Abstract
The circadian clock of the cyanobacterium Synechococcus elongatus can be reconstituted in vitro by the KaiA, KaiB and KaiC proteins in the presence of ATP. The principal clock component, KaiC, undergoes regular cycles between hyper- and hypo-phosphorylated states with a period of ca. 24 h that is temperature compensated. KaiA enhances KaiC phosphorylation and this enhancement is antagonized by KaiB. Throughout the cycle Kai proteins interact in a dynamic manner to form complexes of different composition. We present a three-dimensional model of the S. elongatus KaiB–KaiC complex based on X-ray crystallography, negative-stain and cryo-electron microscopy, native gel electrophoresis and modelling techniques. We provide experimental evidence that KaiB dimers interact with KaiC from the same side as KaiA and for a conformational rearrangement of the C-terminal regions of KaiC subunits. The enlarged central channel and thus KaiC subunit separation in the C-terminal ring of the hexamer is consistent with KaiC subunit exchange during the dephosphorylation phase. The proposed binding mode of KaiB explains the observation of simultaneous binding of KaiA and KaiB to KaiC, and provides insight into the mechanism of KaiB's antagonism of KaiA.
Keywords: electron microscopy, image reconstruction, protein–protein interactions, X-ray crystallography
Introduction
The circadian clock of the cyanobacterial model organism Synechococcus elongatus can be reconstituted in vitro from the three proteins KaiA, KaiB and KaiC in the presence of ATP and Mg2+ (Nakajima et al, 2005). The in vitro system exhibits the two main characteristics of circadian clocks (Dunlap et al, 2004), a regular rhythm with a period of ca. 24 h and temperature compensation. The KaiC protein constitutes the central cog of the clock and forms a hexamer (Mori et al, 2002; Hayashi et al, 2003; Pattanayek et al, 2004) with auto-kinase and auto-phosphatase activity in vitro (Xu et al, 2003; Nishiwaki et al, 2004). KaiC rhythmically oscillates between hypo- and hyper-phosphorylated forms in vivo (Xu et al, 2003; Nishiwaki et al, 2004), and its phosphorylation status is correlated with the period of the clock (Xu et al, 2003). KaiA enhances KaiC phosphorylation and KaiB exert an effect as a antagonist of KaiA (Iwasaki et al, 2002; Williams et al, 2002, Kitayama et al, 2003; Xu et al, 2003; Nishiwaki et al, 2004). The discoveries that the KaiABC clock ticks in the absence of a transcription–translation oscillatory feedback loop (Tomita et al, 2005), and that it can be reconstituted from three proteins (Nakajima et al, 2005) render this molecular timer a unique target for detailed biochemical and biophysical investigations. In particular, a better understanding of the interactions among the three Kai proteins will be key to gaining insight into the ability of this relatively simple system to sustain a robust oscillation between phosphorylated and dephosphorylated states with a 24-h period.
Three-dimensional (3D) structures for the KaiA (Williams et al, 2002; Garces et al, 2004; Uzumaki et al, 2004; Vakonakis and LiWang, 2004; Ye et al, 2004), KaiB (Garces et al, 2004; Hitomi et al, 2005; Iwase et al, 2005) and KaiC (Pattanayek et al, 2004) proteins or domains thereof (KaiA) have been determined by X-ray crystallography (KaiA, KaiB and KaiC) or NMR (KaiA). S. elongatus KaiC (SeKaiC) takes the shape of a double doughnut with 12 ATP-binding sites between N-terminal KaiCI and C-terminal KaiCII domains. Interactions between KaiC subunits and ATP in the CI half involve hydrogen bonds to the nucleobase portion that are absent in the CII half (Pattanayek et al, 2004), consistent with a tighter binding of ATP molecules in the CI compared with the CII ring (Hayashi et al, 2004a). Crystallographic data allowed identification of phosphorylation sites in the CII half, T432 and S431, and the phosphorylated form of KaiC (P-KaiC) exhibits more extensive subunit interactions relative to the non-phosphorylated form (Xu et al, 2004). Interestingly, the dephosphorylation phase of the KaiC hexamer triggered by KaiB binding is accompanied by KaiC subunit exchange (Kageyama et al, 2006), a process that appears to synchronize the phosphorylation state of KaiC particles and is important for sustaining a high-amplitude oscillation (Mori et al, 2007).
The S. elongatus KaiA (SeKaiA) protein forms a domain-swapped dimer, with the C-terminal dimerization and KaiC-interacting domain adopting a four-helix bundle fold (Ye et al, 2004). The latter finding was confirmed by structures of the C-terminal domain of KaiA from Anabaena sp. PCC7120 (Garces et al, 2004) and KaiA from Thermosynechococcus elongatus BP-1 (ThKaiA) (Uzumaki et al, 2004; Vakonakis et al, 2004). In the NMR structure of a complex between the ThKaiA C-terminal domain and a 30mer C-terminal peptide from ThKaiC two peptides are bound inside grooves above the dimerization interface on opposite faces of the KaiA dimer (Vakonakis and LiWang, 2004). However, this observation did not provide an explanation for KaiA's ability to enhance KaiC phosphorylation. On the basis of single-particle EM reconstruction of the negatively stained T. elongatus BP-1 KaiA–KaiC (ThKaiAC) complex and in vitro studies of complex formation between wild-type and mutant KaiA and KaiC proteins, we recently derived a model for the 3D structure of a KaiAC complex with 1:1 stoichiometry (one KaiA dimer:one KaiC hexamer) (Pattanayek et al, 2006). Tethering the KaiA dimer to the KaiC hexamer through the flexible C-terminal peptide from a KaiC subunit potentially allows for a second more transitory interaction between an apical loop from a KaiA monomer and a KaiCII ATP-binding cleft. The KaiAC model involved extending the KaiC residues in the S-shaped loop (aa 485–497) of the KaiC crystal structure to form a flexible linker between the KaiA dimer and the hexameric barrel of KaiC. The model is consistent with all of the structural information of the component proteins (Egli et al, 2007), the observation that a single KaiA dimer is able to interact with the KaiC hexamer and enhance the latter's auto-kinase activity (Hayashi et al, 2004b), and rapid and repeated action of KaiA on KaiC during the phosphorylation phase for up to 6 h after start of incubation (Kageyama et al, 2006).
Little is known at the moment regarding the mechanism of KaiB-induced dephosphorylation of the KaiC hexamer and the binding mode between KaiB and KaiC. Crystal structures of KaiB from Anabaena sp. PCC7120 (Garces et al, 2004), Synechocystis PCC6803 (Hitomi et al, 2005) and T. elongatus BP-1 (T64C–ThKaiB mutant) (Iwase et al, 2005) revealed a thioredoxin-like fold of the monomer. In the crystals and in solution, KaiB forms a tetramer with a positively charged perimeter, a negatively charged centre and a zipper of aromatic rings important for oligomerization (Hitomi et al, 2005). On the basis of altered rhythm caused by mutant KaiBs in vivo, it was concluded that the tetrameric state is important for proper clock function (Hitomi et al, 2005). However, a recent in vitro analysis of Kai protein complexes during the KaiC phosphorylation cycle relying on gel filtration chromatography found that KaiB appeared to bind to KaiC as a dimer (Kageyama et al, 2006). Interestingly, association and dissociation between KaiB and KaiC was slower compared with the KaiA–KaiC interaction, with only about 5–20% of KaiCs bound to KaiB. KaiA binds to non-phosphorylated KaiC, the phosphorylated form of KaiC (P-KaiC), with or without KaiB bound, as well as to a phosphorylation-site double mutant (S431A/T432A; Kageyama et al, 2006). By contrast, KaiB associates most easily with the P-KaiC form. In addition, KaiA does interfere with KaiC subunit exchange but KaiB promotes it. It is clear that KaiB action does not lead to a dissociation of KaiA from KaiC; rather the data are consistent with the existence of a ternary KaiABC complex during the dephosphorylation phase (Kageyama et al, 2006; Mori et al, 2007).
