGFA and VEGF in cancer has only been described in a few studies [26] but not in pancreatic cancer tissue. In our patient samples, the correlation between PDGFA and VEGF was significant at a level of P = .02. The correlation between PDGFA and HIF1a was significant at P = .03, and the survival probability of patients with a high PDGFA expression was significantly lower as of patients with a low expression of the same gene (P = .003).
When using gene expression as an approach to classify tumors, one can always question whether generally more aggressive tumors are metabolically more active or whether the expression of certain genes leads to a more aggressive tumor. Whereas this question cannot, of course, be definitively answered from a correlative study, the fact that these gene expressions did associate with various metrics of tumor aggressiveness strengthens the hypothesis of cause, not effect. Identifying genes that are associated with more aggressive tumors is useful to form a candidate oncogene pool that is available for further work to more definitively address the cause-or-effect question, such as in vitro experiments where the genes in question are transfected into cells. For example, insertion of a mutated p53 into cells has been used to demonstrate that this gene directly causes many different effects, but it had to be identified first as being associated with more aggressive tumors.
Conclusions
The significant impact of a high PDGFA expression on survival probability (P = .003), the significantly higher survival probability of patients with a low bFGF expression (P = .01), and the importance of a high HIF1a expression (P = .009) in comparison to all clinicopathologic parameters suggest that these three genes may contribute to a better understanding of the prognosis of patients with pancreatic cancer and may even play a crucial role for the distribution of patients to multimodal therapeutic regimens. Larger studies including patients treated with actual chemotherapeutics seem to be warranted.
Abbreviations
- HIF1a
hypoxia-inducible factor 1 alpha
- PDGFA
platelet-derived growth factor alpha
- VEGF
vascular endothelial growth factor
- bFGF
basic fibroblast growth factor
- FFPE
formalin-fixed paraffin-embedded
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Levi F, Lucchini F, Negri E, La Vecchia C. Pancreatic cancer mortality in Europe: the leveling of an epidemic. Pancreas. 2003;27:139–142. doi: 10.1097/00006676-200308000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Prenzel KL, Warnecke-Eberz U, Brabender J, Baldus SE, Bollschweiler E, Gutschow CA, Drebber U, Hoelscher AH, Schneider PM. Differential c-erbB-1 and c-erbB-2 mRNA expression in cancer of the pancreas compared with cancer of the papilla of Vater. World J Gastroenterol. 2006;12:437–442. doi: 10.3748/wjg.v12.i3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Couvelard A, O'Toole D, Turley H, Leek R, Sauvanet A, Degott C, Ruszniewski P, Belghiti J, Harris AL, Gatter K, et al. Microvascular density and hypoxia-inducible factor pathway in pancreatic endocrine tumours: negative correlation of microvascular density and VEGF expression with tumour progression. Br J Cancer. 2005;92:94–101. doi: 10.1038/sj.bjc.6602245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun HC, Qiu ZJ, Liu J, Sun J, Jiang T, Huang KJ, Yao M, Huang C. Expression of hypoxia-inducible factor-1 alpha and associated proteins in pancreatic ductal adenocarcinoma and their impact on prognosis. Int J Oncol. 2007;30:1359–1367. [PubMed] [Google Scholar]
- 6.Tao J, Li T, Li K, Xiong J, Yang Z, Wu H, Wang C. Effect of HIF- 1alpha on VEGF-C induced lymphangiogenesis and lymph nodes metastases of pancreatic cancer. J Huazhong Univ Sci Technolog Med Sci. 2006;26:562–564. doi: 10.1007/s11596-006-0520-9. [DOI] [PubMed] [Google Scholar]
- 7.Wang F, Li SS, Segersvard R, Strommer L, Sundqvist KG, Holgersson J, Permert J. Hypoxia inducible factor-1 mediates effects of insulin on pancreatic cancer cells and disturbs host energy homeostasis. Am J Pathol. 2007;170:469–477. doi: 10.2353/ajpath.2007.060489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray MJ, Zhang J, Ellis LM, Semenza GL, Evans DB, Watowich SS, Gallick GE. HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 2005;24:3110–3120. doi: 10.1038/sj.onc.1208513. [DOI] [PubMed] [Google Scholar]
