Abstract
In recent years, it has been shown that helicases are able to perform functions beyond their traditional role in unwinding of double-stranded nucleic acids; yet the mechanistic aspects of these different activities are not clear. Our kinetic studies of Holliday junction branch migration catalysed by a ring-shaped helicase, T7 gp4, show that heterology of as little as a single base stalls catalysed branch migration. Using single-molecule analysis, one can locate the stall position to within a few base pairs of the heterology. Our data indicate that the presence of helicase alone promotes junction unfolding, which accelerates spontaneous branch migration, and individual time traces reveal complex trajectories consistent with random excursions of the branch point. Our results suggest that instead of actively unwinding base pairs as previously thought, the helicase exploits the spontaneous random walk of the junction and acts as a Brownian ratchet, which walks along duplex DNA while facilitating and biasing branch migration in a specific direction.
Keywords: branch migration, helicases, single molecule
Introduction
Homologous recombination is a major pathway in the rescue of stalled or collapsed replication forks caused by DNA damage. A key intermediate in homologous recombination, the DNA four-way (Holliday) junction (Holliday, 1964; Potter and Dressler, 1976), can undergo branch migration by way of sequence homology. The spontaneous branch migration can be described as a one-dimensional random walk where both migration directions are equally likely as long as homology exists, but is strongly inhibited by even a single base-pair heterology (Panyutin and Hsieh, 1993). In physiological conditions with divalent ions, this spontaneous process is significantly slowed down by the junction's ion-induced folding into the stacked X structure (Duckett et al, 1988; Murchie et al, 1989; Panyutin and Hsieh, 1994). To overcome this, helicases, most commonly hexameric ring helicases, are required to catalyse the reaction (Lloyd and Sharples, 1993; Kaplan and O'Donnell, 2002). Branch migration is catalysed in the case of ring-shaped helicases by encircling double-stranded DNA (dsDNA) and translocating along dsDNA by tracking one strand directionally (Kaplan and O'Donnell, 2002).
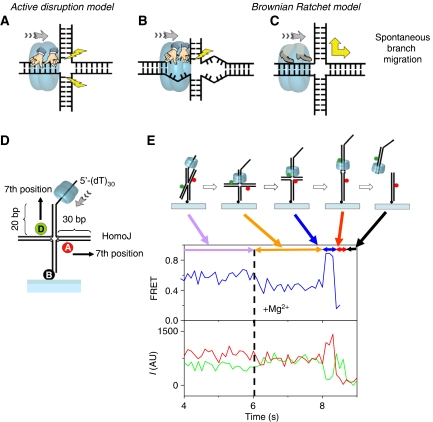
The conventional view of branch migration enzymes, which we call the active disruption model (Figure 1A and B), posits that the translocating helicase pulls the DNA through the central hole such that the pulling force is used to disrupt, or melt, the base pairs at the junction (Figure 1A). In this model, branch migration is more efficient for homologous junctions because pulling on the tracking strand results in simultaneous pulling of the complementary strand, leading to the disruption of base pairs both above and below the branch point (Figure 1A) (Kaplan and O'Donnell, 2002, 2004). If extensive sequence heterology exists, pulling of one strand becomes mechanically decoupled from the other strand, impairing active base-pair disruption (Figure 1B). Indeed, extensive sequence heterology (⩾25 bp) inhibits branch migration catalysed by DnaB (Kaplan and O'Donnell, 2004) and RuvAB (Dennis et al, 2004). To be specific, we further define the active disruption model by two characteristics: kinetics of branch migration is dominated by the helicase, and DNA base disruption is an essential component. Here, we propose an alternative model called the Brownian Ratchet model for helicase-catalysed branch migration (Figure 1C). In this model, the helicase's primary function is in the prevention of junction folding so that the junction remains in the open, unstacked form. Once opened, the junction is free to take branch migration steps that are typically bidirectional and fast, but the unidirectional translocation of the helicase ensures that this branch point walking becomes biased in one direction by preventing backward migration steps. In this paper, we present several lines of evidence in support of the Brownian Ratchet model.
Figure 1.