Using a combination of approaches, including X-ray crystallography, negative-stain and cryo-electron microscopy (EM), gel electrophoresis, mutagenesis and modelling techniques, we have built a 3D model of the S. elongatus KaiB–KaiC (SeKaiBC) complex. The experimental data are consistent with the binding of two KaiB dimers on the C-terminal dome-shaped surface presented by the KaiCII hexamer. The 3D model of this complex together with that of the ThKaiAC complex (Pattanayek et al, 2006) readily suggests mechanisms by which KaiB antagonizes KaiA-stimulated enhancement of KaiC phosphorylation. It also suggests a possible structural explanation for the critical role played by the C-terminal region of KaiB in the KaiC dephosphorylation phase (Iwase et al, 2005). The KaiBC model also explains the observation of simultaneous binding of KaiA and KaiB to KaiC and leads to the idea that all three proteins interact through their C-terminal regions.
Results
X-ray crystal structure of wild-type KaiB from T. elongatus BP-1
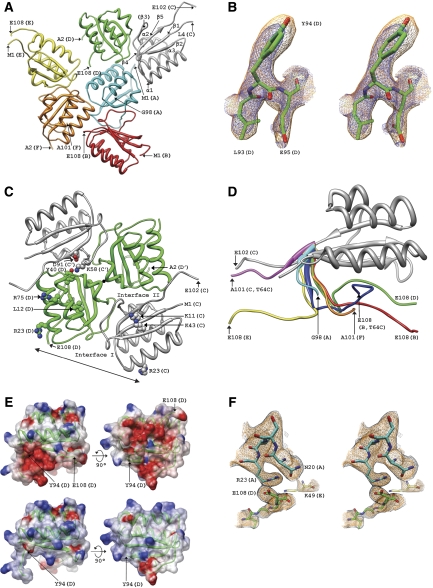
The crystal structure of the full-length wild-type KaiB protein (M1-E108) from T. elongatus BP-1 (ThKaiB) was determined at 2.7 Å by the molecular replacement method, using the previously published structure of the T64C ThKaiB mutant as a model (Iwase et al, 2005). Selected crystal data and refinement statistics are summarized in Table I and stereo diagram illustrating the quality of the final electron density is depicted in Figure 1. Final electron densities around individual N- and C-terminal tails are depicted in Supplementary Figure S1. The structure in space group P21212 reveals six KaiB molecules, termed A–F, per crystallographic asymmetric unit (a.u.; Figure 1A). These form two tetramers of which one is positioned on a crystallographic dyad (Figure 1C). Structures of KaiB tetramers had previously been determined for the proteins from Anabaena (one dimer per a.u.; Garces et al, 2004), Synechocystis (two dimers per a.u.; Hitomi et al, 2005) and a T64C T. elongatus mutant (two dimers per a.u.; Iwase et al, 2005). However, no structures have been reported for the wild-type SeKaiB or ThKaiB (this work) proteins to date. The wild-type ThKaiB structure is the first to include the C-terminal tail, which is either extended or folded under the rest of the KaiB monomer. The core residues (aa 8–94) of the six ThKaiB monomers in the asymmetric unit show close resemblance to each other and to the KaiB structures determined earlier. The r.m.s. deviations for residues 8–94 (87 atom pairs) among independent ThKaiB molecules are 0.52 (A:C), 0.60 (B:C), 0.65 (D:C), 0.56 (E:C) and 0.63 Å (F:C). The r.m.s. deviations between the C monomer of ThKaiB and monomers in the ThKaiB T64C mutant structure are 0.66 and 0.60 Å. The structure confirms the tetramer as the preferred quaternary structural form of KaiB—at least in the absence of other Kai proteins—in a crystalline form. Preference for a tetrameric assembly in the solution state is indicated by dynamic light scattering (Hitomi et al, 2005), gel filtration, chemical crosslinking and analytical ultracentrifugation (Iwase et al, 2005).
Table 1.
Selected crystal and refinement data for ThKaiBa
| Data collection | |
| Space group | P21212 |
| Cell dimensions a, b, c (Å) | 100.13, 191.22, 34.34 |
| Wavelength (Å) | 1.08 |
| Resolution (last shell; Å) | 50–2.78 (2.89–2.78) |
| Unique reflections | 16 993 (1797) |
| Completeness (%) | 98.5 (97.3) |
| R-merge | 0.047 (0.239) |
| I/σ(I) | 30.3 (5.3) |
| Refinement | |
| Working set reflections (F⩾2σF) | 13 727 |
| Test set reflections (F⩾2σF) | 1520 |
| Protein atoms | 4808 |
| Solvent atoms | 63 |
| R-work/R-free (%) | 0.233/0.284 |
| Average B-factors (Å2) | |
| Protein (all residues) | 98 |
| Protein (core residues 8–94) | 93 |
| Solvent | 99 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (deg) | 1.5 |
| Ramachandran analysis (%) | |
| Most favoured | 71.4 |
| Allowed | 25.6 |
| Generously allowed | 3.0 |
| aValues in parentheses refer to the last resolution shell. | |
Figure 1.
Crystal structure of wild-type ThKaiB. (A) The six molecules per crystallographic asymmetric unit illustrating the environments of C-terminal tails in the orthorhombic lattice. The secondary structures, three alpha helices and five beta strands are labelled. Molecules A, B, E and F form a tetramer in a general position and C and D form a second one that is located on a crystallographic dyad. N- and C-terminal residues of the current model are indicated. (B) Stereo diagram illustrating the quality of the omit (2Fo−Fc, 1σ threshold, blue) and Fourier sum electron density (2Fo−Fc, 1σ threshold, orange) in the C-terminal region of the D chain of the ThKaiB crystal structure. (C) The ThKaiB tetramer (molecules C, C′, D and D′) viewed approximately along the crystallographic dyad (indicated by a black dot). Subunits are coloured grey (labelled C and C′) and green (labelled D and D′) and N- and C-terminal residues are labelled. Dimer interfaces (Hitomi et al, 2005) are indicated in Roman numerals and the distance between a pair of Arg residues (R23) across the C/D dimer I interface that matches a similar pair seen in the KaiA dimer (Garces et al, 2004) is shown with a thin line. In addition to the C-terminal tails that contain D and E residues crucial for proper rhythm, side chains of mutants with interesting phenotypes (Ishiura et al, 1998; Hitomi et al, 2005; Iwase et al, 2005) or altered interaction to KaiC (Garces et al, 2004) are highlighted and labelled in one dimer, using the amino-acid numbering previously described for ThKaiB by Iwase et al (2005): K11A (irregular rhythm); L12F (short period, 21 h); R23A (substantially reduced affinity for KaiC); K43A (irregular rhythm); K58A (irregular rhythm); R75F (short period, 21 h); D91G (arrhythmic; mutation disrupts salt bridge between D91 and K58 to orient the latter for a H-bond to Y40 across the dimer II interface). (D) Superimposition of eight ThKaiB molecules based on the crystal structures of the T64C mutant (two molecules, Iwase 2005) and wild-type proteins (six molecules, this work). Residues 8–94 are shown in a single conformation and C-terminal tails beginning with residue 95 are coloured individually, and C-terminal residues are labelled. The colours of individual C-terminal tails match those of molecules in (A). Three of the six C-terminal tails in the crystal structure of wt-ThKaiB were built completely. (E) Electrostatic surface potential of a full-length ThKaiB molecule (molecule D, top panels) and with residues E95–E108 removed (bottom panels) to illustrate the drastic change in the ESP as a result of a lacking C-terminal tail. Electronegative regions are shown in red, neutral regions in white and electropositive regions in blue. (F) Example of the quality of the final electron density around the C terminus of the KaiB D subunit. The stereo diagram shows Fourier sum electron density (2Fo−Fc, 1σ threshold (orange), 0.7σ threshold (grey)) around residue E108 and its neighbouring residues in the crystal lattice. For additional electron density maps and lattice interactions of N- and C-terminal regions see Supplementary Figure S1 and Supplementary Table S1.