- 9.Ho QT, Kuo CJ. Vascular endothelial growth factor: biology and therapeutic applications. Int J Biochem Cell Biol. 2007;39:1349–1357. doi: 10.1016/j.biocel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Batran SE, Atmaca A, Schleyer E, Pauligk C, Hosius C, Ehninger G, Jager E. Imatinib mesylate for targeting the platelet-derived growth factor beta receptor in combination with fluorouracil and leucovorin in patients with refractory pancreatic, bile duct, colorectal, or gastric cancer—a dose-escalation phase I trial. Cancer. 2007;109:1897–1904. doi: 10.1002/cncr.22622. [DOI] [PubMed] [Google Scholar]
- 11.Ghaneh P, Kawesha A, Evans JD, Neoptolemos JP. Molecular prognostic markers in pancreatic cancer. J Hepatobiliary Pancreat Surg. 2002;9:1–11. doi: 10.1007/s005340200000. [DOI] [PubMed] [Google Scholar]
- 12.Bos R, van Diest PJ, de Jong JS, van der Groep P, van der Valk P, van der Wall E. Hypoxia-inducible factor-1alpha is associated with angiogenesis, and expression of bFGF, PDGF-BB, and EGFR in invasive breast cancer. Histopathology. 2005;46:31–36. doi: 10.1111/j.1365-2559.2005.02045.x. [DOI] [PubMed] [Google Scholar]
- 13.Giatromanolaki A, Koukourakis MI, Sivridis E, Turley H, Talks K, Pezzella F, Gatter KC, Harris AL. Relation of hypoxia inducible factor 1 alpha and 2 alpha in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. Br J Cancer. 2001;85:881–890. doi: 10.1054/bjoc.2001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sobin LH, Fleming ID. TNM Classification of Malignant Tumors, fifth edition (1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer. 1997;80:1803–1804. doi: 10.1002/(sici)1097-0142(19971101)80:9<1803::aid-cncr16>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 15.Vallbohmer D, Zhang W, Gordon M, Yang DY, Yun J, Press OA, Rhodes KE, Sherrod AE, Iqbal S, Danenberg KD, et al. Molecular determinants of cetuximab efficacy. J Clin Oncol. 2005;23:3536–3544. doi: 10.1200/JCO.2005.09.100. [DOI] [PubMed] [Google Scholar]
- 16.Lord RV, Salonga D, Danenberg KD, Peters JH, DeMeester TR, Park JM, Johansson J, Skinner KA, Chandrasoma P, DeMeester SR, et al. Telomerase reverse transcriptase expression is increased early in the Barrett's metaplasia, dysplasia, adenocarcinoma sequence. J Gastrointest Surg. 2000;4:135–142. doi: 10.1016/s1091-255x(00)80049-9. [DOI] [PubMed] [Google Scholar]
- 17.Kuramochi H, Hayashi K, Uchida K, Miyakura S, Shimizu D, Vallbohmer D, Park S, Danenberg KD, Takasaki K, Danenberg PV. Vascular endothelial growth factor messenger RNA expression level is preserved in liver metastases compared with corresponding primary colorectal cancer. Clin Cancer Res. 2006;12:29–33. doi: 10.1158/1078-0432.CCR-05-1275. [DOI] [PubMed] [Google Scholar]
- 18.Schneider S, Uchida K, Brabender J, Baldus SE, Yochim J, Danenberg KD, Salonga D, Chen P, Tsao-Wei D, Groshen S, et al. Downregulation of TS, DPD, ERCC1, GST-Pi, EGFR, and HER2 gene expression after neoadjuvant three-modality treatment in patients with esophageal cancer. J Am Coll Surg. 2005;200:336–344. doi: 10.1016/j.jamcollsurg.2004.10.035. [DOI] [PubMed] [Google Scholar]
- 19.Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I, et al. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res. 2004;10:3291–3300. doi: 10.1158/1078-0432.CCR-03-0409. [DOI] [PubMed] [Google Scholar]
- 20.Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 21.Johnson MR, Wang K, Smith JB, Heslin MJ, Diasio RB. Quantitation of dihydropyrimidine dehydrogenase expression by real-time reverse transcription polymerase chain reaction. Anal Biochem. 2000;278:175–184. doi: 10.1006/abio.1999.4461. [DOI] [PubMed] [Google Scholar]
- 22.Johnston PG, Lenz HJ, Leichman CG, Danenberg KD, Allegra CJ, Danenberg PV, Leichman L. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumors. Cancer Res. 1995;55:1407–1412. [PubMed] [Google Scholar]
- 23.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krieg M, Haas R, Brauch H, Acker T, Flamme I, Plate KH. Upregulation of hypoxia-inducible factors HIF-1alpha and HIF-2alpha under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function. Oncogene. 2000;19:5435–5443. doi: 10.1038/sj.onc.1203938. [DOI] [PubMed] [Google Scholar]
- 25.Sosman JA. Targeting of the VHL-hypoxia-inducible factor-hypoxia-induced gene pathway for renal cell carcinoma therapy. J Am Soc Nephrol. 2003;14:2695–2702. doi: 10.1097/01.asn.0000091589.10594.66. [DOI] [PubMed] [Google Scholar]
- 26.Worden B, Yang XP, Lee TL, Bagain L, Yeh NT, Cohen JG, Van Waes C, Chen Z. Hepatocyte growth factor/scatter factor differentially regulates expression of proangiogenic factors through Egr-1 in head and neck squamous cell carcinoma. Cancer Res. 2005;65:7071–7080. doi: 10.1158/0008-5472.CAN-04-0989. [DOI] [PubMed] [Google Scholar]