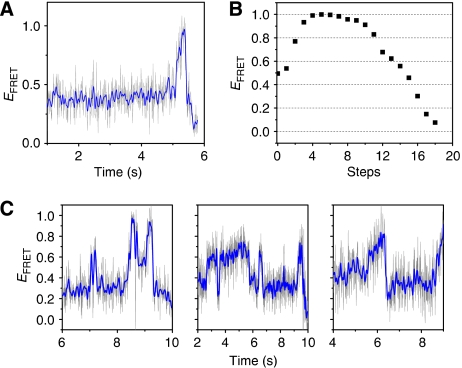
Helicase-catalysed branch migration. (A–C) Schematic representation of the models discussed in this work. (A) Active disruption by simultaneous unwinding of two arms. (B) For the active disruption model, extensive heterology can lead to inefficient unwinding, as only one arm is being pulled. (C) The Brownian ratchet model involves junction opening and rapid spontaneous branch migration, which is biased by the directional movement of the helicase. (D) Schematic representation of substrate and initial binding site. (E) Typical single-molecule FRET intensity and FRET efficiency (blue curve) time traces for a branch migration reaction. Acceptor (A in the figure, Cy5) is in red and donor (D in the figure, Cy3) in green. The fraction of molecules that show complete reaction is 57% (301 molecules out of 529 molecules in four independent experiments), which is comparable to results in bulk solution. The segments and the scheme represent the different stages of the reaction.
Results
We have studied the mechanism of branch migration using T7 gp4, a 5′-to-3′ helicase-primase (T7 helicase) (Egelman et al, 1995; Patel and Hingorani, 1995; Picha and Patel, 1998; Picha et al, 2000; Jeong et al, 2004) as a model protein. T7 helicase requires a single-stranded DNA (ssDNA) overhang to initiate migration; hence, branch migration can be initiated in a controlled manner using T7 helicase and a Holliday Junction substrate that contains a single 5′ ssDNA tail attached to one arm of the junction (Figure 1D). Stable assembly of the helicase ring on the ssDNA overhang (Hingorani and Patel, 1993) requires the presence of dTTP but not Mg2+ (Picha and Patel, 1998), and hence the helicase can be pre-assembled on the substrate, bypassing the slow assembly steps, and branch migration can be triggered by adding Mg2+ to such a pre-assembled ring (Figure 1E). The requirement of an ssDNA overhang not only allows us to load the helicase to a defined arm, but also to prevent additional helicases from loading onto the other arms. No branch migration was observed without an ssDNA tail (data not shown). The branch migration substrate contained two horizontal DNA arms, both 30 bp long, which were homologous except for the first base pair (base-pair positions in this paper are measured with respect to the junction). The substrate (HomoJ) was labelled with Cy3 (donor, D) and Cy5 (acceptor, A) at the seventh position each by means of amino-dT's. The heterologous base pair at the first position creates a barrier that prevents resolution of the junction by spontaneous branch migration and sets a well-defined starting point for the reaction. Using FRET one can resolve reaction progression to within a few base pairs, about an order of magnitude higher than in previous single-molecule studies of RuvAB (Amit et al, 2004; Dawid et al, 2004; Dennis et al, 2004), enabling us to examine the effect of small changes in DNA sequence. For single-molecule FRET measurements, the DNA molecules were tethered, through biotin-streptavidin linkers, to quartz surfaces coated with polymer (polyethyleneglycol) to eliminate nonspecific protein binding to the underlying surface (Ha et al, 2002; Blanchard et al, 2004; Murphy et al, 2004). For bulk solution measurements, the substrates were radiolabelled and time courses were measured using the rapid chemical quenched-flow apparatus.
Figure 1E shows a typical branch migration reaction of HomoJ as the intensity (and FRET efficiency) time traces of the donor and acceptor signals for a single junction. The immobilized substrates in a sample cell were incubated with the helicase and dTTP in the absence of Mg2+. This allows binding of the helicase to the single-strand portion of the substrate but dTTP hydrolyses is not possible. The reaction is initiated by flowing Mg2+ into the sample cell. We observed a slight FRET decrease when the reaction was initiated followed by FRET increase to near 100% and then by FRET decrease as branch migration proceeded, and finally loss of fluorescence upon reaction completion and donor strand release from the surface. Control experiments omitting protein or Mg2+ showed that the contribution of donor bleaching was only about 6% in the first 20 s of all reactions studied here (data not shown). As free protein in solution was displaced from the sample cell when Mg2+ (and dTTP) was added by means of the flow system, reaction from only the pre-bound proteins was observed.