The C-terminal tail of KaiB was recently shown to be crucial for the function of the protein. In vivo experiments using the S. elongatus PCC 7942 strain demonstrated that a C-terminal deletion comprising residues E95 to F102 resulted in loss of rhythm (Iwase et al, 2005). Similarly, the E95Q/D98N/D100N/D101N SeKaiB quadruple mutant abolished rhythmicity, indicating that the presence of several acidic residues in the C-terminal tail, which is a hallmark of all KaiB proteins, is absolutely required for proper function (for sequence alignments, see Hitomi et al, 2005; Iwase et al, 2005). The availability of multiple independent molecules in a single structure offers the possibility to examine the conformational flexibility of the C-terminal tail. The superimposition reveals considerable deviations between individual C termini (Figure 1D). In the majority of ThKaiB monomers, the C-terminal tails display a curved conformation and fold back such that they lie near the surface of the thioredoxin-fold adopted by the core residues (Figures 1D and E). The individual conformations are undoubtedly influenced by nearest neighbour interactions in the crystal. However, the structure reveals the wide range of conformations that are accessible to the C-terminal tail. Interestingly, in all cases the negatively charged C-terminal tail forms salt bridges with Arg or Lys residues from the same molecule and/or one or more symmetry mates, similar to those shown in Figure 1F for one of the ThKaiB molecules. A full account of the interactions involving acidic residues of KaiB C-terminal tails is provided in Supplementary Table S1.
Judging from the conformations of the C termini and the electrostatic properties of their environments in the crystal, we conclude that the tail region is malleable and likely to be involved in electrostatic interactions with partner proteins including KaiC. Furthermore, it is noteworthy that the tails of KaiB proteins from thermophilic strains are typically longer and contain more acidic residues than those from mesophiles. For example, the ThKaiB tail (E95–E108) features three Asp and four Glu residues, whereas the SeKaiB tail (G95–F102) features three Asp residues and a single Glu. In T. vulcanus, no fewer than 8 of the C-terminal 12 residues are negatively charged. Calculations of the electrostatic surface potentials for ThKaiB with and without the C-terminal residues manifest fundamentally different values of the potential (Figure 1E). The importance of the C-terminal region of KaiB was demonstrated by a series of deletion mutants where SeKaiB was truncated one residue at a time starting with SeKaiB:1–100 and ending with SeKaiB:1–94. When these C-terminally truncated proteins were tested in rhythm assays in S. elongatus kaiB-null host cells, the results indicated a strongly weakened and destabilized rhythm for the SeKaiB:1–94 protein (Iwase et al, 2005).
Gel-shift-binding assays with full-length SeKaiC, SeKaiB and ThKaiB proteins
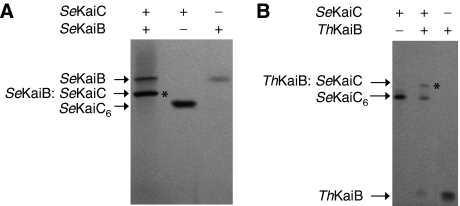
We used native polyacrylamide gel electrophoresis (PAGE) to assay binding between SeKaiB and ThKaiB with SeKaiC. Both Se proteins were expressed in Escherichia coli using constructs with N-terminal GST tags that were cleaved off prior to PAGE, following previously described procedures with slight modifications (Nishiwaki et al, 2004; Mori et al, 2007). The ThKaiB protein used in the binding assay is identical to the one that served the crystal structure determination and features an N-terminal (His)6-tag. The appearance of bandshift gels did not fundamentally change, irrespective of whether SeKaiC proteins with N- or C-terminal (His)6-tags (data not shown) or the tag-free (GST-off) version were used.
Our data demonstrate that both SeKaiB and ThKaiB are able to bind to SeKaiC in the presence of ATP (Figures 2A and B, respectively). Previous observations by our laboratories (Mori et al, 2007) and others (Kageyama et al, 2006) demonstrated that, unlike KaiA, KaiB binds preferably to the P-KaiC hexamer. Binding of ThKaiB to the SeKaiC hexamer is consistent with the observation that ThKaiB partially complements the null KaiB mutant of S. elongatus in vivo, generating a faint rhythm (Iwase et al, 2005). Similarly, KaiB from Anabaena partially complemented the null mutant, whereas KaiB from Synechocystis sp. strain PCC 6803 was able to fully complement it (Iwase et al, 2005).
Figure 2.
Native PAGE assays for KaiBC complex formation. Binding between (A) full-length SeKaiB and SeKaiC proteins and (B) ThKaiB and SeKaiC proteins. A ‘+' indicates presence of and a ‘−' indicates absence of protein (KaiB dimer or SeKaiC hexamer ([SeKaiC]6)). The bands corresponding to KaiBC complexes are indicated with asterisks. Note the considerably different shifts of the SeKaiB and ThKaiB proteins, which are likely due to deviating net charges. ThKaiB has a higher number of acidic residues in the C-terminal tail. The protein amounts used were 30 pmol (ThKaiB and SeKaiC) and 22 pmol (SeKaiB). The buffer for all proteins was 20 mM Tris (pH 7.8), 150 mM NaCl and 1 mM DTT. Only SeKaiC and ThKaiC were supplemented with 5 mM MgCl2 and 1 mM ATP.
Binding of KaiB to KaiC fragments
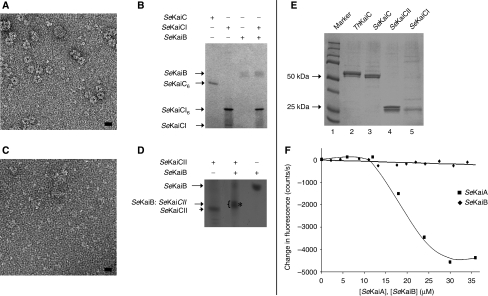
The crystal structure of SeKaiC revealed two serially arranged domains (N-terminal KaiCI and C-terminal KaiCII), connected by a 15-amino-acid linker (Pattanayek et al, 2004). Upon oligomerization, these give rise to a double-doughnut-shaped hexamer (featuring KaiCI and KaiCII halves) with a constricted waist in the region of the linkers. KaiA was found to bind to flexible C-terminal peptides (Vakonakis and LiWang, 2004) and may bind more transiently to the KaiCII dome with its six ATP-binding clefts (Pattanayek et al, 2006). However, it is currently not known whether KaiB binds to the KaiCI or the KaiCII half. To examine whether KaiB binds to KaiCI or KaiCII, we expressed the two KaiC domains from S. elongatus separately as GST fusion proteins (CI, residues 1–250, and CII, residues 253–519). The goal was to assay potential formation of a hexameric ring from each of the two fragments to simulate the two halves of the KaiC hexamer. Following affinity chromatography and cleavage of the GST portion, we used native PAGE and negative-stain EM to assay hexamerization of SeKaiCI and SeKaiCII, and tested KaiB binding to either individually expressed KaiC domain. KaiCI in the presence of ATP forms rings by negative-stain EM (Figure 3A). These rings are of the same diameter (100 Å) as observed previously for the hexamer of full-length KaiC (Pattanayek et al, 2006). Native PAGE indicates that KaiCI fails to bind SeKaiB (Figure 3B) or ThKaiB (Supplementary Figure S2A).
Figure 3.
Oligomerization of KaiC CI and CII fragments and KaiB-binding assays. (A) A negative-stain electron micrograph of the SeKaiCI fragment (aa 1–250) in the presence of ATP. Note the presence of rings ∼100 Å in diameter. The scale bar represents 100 Å. (B) Native PAGE showing that the SeKaiCI fragment oligomerizes in the presence of ATP, but does not bind to SeKaiB. A ‘+' indicates the presence of and a ‘−' indicates the absence of protein; the protein amounts used were 30 pmol (SeKaiC and SeKaiCI) and 24 pmol (SeKaiB). (C) A negative-stain electron micrograph of the SeKaiCII fragment (aa 253–519) in the presence of ATP. Note the absence of hexameric rings; the scale bar represents 100 Å. (D) Native PAGE showing that the SeKaiCII binds to SeKaiB. A ‘+' indicates the presence of and a ‘−' indicates the absence of protein; 24 pmol SeKaiB and 15 pmol SeKaiCII were used. The band corresponding to a complex between SeKaiB and the SeKaiCII fragment is indicated with an asterisk. (E) SDS–PAGE assay of phosphorylation status of KaiC proteins and SeKaiCI and SeKaiCII. The double bands for ThKaiC, SeKaiC and SeKaiCII indicate that these proteins have both a phosphorylated and an unphosphorylated states. In contrast, the single band for SeKaiCI (lane 5) indicates that it is not phosphorylated. (F) Fluorescence spectra for mixtures of SeKaiA or SeKaiB and a 25mer peptide corresponding to the C-terminal end of SeKaiCII (P494–S519) and labelled with Trp at its C terminus. The concentration of the peptide in the assays was 85 μM.
In contrast, the KaiCII fragment did not hexamerize under the same conditions (Figure 3C). This replicates in a second cyanobacterial species the earlier finding by Ishiura et al. who studied hexamer formation with the CI and CII halves from ThKaiC and noted the absence of hexamers for CII. Instead, gel filtration chromatography indicated the presence of either tetramers or pentamers (Hayashi et al, 2006). EM indicates that CII forms irregular oligomers that are smaller than hexamers (Figure 3C). Despite the irregular CII oligomer size, the native PAGE assay reveals that KaiB binds to KaiCII (Figure 3D, SeKaiB, and Supplementary Figure S2B, ThKaiB). It is noteworthy that unlike KaiCI, KaiCII is phosphorylated (Figure 3E) and the protein can be dephosphorylated with λ-phosphatase (Supplementary Figure S3). These data provide evidence that similarly to KaiA, KaiB interacts with the C-terminal ring of the KaiC homo-hexamer.
The C-terminal domains of the KaiA dimer bind individual KaiC C-terminal peptides (Vakonakis and LiWang, 2004) and enhance phosphorylation of the hexamer through rapid and repeated action on subunits (Kageyama et al, 2006). To examine whether KaiB also binds the C-terminal KaiC peptide, we tagged a 25mer C-terminal SeKaiC peptide with Trp and measured fluorescence for mixtures of SeKaiB (which lacks Trp) and the KaiC peptide at various ratios (Figure 3F). This fluorescence assay indicates that SeKaiB does not exhibit any affinity for the isolated SeKaiC C-terminal region (residues 495–519). In contrast, the fluorescence assay with SeKaiA and the Trp-labelled SeKaiC peptide confirms KaiA binding to the KaiC C-terminal region (Figure 3F).
Negative-stain and cryo-EM analysis of the SeKaiBC complex
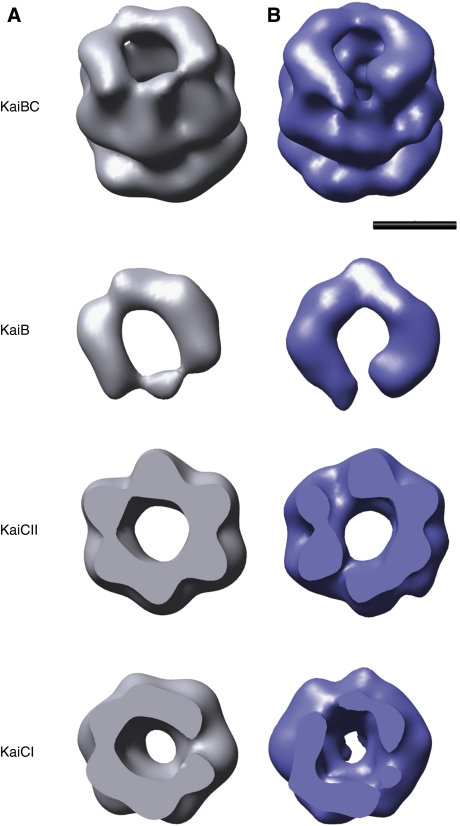
We have previously used EM to analyse the interactions among the KaiABC proteins during the in vitro circadian cycle (Mori et al, 2007). To help interpret EM images of the observed complexes, we collected 4130 particle images of the negatively stained SeKaiBC complex (Mori et al, 2007). To this we added 4590 particles images of SeKaiBC selected by meta-class sorting from SeKaiABC cycling reactions (Mori et al, 2007), for a total dataset of 8720 particle images. Here, we present a refined 3D structure at ∼19-Å resolution based on these data (Figure 4A). The structure reveals a double-ringed hexameric barrel for KaiC with an additional third layer of density at one end. Assignment of CII as the KaiB contact domain is consistent with our KaiB native PAGE-binding assays (Figure 3 and Supplementary Figure S2). As the reaction mixtures were not purified to defined stoichiometric complexes, the KaiBC structure could represent the average of a heterogeneous population with zero, one, two or three KaiB dimers bound per KaiC hexamer. Therefore, we used the Multirefine program in EMAN to sort the dataset based on five starting model structures (Supplementary Figure S4). These five structures included KaiC alone, as well as hypothetical representations of KaiC with one KaiB dimer, KaiC with two KaiB dimers bound in two different manners, and KaiC with three KaiB dimers. One of the models with two KaiB dimers had the dimers positioned parallel to each other on either side of the central channel, and the other had the two dimers positioned close to each other in a ‘V' formation. Multirefine sorted the particle images into five sub-datasets ranging from 1400 to 2000 particle images each. These five sub-datasets were then separately refined with a Gaussian sphere as the starting model. The sub-dataset that was grouped with the model containing two parallel KaiB dimers refined to the highest resolution (19 Å) as assessed by Fourier shell correlation (FSC) and resulted in a final structure most closely resembling its starting model (Figure 4B). This KaiBC structure is based on 22% of the data and is quite similar to the average KaiBC structure with three density layers.
Figure 4.
Negative-stain EM structures of the SeKaiBC complex. (A) Three-dimensional reconstruction of the KaiBC complex based on 7415 particle images (selected from the total dataset of 8720) processed without imposed symmetry. The full reconstruction (top) and its three density layers assigned to KaiB, the ring of KaiC CII domains and the ring of KaiC CI domains are shown. (B) Three-dimensional reconstruction of the KaiBC complex based on 1106 particle images (selected from a sub-dataset of 1880 grouped with the model containing two parallel KaiB dimers) processed without imposed symmetry. Both structures have a resolution of 19 Å and are shown filtered to 19 Å resolution. The isosurface values are set to enclose 100% of the expected volume for a KaiC hexamer and two KaiB dimers. The scale bar represents 50 Å.
Comparison of KaiBC class sum images of the full dataset with two-dimensional projections of the structure serves as a check of the veracity of the structure and also indicates that the structure captures molecular features discernible in the class sum images (Supplementary Figure S5). In particular, the class sum images representing side views of the KaiBC complex clearly show a third layer of density that is also evident in projections of the structure. We also repeated the EM analysis with unstained cryo-preserved samples and collected a cryo-EM dataset of 10 162 particle images. Unfortunately, this cryo-EM dataset exhibited a preferential orientation with most of the particles aligned with the central channel of KaiC along the direction of the electron beam. Refinement of the cryo-EM dataset produced a ∼24-Å resolution structure. Because of lower image contrast, heterogeneity and preferential orientation of the particles, the cryo-EM structure displayed fewer features than were apparent in the negative-stain structures. Nevertheless, both the cryo-EM class sum images representing side views and the cryo-EM reconstruction confirm the presence of a third layer of density in the KaiBC complex (Supplementary Figure S5).
Extension of the KaiC S-shaped loops enlarges the central channel
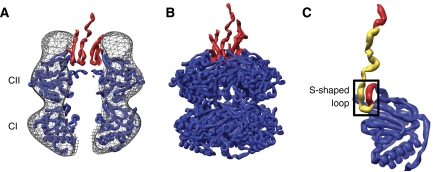
The KaiC crystal structure reveals that each subunit is composed of two tandemly oriented domains, CI and CII, with a flexible C-terminal peptide tail protruding from the CII domain. A portion of the KaiC C-terminal region forms an S-shaped loop (aa 485–497) that inserts into the CII end of the central channel (Figure 5). The C-terminal peptide and S-shaped loop include residues that have been shown to bind KaiA (Vakonakis and LiWang, 2004). We have noted previously that extension of the KaiC S-shaped loops is likely required for the KaiAC interaction (Pattanayek et al, 2006). In the KaiBC negative-stain EM structures, the CII ring has a larger central channel diameter than the CI ring (Figure 4). This observation can be explained if the S-shaped loops are extended from the central channel of the CII ring in the KaiBC complex. Aligning the CII domain ring of the KaiC crystal structure with the middle density layer in the KaiBC EM structures shows that no density is present for the S-shaped loops within the central channel. Thus, we propose that the S-shaped loops are extended in the KaiBC complex. The KaiC C-terminal tails, including the extended S-shaped loops, are likely to be flexible and thus density for them would not be expected in the EM reconstructions.