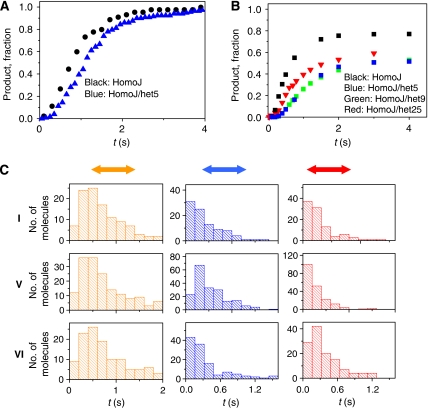
In contrast to the rapid branch migration of HomoJ, branch migration of a fully heterologous junction with 30 bp arms (HeteroJ) was negligible (data not shown). The active disruption model predicts that the heterologous substrate should be resolved, because helicase is actively disrupting the base pairs. On the other hand, the Brownian Ratchet model predicts that branch migration will be severely inhibited in the heterologous substrate. Because the heterology in the above substrate extends to 30 bp, one can argue that the two strands will be mechanically uncoupled and hence the interpretation of the results of the above experiment is more complicated and cannot distinguish between the two models. Therefore, we designed alternate substrates with one or two base-pair heterology. The active model predicts that a short stretch of heterology, for example, one or two base pairs, should not have any effect because the two strands in the loading arm are still mechanically coupled. To explore the effect of minimal heterology on the kinetics of branch migration, we introduced a one base-pair heterology in the HomoJ by switching a GC base pair at the fifth position in one of the arms to CG (HomoJ/het5). The time it takes to complete the branch migration reaction was determined from analyses of the single-molecule trajectories (such as in Figure 1E) as the time when the fluorescence signal disappears (end of red double arrow in Figure 1E) after initiation with Mg2+. To make a comparison of single-molecule kinetics with that in bulk solution, we counted the total number of molecules that completed the reaction at a given time (Figure 2A, normalized data), indicating product formation. This analysis reveals that even a single base-pair heterology lengthens the reaction time by about 300 ms (obtained from the difference in the average times that it takes a molecule to complete the reaction for each substrate). Quenched-flow measurements on substrates without fluorescent labels also showed that an extra heterology in the fifth or ninth position delays the reaction (Figure 2B), and with more moderate effect when the heterology is near the end of the arm (25th position). Direct comparison of the kinetics of product formation from single-molecule measurements and quenched-flow experiments must take into consideration that in this latter type of experiments the reaction is stopped by adding SDS and EDTA. Addition of EDTA stops dTTP hydrolyses, but it also sequesters Mg2+. In the single-molecule experiments, Mg2+ is present at all times. The more moderate effect of the heterology in the 25th position shown for quenched-flow experiments is possibly related to the fact that the heterology is close to the DNA end and the low stability of the duplex DNA with only five base pairs results in resolution by spontaneous branch migration of a fraction of molecules after the enzyme-catalysed reaction is stopped. The marked effect of a single base-pair heterology in the resolution of Holliday junctions indicates that the helicase is not destabilizing the base pairs at the junction to a significant extent. Instead, it suggests that spontaneous branch migration, which is likewise significantly impacted by a single mismatch, has a significant role in the helicase-catalysed migration process (Panyutin and Hsieh, 1993).
Figure 2.
Kinetics of the enzyme-catalysed branch migration reaction and effect of heterologies. (A) Product formation versus time for single-molecule experiments measured as the number of molecules that completed the reaction at a given time, normalized by the total number of molecules that complete the reaction and (B) bulk solution experiments. (C) Effect of heterologies on branch migration reactions. Histograms of the times that molecules spend in the pre-high (left panel), high (central panel), and low (right panel) FRET states, indicated in orange, blue, and red, respectively (each stage is also indicated in Figure 1E). The results for HomoJ, HomoJ/het5, and HomoJ/het16/het17 (from top to bottom) are shown. Roman numbers on the left indicate substrate schematics in Figure 3A.