Figure 5.
Analysis of the KaiC portion of the KaiBC negative-stain EM density. (A) The SeKaiC crystal structure (PDB ID 2GBL) (Pattanayek et al, 2006) fit within the average KaiBC structure from Figure 4A. The fit was optimized for the ring of KaiC CII domains. Only a slab of density ∼20-Å thick around the central channel is shown. The KaiC C-terminal tails and the S-shaped loops, aa 485–519, are shown in red. (B) The full SeKaiC crystal structure with the C-terminal regions (aa 485–519) in red. Note that the C-terminal tails are flexible and the crystal structure includes complete C-terminal tail coordinates for only two of the six subunits. (C) Enlarged view of the CII domain and C-terminal region of a subunit (chain A) from the KaiC crystal structure. The S-shaped loop (aa 485–497) is indicated by a box. The residues implicated by NMR (Vakonakis and LiWang, 2004) to bind KaiA (aa 490–513) are shown in yellow.
The KaiC crystal structure indicates that the S-shaped loops participate in a hydrogen bond network. In particular, T495 makes an interaction with E487 in the S-shaped loop of another subunit. We speculate that extension of the S-shaped loops disrupts the hydrogen bond network, weakens the interaction between CII domains in the context of the hexamer and may lead to an increased separation of the CII domains relative to their orientations in the KaiC crystal structure.
A model of the KaiBC complex
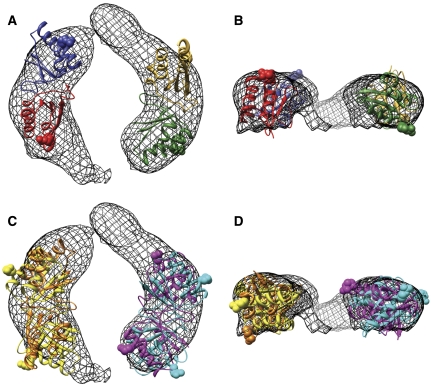
We attribute the third layer of density observed in the SeKaiBC negative stain and cryo-EM structures to KaiB. The shape and volume of this density suggest that the primary binding mode of KaiB is as a dimer (Figure 6), rather than as a compact tetramer. Recent data from the Kondo lab based on size exclusion chromatography also indicate dimeric KaiB binding to KaiC hexamer (Kageyama et al, 2006). Sorting of the negative-stain EM dataset against five hypothetical models indicates that the most uniform sub-population of the dataset has two KaiB dimers bound flanking the central channel of the KaiC hexamer without occluding the channel. Fitting of the wild-type ThKaiB dimer coordinates (chains C and D, core residues) into this density indicates that two dimers fit well (Figure 6). However, the rotation of the dimer about the long axis is ambiguous, and thus the surface of KaiB that interacts with KaiC is not defined by this EM structure. We note that the Arg pair, which was postulated by Garces et al (2004) to bind to KaiC, may be located at the interface with KaiC or alternatively may be directed away from KaiC. The C-terminal tails of KaiB (aa 95–108) are long enough that regardless of the orientation of the KaiB dimer with respect to KaiC at least one of the two flexible tails should be able to interact with the CII ring of KaiC, but not with the CI ring. It is also possible that extended C-terminal tails of KaiB might inhibit binding of two KaiB dimers next to each other on the CII surface of KaiC.
Figure 6.
Fitting of KaiB dimers within the KaiBC EM density. (A) Two KaiB dimers (core residues, aa 8–94) from the wild-type ThKaiB crystal structure are shown within the third layer of density (mesh) of the KaiBC structure from Figure 4B. This KaiBC structure is based on the sub-dataset grouped with the model containing two parallel KaiB dimers. The top-scoring fit for a KaiB dimer on either side of the central channel is shown (Laplacian correlation values of 0.31 for both). The arginine pair (R23), which has been suggested to be involved in the interaction with KaiC (Garces et al, 2004), is shown with atoms in a sphere representation. (B) A 90°-rotated view. (C) Two additional high-scoring fits (Laplacian correlation values of 0.28–0.30) for each KaiB dimer position within the same EM density. (D) A 90°-rotated view. Note that the orientation of the arginine pair (atom spheres) with respect to KaiC is not well defined.
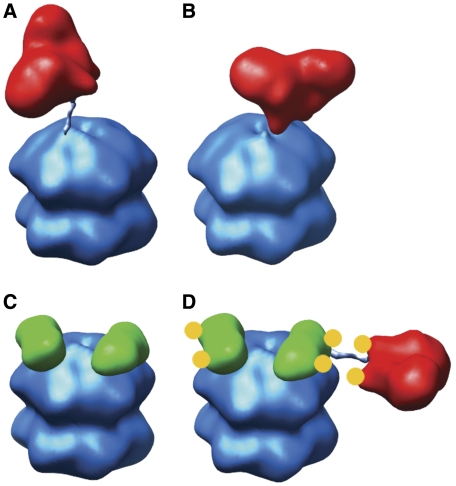
This model of the KaiBC complex suggests a way in which KaiB may serve to antagonize the effect of KaiA. KaiB may exert an effect as a shield and prevent an ‘engaged' binding mode between KaiA and KaiC (Figure 7), which we have previously postulated between the KaiA apical loop and the KaiC ATP-binding cleft (Pattanayek et al, 2006). As a shield KaiB would not disrupt binding of the KaiA dimer to a C-terminal KaiC peptide, which is still largely accessible in our KaiBC model, but rather block a transient interaction between KaiA and the ATP-binding cleft of the KaiCII C-terminal dome region. Furthermore, the stable interaction of KaiB with KaiC may create a sub-population of clock complexes that are competent to bind KaiA, but incompetent to autophosphorylate. A high proportion of autophosphorylation-incompetent KaiBC complexes could exert an effect as a sink for KaiA. This would further accelerate the dephosphorylation of the population by starving the free KaiC hexamers of KaiA in the cycling reaction.
Figure 7.
Models of KaiAC, KaiBC and KaiABC complexes. (A) The KaiAC ‘tethered' model from Pattanayek et al (2006). Note that a KaiA dimer (red) is shown bound to an extended C-terminal tail of KaiC (blue). (B) The KaiAC ‘engaged' model from Pattanayek et al (2006) with the apical loops of a KaiA dimer shown interacting with the surface of the KaiC hexameric barrel. (C) A KaiBC model based on the negative-stain EM structure of KaiBC shown in Figure 4B with two KaiB dimers (green) shown bound to the CII side of the KaiC hexameric barrel. Only the KaiB core residues are represented, without the N- and C-terminal tails. (D) A KaiABC model with KaiA and KaiB dimers in orientations resembling those in the class IV KaiABC particle images from negative-stain EM (see Figure 1D in Mori et al, 2007). The arginine pairs (R249 in SeKaiA and R23 in ThKaiB), which have been suggested to compete for a common KaiC-binding site (Garces et al, 2004), are indicated with yellow circles. In our KaiABC model, the arginine pairs are oriented such that the KaiB arginines could repel KaiA away from KaiC by electrostatic repulsion. All models are based on crystal structures of the component proteins, SeKaiA (PDB 1R8J) (Ye et al, 2004), wild-type ThKaiB (this work) and SeKaiC (PDB ID 2GBL) (Pattanayek et al, 2006), which are shown in 25-Å filtered representations.
Discussion
We present the crystal structure of the native form of the T. elongatus BP-1 KaiB protein. Previously, the structure of a mutant form of ThKaiB (T64C) was determined (Iwase et al, 2005). Both the native and mutant structures show essentially the same structure for the core of the molecule (aa 8–94). The native KaiB structure, however, provides complete atomic models for C-terminal tails (aa 95–108) of multiple monomers within the asymmetric unit (Figure 1). Superposition of molecules from both the native and mutant ThKaiB asymmetric units reveals a high degree of flexibility for the C-terminal tail. The KaiB C-terminal tail has been shown to be important for sustained KaiC oscillation in rhythm assays (Iwase et al, 2005). We propose a way in which KaiB's action on KaiC may involve the flexible and negatively charged C-terminal tail of KaiB.
Native PAGE analysis indicates that both SeKaiB and ThKaiB bind to SeKaiC in the presence of ATP (Figure 2). We show by native PAGE and EM that an expressed KaiC CI fragment (aa 1–250) hexamerizes in the presence of ATP; however, no binding of the KaiC CI fragment to KaiB was detected. A fluorescence assay with the expressed C-terminal tail of KaiC also indicates no binding with KaiB. However, the CII half expressed separately exhibited phosphorylation and interacted with KaiB (Figure 3). These results demonstrate that KaiB binds to the CII side of the KaiC hexameric barrel and forms the third layer of density observed in our EM structure of the SeKaiBC complex (Figure 4).
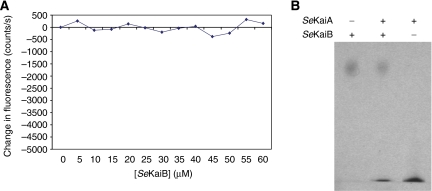
We have noted in the KaiBC EM structures that the central channel of the CII ring of the KaiC double doughnut appears larger than that of the CI ring. This can be explained if the S-shaped loops of KaiC are extended. In the KaiC crystal structure, these loops insert into the CII end of the central channel and form a hydrogen bond network. As the CII domain lacks the ability to hexamerize independently similar to the CI domain, extension of the S-shaped loops may weaken the interaction between CII domains in the context of the hexamer and allow for separation or conformational change of the CII domains relative to their orientations in the KaiC crystal structure. As phosphorylation of KaiC subunits occurs through intersubunit interactions (Pattanayek et al, 2004; Xu et al, 2004; Hayashi et al, 2006), weakening of the interaction between CII domains may reduce the phosphotransfer capability of KaiC. It is also possible that CII domain separation may promote subunit exchange, which in turn may promote net dephosphorylation of KaiC (Kageyama et al, 2006; Mori et al, 2007). Our modelling of the KaiBC complex also suggests that perhaps the negatively charged KaiB C-terminal tails interact with the ATP-binding cleft between KaiC CII domains and disrupt ATP binding. However, these possibilities would all suggest that KaiB would have a positive effect on net KaiC dephosphorylation, which has not been observed. Another possible mechanism for the action of KaiB suggested by the KaiBC EM structure is that bound KaiB may sterically hinder KaiA from interacting with the ATP-binding regions between KaiC CII subunits (Pattanayek et al, 2006). This obstruction model is supported by the fact that KaiA and KaiB do not exhibit any affinity for each other judging from fluorescence (Figure 8A) or native PAGE (Figure 8B) of SeKaiA/SeKaiB mixtures.
Figure 8.
Probing the potential binding between SeKaiA and SeKaiB. (A) Fluorescence and (B) native PAGE-binding assays with full-length SeKaiA and SeKaiB proteins. For gel electrophoresis the protein amounts used were 23 pmol (SeKaiA) and 20 pmol (SeKaiB), and the buffer was 20 mM Tris (pH 7.8), 150 mM NaCl and 1 mM DTT. The concentration of KaiA protein in the fluorescence experiment was 50 μM.
Comparison of our models of the KaiAC (Pattanayek et al, 2006) and KaiBC complexes suggests that bound KaiB dimers would block the proposed transient interaction of KaiA with the KaiCII dome surface in our KaiAC engaged model (Figure 7). However, both KaiA and KaiB may bind to the KaiC hexamer when the KaiA dimer is bound to an extended C-terminal tail of KaiC and is not interacting with the KaiC hexameric barrel, as in our KaiAC-tethered model. In the KaiAC-tethered model, the KaiA dimer is ∼35 Å above the hexameric barrel of KaiC. Here, we propose a model for KaiABC with two KaiB dimers on the CII side of KaiC blocking KaiA from interacting with the CII dome surface of KaiC and with one (or more) KaiA dimers bound to extended C-terminal tails of KaiC and oriented to the side of the complex to resemble class IV KaiABC particle images from negative-stain EM (Mori et al, 2007). We note that the molecular weight of a complex with one KaiA dimer, two KaiB dimers, and one KaiC hexamer is consistent with the size of KaiABC protein assemblies reported by Kageyama et al (2003).
KaiB is known to bind to the hyper-phosphorylated form of KaiC. KaiABC ternary complexes are observed by blue native gels, size exclusion chromatography and negative stain EM predominantly during the dephosphorylation phase of the clock cycle (Kageyama et al, 2006; Mori et al, 2007). It is an open question exactly what triggers binding of KaiB to KaiC but it seems that KaiA is not necessary, as we and others have formed KaiBC complexes in the absence of KaiA. Perhaps the initiation of KaiB binding to KaiC is triggered by a conformational change and/or an electrostatic change in the KaiC dome region on the CII side of KaiC that is unique to a hyper-phosphorylated state. Assuming that KaiA and KaiB bind to distinct sites on KaiC as our KaiBC model suggests, then the KaiB-binding modes in the BC and ABC complexes might be indistinguishable (Figure 7). Pai and associates pointed out arginine pairs in the structures of the KaiA and KaiB dimers (Figures 1C and 6) with similar spatial separations and thought it possible that KaiA and KaiB might compete for a common binding site on KaiC (Garces et al, 2004). The Arg pair of KaiA may be involved in a transient interaction with KaiC (see Figure 9 in Pattanayek et al, 2006), and it is possible that the Arg pair of KaiB is involved in the interaction with KaiC. However, our data suggest an alternative possible role for these Arg pairs. It is possible that the Arg pair of KaiB might be oriented away from KaiC and thus could repel KaiA away from the hexameric barrel of KaiC by electrostatic repulsion with the similarly spaced Arg pair on KaiA (Figure 7D).
Our model of the 3D structure of the SeKaiBC complex adds significantly to the understanding of the structure and function of the cyanobacterial KaiABC circadian clock. In this model of the KaiBC complex, KaiB binds KaiC at the CII side, and the flexible C-terminal tail of KaiB may interact with the ATP-binding clefts between adjacent KaiC subunits. In addition to earlier insights on the enhancement of KaiC phosphorylation by KaiA (Vakonakis and LiWang, 2004; Pattanayek et al, 2006), the present work now suggests plausible models for the antagonistic effects exerted by KaiB to counteract the action of KaiA. A more detailed understanding of the interactions between KaiB and KaiC will have to await higher resolution structures revealing the binding interface between the two proteins at an atomic level.
The KaiBC structural model, together with structural information on the KaiAC complex (Vakonakis and LiWang, 2004; Pattanayek et al, 2006), leads to the idea that the cyanobacterial circadian clock is driven by the interplay between the C-terminal regions of these three proteins. The C-terminal (CII) half of the KaiC has phosphotransferase activity and harbours serine and threonine residues whose phosphorylation is enhanced by KaiA binding. The C-terminal domain of KaiA binds to the extended C-terminal tail of a KaiC subunit and may also bind transiently to the C-terminal half of the KaiC hexameric barrel. Here, we propose that the negatively charged C-terminal tail of KaiB may interact with the ATP-binding site between KaiCII subunits, displace ATP and launch KaiC subunit exchange that is required for sustaining a stable high-amplitude oscillation of the clock.
Materials and methods
Expression and purification of wild-type KaiB and KaiC proteins and KaiCI and KaiCII
SeKaiC and ThKaiC proteins with C- or N-terminal (His)6-tags were produced in E. coli as previously described (Mori et al, 2002; Pattanayek et al, 2004) and purified by affinity and gel filtration chromatography. The GST–SeKaiA, GST–SeKaiB and GST–SeKaiC fusion proteins were expressed as described in Nishiwaki et al (2004) and Mori et al (2007). The (His)6–ThKaiB protein was produced following the same protocol as that used for SeKaiC–(His)6 (Pattanayek et al, 2004). Briefly, (His)6–ThKaiB was overexpressed in E. coli cells (BL21) and purified using metal affinity chromatography (Ni-NTA resin; Sigma), followed by gel filtration chromatography on a Superdex200 HR10/30 column (Amersham Biosciences). The protein was eluted as a single peak and the elution buffer was 50 mM HCl (pH 7.8), 100 mM NaCl, 1 mM EDTA and 1 mM DTT. The same methods were used for expression and purification of the SeKaiCI and SeKaiCII halves. The above buffer was supplemented with 5 mM MgCl2 and 1 mM ATP for the purification of the SeKaiC half- and full-length proteins. The purity of the proteins was estimated by SDS–PAGE using mini-gels (Bio-Rad System). All proteins were analysed by tryptic digestion followed by MALDI-TOF mass spectrometry.