To locate the physical position in the substrate of the kinetic barrier imposed by the heterology, we divided the reaction time course (Figure 1E) of individual molecules into three segments and measured the duration of each segment for hundreds of molecules undergoing reaction: segment 1 before high FRET (yellow arrow), segment 2 with high FRET (blue arrow), and segment 3 after high FRET (red arrow) (Figure 1E). These segments correspond to different degrees of ‘proximity' of the fluorescent dyes. The division into three regions is arbitrary and was chosen so that the time durations can be measured reliably within our signal-to-noise levels. The high FRET segment is defined as contiguous data points with FRET values above 0.75, approximately corresponding to the first 2–11 bp steps of reaction, based on the open junction crystal structure in complex with RuvA (Ariyoshi et al, 2000). To determine the time intervals for the different stages of the reactions, individual traces were analysed (one by one) using a Matlab code for visualizing, calculating FRET efficiency, and storing time values by mouse clicking. Increase in acceptor brightness when Mg2+ ions are flowed inside the cell sample provides the starting point (time t=0). Two factors affect the accuracy in the determination of FRET efficiency intervals, noise and slope of the FRET efficiency change. Intervals were determined by clicking on the individual time traces; under typical experimental conditions, the beginning and the end of each interval were accurate within one data point (based on the noise level and slope of FRET efficiency change in the interval) that corresponds to ±30 ms interval. The reproducibility of the time intervals statistics distributions was tested by repeating each experimental condition at least three times.
The time spent in the different segments from 120–200 molecules for each junction sequence is shown in Figure 2C. As compared to the HomoJ substrate (I), introduction of a heterologous base pair at the fifth position (Figure 3A, substrate V, HomoJ/het5 with dyes at the seven position) increases the duration of segment 2, but not the durations of other segments. As high FRET is expected when the donor and acceptor are within 4 bp of each other; the data suggest that reaction stalls when the heterology has reached the branch point. To test this interpretation further, we measured a variant of HomoJ with consecutive heterologous base pairs in the 16th and 17th positions (Figure 3A, substrate VI, HomoJ/het16/het17 with dyes at the seventh position). Dwell time analysis shows that only segment 3 experiences significant effect of heterology (Figure 2C). In this substrate, the donor and acceptor would move past each other and reach an 18 bp separation by the time the additional heterologous base pairs reach the branch point. These results suggest that the slowed branch migration reaction is due to a local kinetic barrier imposed by sequence heterology, as opposed to a global effect.
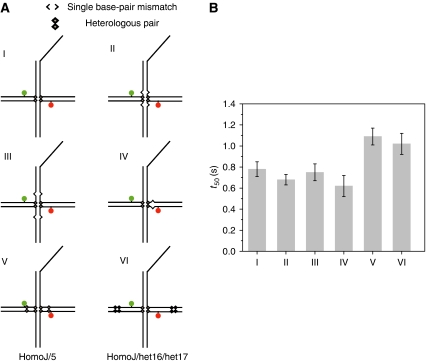
Figure 3.
Effect of mismatches (bubbles) and heterologies on the reaction kinetics. (A) Substrates used to study the effect of heterologies and bubbles. (B) Effect of mismatches and heterologies on reaction rates represented by the average time (t50) to complete 50% of the final product formation for several substrates under the same experimental conditions. Error bars represent the variability in the determination of t50 for at least three repetitions for each substrate.
It is also possible that the local kinetic barrier arises because the single base-pair heterology becomes a bubble (single base-pair mismatch) after the heterology has moved past the branch point. To test if such a bubble would affect the helicase progression, we introduced bubbles at various locations (Figure 3A). Results from single-molecule studies are summarized in Figure 3B, which compares the time it takes for 50% of the molecules to react to completion. Bubbles themselves did not slow the overall reaction time regardless of their positions, including when they were introduced on the loading arm, whereas extra heterologies did. We can conclude from the combined results that the single base-pair heterology slows down the overall reaction because it stalls the branch point migration, not because of subsequent bubble formation inhibits the reaction.
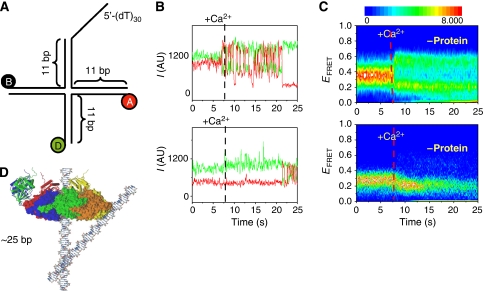
Spontaneous branch migration is strongly inhibited under physiological conditions (Panyutin and Hsieh, 1994) because the Holliday junction forms the stacked X structure (Joo et al, 2004). Therefore, spontaneous branch migration can have a major role under physiological conditions only if the helicase keeps the junction in the open, unstacked form. A DNA substrate (MonoJ; Figure 4A) fluorescently labelled on adjacent arms can be used to observe such transitions between different stacked conformations (McKinney et al, 2005) with and without the helicase. In these experiments, we used Ca2+ instead of Mg2+ and a heterologous sequence to assure that the helicase remains bound to the substrate for an extended time for observation (due to a decrease in the branch migration rate). The addition of Ca2+ preserves the junction folding dynamics, but it dramatically reduces the dTTPase rate, hence reducing the extent of branch migration reaction and allows looking at the folding conformational dynamics of the junction for longer periods. In the absence of protein, the junction is unfolded before Ca2+ addition (low FRET), but begins to fluctuate between high and low FRET states representing the two major stacked X conformations upon Ca2+ addition (Figure 4B and C, top panels). No such fluctuations were observed with the protein present (Figure 4B and C, bottom panels) except at later times, likely due to protein dissociation. Superposition of the crystal structures of T7 helicase and a Holiday junction (Figure 4D) supports the idea that steric hindrance could significantly affect the junction's folding dynamics. The protein keeps the junction in its open form, which should facilitate spontaneous branch migration at a rapid rate.
Figure 4.
(A) DNA substrate (MonoJ) used for measuring the effect of helicase binding and translocation on junction folding. (B, C) Stacking conformer transitions of a junction (MonoJ) with (top panels) and without (bottom panels) protein. (B) Single-molecule time traces on addition of Ca2+ at about 7 s. (C) The density plot shows a synchronized behaviour of ∼120 molecules at a time (Blanchard et al, 2004). The colour scale is proportional to the number of molecules with a given FRET value at a given time. Protein (bottom panels) was added at a concentration of 150 nM in the presence of dTTP (2 mM) and allowed to incubate as in experiments in Figure 1E before the addition of Ca2+. (D) Overlap of crystal structures of a Holliday junction and T7 helicase (Toth et al., 2003) indicating the relative size of both structures (figure courtesy of Jin Yu).
If spontaneous branch migration indeed occurs during the helicase-catalysed reaction, it may be possible to detect forward and backward movements of the junction during the reaction. If the helicase moves fast with respect to the spontaneous branch migration stepping rate, it will always reach the branch point to capture the forward steps before significant migration in the backward direction has occurred. The data under our standard conditions are consistent with unidirectional movements of the branch point, that is, FRET increase followed by its decrease (Figure 5A), which is consistent with a monotonic progression of the branch point. A step-by-step estimate of FRET efficiency values (Figure 5B) calculated using available crystal structure of a Holiday junction in the open form (Ariyoshi et al, 2000) shows this monotonic increase followed by a monotonic decrease in FRET efficiency. However, in experiments carried out at a lower temperature (17°C) and with higher time resolution (8 ms), we observed more complex behaviour in the FRET efficiency trajectories including multiple occurrences of FRET increase and decrease (Figure 5C). Such time trajectories are consistent with the random walk of the junction when the helicase is unable to catch up with the branch point. This behaviour was observed in a wide range of Mg+2 ion concentration (data not shown). Given the fast observed dynamics, and the wide range of FRET efficiency values observed for individual trajectories, it is highly unlikely that these excursions correspond to folding dynamics of the junction themselves or spontaneous branch migration in the absence of protein. Decreasing the temperature reduces both the translocation rate of the helicase and the stepping rate of the spontaneous branch migration and allowed us to observe the random excursions of the branch point.