Peptides
The SeKaiC C-terminal peptide with sequence n-TRITVDEKSE LSRIVRGVQE KGPESW-c was purchased from EZBiolab (Westfield, IN) and purity was greater than >95% following HPLC purification. A single Trp residue was added at the C-terminal end of the sequence for fluorescence spectroscopy.
X-ray crystallography
Crystals for the ThKaiB protein were grown by mixing equal volumes of protein (concentration 3 mg/ml in 20 mM Tris pH 7.8, 100 mM NaCl, and 1 mM BME) and reservoir solutions (10% PEG 3350, 10% DMSO and 0.1 M acetate buffer pH 5.0) at 293 K, using the hanging drop vapour-diffusion method. Micro crystals grew in a week and diffraction-quality crystals of rectangular shape grew in 2–3 weeks.
Diffraction data were collected on the DND-CAT beamline sector 5 of the APS (Argonne, IL) and processed with HKL2000 (Otwinowski and Minor, 1997). The crystals belong to orthorhombic space group P21212, with unit-cell parameters a=100.13, b=191.22 and c=34.34 Å, and diffract to 2.7 Å. The asymmetric unit of ThKaiB crystals contains six monomers or three dimers (Figure 1D; monomers are labelled A–F).
The structure was determined by the molecular replacement method using the program PHASER (CCP4, 1994) and the crystal structure of the T64C–ThKaiB mutant with PDB-ID code 1VGL (Iwase et al, 2005) as a search model. The programs TURBO-FRODO (Cambillau and Roussel, 1997) or O (Jones, 2004) were used for map display and model building. All refinements were carried out with the program CNS (Brünger et al, 1998) using omit maps to assess the correctness of chain tracing. The final R-work and R-free are 0.234 and 0.284, respectively, using all F⩾2σ(F) data to 2.78-Å resolution and setting aside 8.7% randomly chosen reflections for calculating the R-free. Selected crystal data and refinement parameters are listed in Table I. The r.m.s. deviations from standard values for bond lengths and angles are 0.008 Å and 1.5°, respectively. In the Ramachandran plot, 71.4% of residues fall into the most favoured regions, 25.6% into allowed regions and 3.0% into generously allowed regions.
Gel electrophoresis
Native PAGE was carried out on a PhastSystem (Pharmacia LKB) using PhastGel Gradient 4–15 or 8–25% gels and PhastGel Native Buffer Strips (Amersham Biosciences). The gels were stained with 0.1% PhastGel Blue R solution in 10% acetic acid and 30% methanol and destained with 30% methanol and 10% acetic acid. SDS–PAGE was performed using the mini-gel system from Bio-Rad and ready-made 10% Tris gels (Bio-Rad Laboratories). Bio-Safe Coomassie G250 solution was used for staining.
Fluorescence spectroscopy
Fluorescence spectra for mixtures between C-terminal SeKaiCII peptide and either full-length SeKaiA or SeKaiB and for mixtures between the SeKaiA and SeKaiB proteins were recorded using a Fluorescence Spectrophotometer (Fluorolog-3; Jobin Yvon-Spex Instruments SA Inc., New Jersey, NY). Steady-state techniques were employed to monitor changes in the fluorescence intensity of KaiC peptide upon addition of KaiB or KaiA (C peptide only) or of KaiA upon addition of KaiB. Titrations were performed while monitoring the change of the tryptophan fluorescence of KaiC peptide or KaiA (λex=285 nm; λem=340 nm). The temperature of the cell compartment was kept at 25±0.1°C, using a circulating water bath. Results are based on two independent measurements and data were corrected for dilution and blank solutions. The buffer was 20 mM Mops (pH 7.8) and 100 mM NaCl was used as blank solution.
Electron microscopy and image processing
Complexes of SeKaiB and SeKaiC were formed for electron microscopy studies as previously described (Mori et al, 2007). Particle images (4130) were obtained from negatively stained KaiB/KaiC mixtures and 10 162 particle images were obtained from cryo-preserved KaiB/KaiC mixtures. To augment the particle number in the negative-stain dataset, 4590 particle images of the KaiB–KaiC complex from cycling KaiA, KaiB and KaiC reactions were added to the dataset. These particles were partitioned from a larger 160 000 particle dataset by the metaclassification procedure described in Mori et al (2007). This resulted in a total negative-stain EM dataset of 8720 particle images. Electron micrographs of negatively stained (0.75% uranyl formate) and cryo-preserved samples were collected on an FEI Tecnai 12 (120 kV, LaB6 filament) electron microscope equipped with a Gatan UltraScan 1000 (2K × 2K) pixel CCD camera and a Gatan 626 cryo-holder. The nominal magnification was × 67 000, which results in an actual magnification of × 98 000 at the level of the CCD camera. The defocus range for the negative-stain dataset was −0.7 to −1.5 μm and for the cryo-EM dataset −1.0 to −2.0 μm. The Boxer routine in EMAN (Ludtke et al, 1999) was used for selection of individual particle images.
Both the negative-stain and cryo-EM datasets were processed without imposed symmetry. The IMAGIC software package (van Heel et al, 1996) was used for image classification and angular reconstitution to produce an initial 3D reconstruction based on a subset of the negative-stain EM dataset. This reconstruction was used as a starting model for refinement with the EMAN program Refine. Refinement proceeded until convergence was reached for the independently processed negative-stain and cryo-EM datasets. A 3D reconstruction was generated from 7415 negative-stain EM particle images. A separate reconstruction was generated from 8140 cryo-EM particle images. Not all of the particle images in the two datasets were included in these reconstructions as a consequence of the EMAN classkeep parameter, which was set at 0.7. The resolutions of the negative-stain and cryo-EM structures are 19 and 24 Å, respectively, at the FSC 0.5 thresholds as assessed by the EMAN Eotest program.
The negative-stain EM dataset was subjected to a further sorting and refinement procedure beginning with the EMAN program Multirefine. Five starting models were created from the SeKaiC (PDB 2GBL) and wild-type ThKaiB crystal structures using UCSF Chimera (Pettersen et al, 2004) and converted to 20-Å filtered density representations using Pdb2mrc in EMAN. These five models corresponded to a KaiC hexamer (C6), a KaiC hexamer with a single dimer of KaiB bound at the CII side of KaiC (C6B2), a KaiC hexamer with two dimers of KaiB bound at the CII side of KaiC and in a parallel configuration (C6B4P), a KaiC hexamer with two dimers of KaiB bound at the CII side of KaiC and in a V-shaped configuration (C6B4V), and a KaiC hexamer with three dimers of KaiB bound at the CII side of KaiC (C6B6) (Supplementary Figure S4).
Partitioning of the particle images occurred over five iterative rounds of Multirefine and resulted in five sub-datasets (C6=1976 particles, C6B2=1587 particles, C6B4P=1880 particles, C6B4V=1471 particles and C6B6=1806 particles). These sub-datasets were further refined with five rounds of Refine (with parameters classiter=5, classkeep=0.7, sym=c1, hard=25 and phasecls) yielding structures with resolutions of C6=22 Å, C6B2=23 Å, C6B4P=21 Å, C6B4V=24 Å and C6B6=23 Å at the FSC 0.5 thresholds. To remove any potential model bias from the structures, the sub-datasets were each refined for an additional 25 rounds starting with a Gaussian sphere model (radius=46 Å) created with Makeinitialmodel.py in EMAN. This produced final structures with resolutions of C6=20 Å, C6B2=22 Å, C6B4P=19 Å, C6B4V=22 Å and C6B6=20 Å at the FSC 0.5 thresholds.
The quantitative docking tool CoLoRes in the Situs package (Chacon and Wriggers, 2002) was used with the Laplacian correlation function for docking atomic models of the wild-type KaiB dimer (core residues, aa 8–94) within EM density assigned to KaiB. The third layer of density was segmented from the final C6B4P structure using the ‘Volume Eraser' tool of UCSF Chimera (Pettersen, 2004) to remove density assigned to the hexameric barrel of KaiC.
The fitting of the SeKaiC crystal structure (PDB 2GBL) within the EM density shown in Figure 5 was first performed interactively in Chimera and then refined for the KaiC CII domain residues (aa 250–484) with the ‘Fit Model in Map' tool of Chimera. Graphics figures were produced with Chimera.
Coordinates
Final coordinates and structure factors for the crystallographic model of T. elongatus KaiB have been deposited in the Protein Data Bank (http://www.rcsb.org): PDB ID code 2QKE.
Supplementary Material
Supplementary Information
Acknowledgments
Support for clock research at Vanderbilt University by the National Institutes of Health (R01 GM73845 to ME, R01 GM67152 to CHJ and F32 GM71276 to DRW) is gratefully acknowledged. We thank Dr David Friedman of the Vanderbilt Proteomics Facility for help with mass spectrometric characterization of all proteins. The DuPont–Northwestern–Dow Collaborative Access Team (DND-CAT) Synchrotron Research Center at the Advanced Photon Source (Sector 5) is supported by EI DuPont de Nemours & Co., The Dow Chemical Company, the National Science Foundation and the State of Illinois. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38.
References
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Cambillau C, Roussel A (1997) Turbo Frodo, Version OpenGL.1. Marseille, France: Université Aix-Marseille II [Google Scholar]
- CCP4 (1994) Collaborative computing project number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Chacon P, Wriggers W (2002) Multi-resolution contour-based fitting of macromolecular structures. J Mol Biol 317: 375–384 [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, DeCoursey PJ (2004) Chronobiology: Biological Timekeeping. Sunderland, MA: Sinauer [Google Scholar]
- Egli M, Pattanayek R, Pattanayek S (2007) Protein–protein interactions in the cyanobacterial KaiABC circadian clock. In Models, Mysteries and Magic of Molecules, Boeyens JCA, Ogilvie JF (eds), pp 287–303. Dordrecht, Netherlands: Springer Science [Google Scholar]
- Garces RG, Wu N, Gillon W, Pai EF (2004) Anabaena circadian clock proteins KaiA and KaiB reveal potential common binding site to their partner KaiC. EMBO J 23: 1688–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F, Ito H, Fujita M, Iwase R, Uzumaki T, Ishiura M (2004b) Stoichiometric interactions between cyanobacterial clock proteins KaiA and KaiC. Biochem Biophys Res Comm 316: 195–202 [DOI] [PubMed] [Google Scholar]
- Hayashi F, Itoh N, Uzumaki T, Iwase R, Tsuchiya Y, Yamakawa H, Morishita M, Onai K, Itoh S, Ishiura M (2004a) Roles of two ATPase-motif-containing domains in cyanobacterial circadian clock protein KaiC. J Biol Chem 50: 52331–52337 [DOI] [PubMed] [Google Scholar]
- Hayashi F, Iwase R, Uzumaki T, Ishiura M (2006) Hexamerization by the N-terminal domain and intersubunit phosphorylation by the C-terminal domain of cyanobacterial circadian clock protein KaiC. Biochem Biophys Res Comm 318: 864–872 [DOI] [PubMed] [Google Scholar]
- Hayashi F, Suzuki H, Iwase R, Uzumaki T, Miyake A, Shen J-R, Imada K, Furukawa Y, Yonekura K, Namba K, Ishiura M (2003) ATP-induced hexameric ring structure of the cyanobacterial circadian clock protein KaiC. Genes Cells 8: 287–296 [DOI] [PubMed] [Google Scholar]
- Hitomi K, Oyama T, Han S, Arvai AS, Getzoff ED (2005) Tetrameric architecture of the circadian clock protein KaiB: a novel interface for intermolecular interactions and its impact on the circadian rhythm. J Biol Chem 280: 18643–18650 [DOI] [PubMed] [Google Scholar]
- Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T (1998) Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281: 1519–1523 [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T (2002) KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA 99: 15788–15793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase R, Imada K, Hayashi F, Uzumaki T, Morishita M, Onai K, Furukawa Y, Namba K, Ishiura M (2005) Functionally important substructures of circadian clock protein KaiB in a unique tetramer complex. J Biol Chem 280: 43141–43149 [DOI] [PubMed] [Google Scholar]
- Jones TA (2004) Interactive electron-density map interpretation: from INTER to O. Acta Crystallogr D Biol Crystallogr 60: 2115–2125 [DOI] [PubMed] [Google Scholar]
- Kageyama H, Kondo T, Iwasaki H (2003) Circadian formation of clock protein complexes by KaiA, KaiB, KaiC, and SasA in cyanobacteria. J Biol Chem 278: 2388–2395 [DOI] [PubMed] [Google Scholar]
- Kageyama H, Nishiwaki T, Nakajima M, Iwasaki H, Oyama T, Kondo T (2006) Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol Cell 23: 161–171 [DOI] [PubMed] [Google Scholar]
- Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T (2003) KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J 22: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- Mori T, Saveliev SV, Xu Y, Stafford WF, Cox MM, Inman RB, Johnson CH (2002) Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc Natl Acad Sci USA 99: 17203–17208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Williams DR, Byrn M, Qin X, Egli M, Mchaourab H, Stewart PL, Johnson CH (2007) Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol 5: 841–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T (2005) Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308: 414–415 [DOI] [PubMed] [Google Scholar]
- Nishiwaki T, Satomi Y, Nakajima M, Lee C, Kiyohara R, Kageyama H, Kitayama Y, Temamoto M, Yamaguchi A, Hijikata A, Go M, Iwasaki H, Takao T, Kondo T (2004) Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc Natl Acad Sci USA 101: 13927–13932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pattanayek R, Wang J, Mori T, Xu Y, Johnson CH, Egli M (2004) Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol Cell 15: 375–388 [DOI] [PubMed] [Google Scholar]
- Pattanayek R, Williams DR, Pattanayek S, Xu Y, Mori T, Johnson CH, Stewart PL, Egli M (2006) Analysis of KaiA–KaiC protein interactions in the cyanobacterial circadian clock using hybrid structural methods. EMBO J 25: 2017–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H (2005) No transcription–translation feedback in circadian rhythm of KaiC phosphorylation. Science 307: 251–254 [DOI] [PubMed] [Google Scholar]
- Uzumaki T, Fujita M, Nakatsu T, Hayashi F, Shibata H, Itoh N, Kato H, Ishiura M (2004) Crystal structure of the C-terminal clock-oscillator domain of the cyanobacterial KaiA protein. Nat Struct Mol Biol 11: 623–631 [DOI] [PubMed] [Google Scholar]
- Vakonakis I, LiWang AC (2004) Structure of the C-terminal domain of the clock protein KaiA in complex with a KaiC-derived peptide: implications for KaiC regulation. Proc Natl Acad Sci USA 101: 10925–10930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakonakis I, Sun J, Wu T, Holzenburg A, Golden SS, LiWang AC (2004) NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: implications for the KaiA–KaiC interaction. Proc Natl Acad Sci USA 101: 1479–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M (1996) A new generation of the IMAGIC image processing system. J Struct Biol 116: 17–24 [DOI] [PubMed] [Google Scholar]
- Williams SB, Vakonakis I, Golden SS, LiWang AC (2002) Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proc Natl Acad Sci USA 99: 15357–15362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Johnson CH (2003) Cyanobacterial circadian clockwork: roles of KaiA, KaiB, and the kaiBC promoter in regulating KaiC. EMBO J 22: 2117–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Pattanayek R, Pattanayek S, Egli M, Johnson CH (2004) Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proc Natl Acad Sci USA 101: 13933–13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Vakonakis I, Ioerger TR, LiWang AC, Sacchettini JC (2004) Crystal structure of circadian clock protein KaiA from Synechococcus elongatus. J Biol Chem 279: 20511–20518 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information