Figure 5.
(A) Single-molecule FRET efficiency time traces for the catalysed branch migration reaction obtained at 23°C with 8 ms time resolution showing the junction moving unidirectionally. (B) Step-by-step FRET efficiency estimated from the crystal structure of an open Holiday junction (Ariyoshi et al, 2000) assuming R0=55 Å. (C) FRET efficiency trajectories at 17°C with 8 ms time resolution showing fluctuations consistent with random excursions of the junction. High time resolution data are from HomoJ and HomoJ/L16, identical to HomoJ but labelled at the 16th positions. Grey lines are raw data and blue lines are 5 points averaged data. Experiments were performed by following the same procedure as experiments described in Figure 1E.
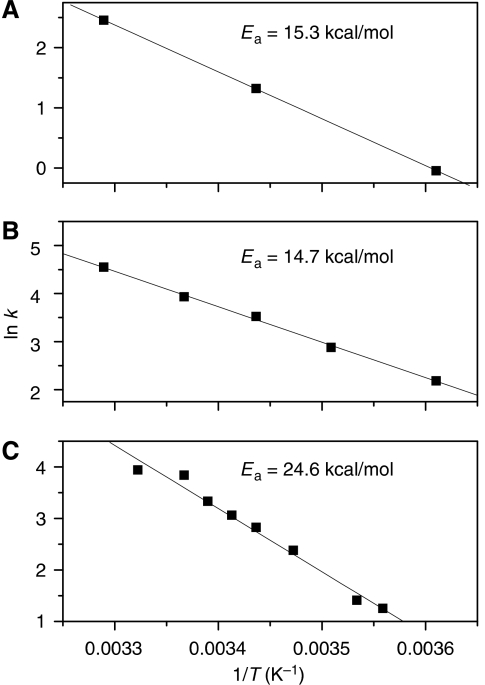
To further explore the effect of temperature on the overall reaction rate, we measured the temperature dependence of the branch migration activity of T7 in bulk phase and compared with the temperature dependence of the dTTPase activity and DNA unwinding activity (Figure 6). The unwinding reaction has an identical activation energy (Figure 6A), 15 kcal/mol, as the dTTPase (Figure 6B) reaction whereas the catalysed branch migration has much higher activation energy of 25 kcal/mol (Figure 6C), closer to that of spontaneous branch migration reaction (Panyutin and Hsieh, 1994), suggesting that junction reorganization, not base disruption, is a major rate-limiting process.
Figure 6.
Temperature dependence of the reaction rate for T7 helicase determined from bulk solution experiments. (A) Pre-steady-state DNA unwinding rate. (B) M13 ssDNA-stimulated dTTPase rate. (C) Branch migration reaction.
The simplest model that explains our data is the Brownian ratchet model where a random walk of the junction is aided and biased by the helicase. When two base pairs are broken, one each from the two downstream arms, they can either reform as they were or reorganize to move the branch point by a single base pair. If the broken base pairs are heterologous, the latter reaction, which causes an energetically costly product due to bubble formation, would lead to a rapid back step. Therefore, branch migration over a heterologous base pair would not occur until the translocating helicase traps the transient branch point. The active model in its pure form assumes that the branch migration kinetics is essentially controlled by the helicase, which will keep pushing forward at its own translocation rate. The Brownian ratchet model for catalysed branch migration of homologous junctions proposes that the overall kinetics is dictated both by the helicase translocation rate and the spontaneous branch migration rate.
Discussion
It has been shown that helicases are able to exert considerable forces while translocating on the DNA (Morris and Raney, 1999) and the possibility of disrupting the junction through torque or force should be considered if there is significant sequence heterology such that the helicase spends considerable time right at the junction. The helicase is likely to make active contributions to overcome heterologous barriers, as has been suggested for the strand separation reaction (Donmez et al, 2007; Johnson et al, 2007). However, branch migration in the bacterial homologous recombination likely occurs between nearly, if not completely, identical sequences. In these physiologically relevant contexts, the spontaneous branch migration stepping would be faster than helicase translocation such that the helicase rarely encounters a static junction to exert force on. Our experiments strongly suggest that for homologous junctions all the essential aspects of the reaction and the experimental observations are accounted for by the Brownian ratchet model. Conversely, many of the experimental observations cannot be explained by a purely active model. Because a ring helicase alone can unfold the junction without contacting the central bases, the main role of RuvA in RuvAB may not be in the junction opening. RuvB can catalyse branch migration of homologous junction but requires RuvA for branch migration of DNA with extensive sequence heterology (Adams and West, 1996). In such cases, RuvA may prevent the dissociation of RuvB such that RuvB exerts the necessary force to overcome heterology.
Our experiments cannot determine unequivocally if there is a component of the energy obtained from dTTP hydrolyses that is being utilized in base disruption at the junction. Our results show conclusively though that even under slow hydrolysis rates and presence of divalent ions, helicase binding strongly affects junction conformational dynamics. Our experiments also show the presence of random excursions of the branch point, a stronger temperature dependence of the reaction rate than for dTTPase rate or unwinding kinetics and that a single base-pair heterology has a profound effect on the reaction rate. We have also shown that the formation of bubbles (expected after migration of heterologous pairs) does not slow down the reaction in itself and that reduction of reaction rate due to heterologies is local. All these constitute strong evidence that spontaneous branch migration has a fundamental role in the enzyme-catalysed branch migration reaction, that base-pair disruption is not limiting for the overall reaction and that the main role of the helicase is to allow the junction to undergo spontaneous branch migration.
Previous single-molecule studies (Dawid et al, 2004) have determined that the rate of RuvAB-catalysed branch migration for homologous junctions is 20–30 bp/s, a rate comparable to the 27 bp/s average rate obtained in our experiments (to eliminate the effect of the heterologous barrier on the homologous branch migration rate, we determine the average duration for the second and third segments of the reaction in Figure 2C). It has been shown that the presence of a single heterology inhibits three-fold the initial rate of branch migration by Rad54 (Bugreev et al, 2006), an enzyme with branch migration activity. These results suggest that the Brownian ratchet model may be a general mechanism for branch migration enzymes.
Lifting the conformational ban on branch migration coupled with unidirectional translocation may be a general mechanism for ring helicase-catalysed branch migration and should be applicable to branch migration by two helicase rings operating on two opposing arms with up to a factor of 2 increase in branch migration rate. This mechanism is appealing in its generality, as it does not invoke any specific interaction of the helicase with the junction. The simplicity and power of our model together with the recent observations of alternative roles of helicases shows that unidirectional translocation itself is an essential component that allows these enzymes to perform a variety of functions (Jankowsky et al, 2001; Krejci et al, 2003; Veaute et al, 2003; Byrd and Raney, 2004; Myong et al, 2005) and suggests how higher degrees of specialization can arise from a general common mechanism. Finally, we note that the hexameric helicase in T4 phage is involved in recombination-dependent replication (Kreuzer, 2000) and the T7 helicase may have a similar role in DNA replication. In addition, T7 helicase may have a role in restarting arrested replication forks by converting blocked replication forks into double-stranded tailed structures, which requires branch migration activity (Heller and Marians, 2006).
Materials and methods
Protein, nucleotides, buffer, and oligonucleotides
The gp4A′ (T7 helicase) is an M64L mutant of T7 helicase-primase protein that was overexpressed in Escherichia coli and purified as described previously (Patel et al, 1992; Hingorani and Patel, 1996). dTTP was purchased from Sigma Chemicals. Buffer A (50 mM Tris/Cl (pH 7.6), 40 mM NaCl, and 10% (v/v) glycerol) was used unless specified otherwise. Quenching solution consisted of 100 mM EDTA, 0.4% (v/v) SDS, and 20% glycerol. The oligodeoxynucleotides were purchased from Intergrated DNA Technologies (Coralville, IA).
Bulk kinetic measurements
Quenched-flow experiments of branch migration were performed using the same procedures described for the DNA unwinding experiments by the T7 helicase (Jeong et al, 2004). T7 helicase (200 nM), EDTA (5 mM), dTTP (2 mM), and 5′-32P-end-labelled DNA (5 nM) were rapidly mixed with an equal volume of MgCl2 (13 mM), dTTP (2 mM), and dT100 trap DNA (3 μM) in the rapid chemical quench-flow instrument. After pre-determined times, the reactions were stopped with the quenching solution, and the DNAs were resolved by electrophoresis on a 4–20% gradient non-denaturing polyacrylamide gel. The radioactivity was quantified using the Phosphorimager (Molecular Dynamics Inc.).
The temperature dependence of unwinding and branch migration was measured on the rapid chemical quenched flow, as previously described. The unwinding kinetics were obtained using a forked substrate carrying a dT35 5′-tail to load the helicase, a dT15 3′-tail to exclude the displaced strand from the central channel, and a 30 bp duplex region. Kinetic time courses of unwinding were obtained at 4, 22, and 30°C. Kinetic time courses of branch migration were obtained using HomoJ Holliday junction substrate at 8, 10, 15, 18, 20, 22, 24, and 28°C.
The temperature dependence of steady-state dTTP hydrolysis was measured as previously described (Liao et al, 2005). T7 helicase (20 nM), EDTA (5 mM), dTTP (2 mM), and single-stranded phage m13 DNA (1.5 nM) were mixed with an equal volume of MgCl2 (13 mM) and dTTP (2 mM, spiked with [α-32P]dTTP). After pre-determined times, the reactions were stopped with 4 N formic acid and the products were resolved by thin-layer chromatography. The radioactivity was quantified using the Phosphorimager (Molecular Dynamics Inc.). The kinetic time courses of dTTPase were obtained at 4, 12, 18, 25, and 30°C.
An Arrhenius plot was generated for each of the three activities, by plotting the natural logarithm of the reaction rates (ln k) as a function of the inverse of temperature (in kelvin). The activation enthalpy of the reaction (Ea) was obtained by multiplying the slope of the line with R (the universal gas constant, assumed to be 1.987 cal/K/mol).
Single-molecule experiments
The sample cell consists of a channel between a quartz slide and a glass coverslip with approximate dimensions of 5 × 20 × 0.1 mm3 and two 1 mm diameter holes on the slide for flow exchange. A syringe needle is glued to the inlet hole and connected with tubing to a syringe mounted on a syringe pump controller. This system allows for synchronized initiation of reactions with simultaneous observation of about 200 molecules with a CCD camera (iXon, Andor). The experiments were performed with 30 ms time resolution using total internal reflection excitation with a 532 nm laser and dual view imaging. First, junctions are immobilized to a polymer-coated surface by means of biotin-streptavidin. Next, 300 nM (hexamer) T7 helicase, 2 mM dTTP, and Mg2+-free solution is added and incubated for 10 min. Experiments were carried out on buffer A with the addition of 0.4% w/v glucose, enzymatic oxygen scavenging system, and 1% v/v 2-mercaptoethanol or Trolox® as triplet quencher for single-molecule imaging (Rasnik et al, 2006). After the pre-incubation period, data were taken for about 5 s before the flow system delivers a buffer containing both dTTP and Mg+2 but no protein at a rate of 1 ml/min. The effective starting point of the reaction is determined by the slight increase in the average brightness of the fluorescent dyes with Mg+2.
DNA sequences and labelling
DNA strands were obtained from Integrated DNA Technologies and further purified using PAGE. For strands requiring internal labelling with fluorescent dyes (Cy3 or Cy5), the amino-modified form of the oligonucleotide was purchased (internal amino modifier C6 dT (iAmMC6T)).
Internal labelling of the single-strand amino-modified oligonucleotides was performed using monoreactive Cy3 (b strand) or Cy5 (r strand) NHS-ester (Amersham Biosciences) as follows: 0.2 mg of the monoreactive fluorescent dye was dissolved in 15 μl DMSO and mixed with 1–4 mM DNA (final concentration of amino groups) in 85 μl of 0.1 M sodium tetraborate buffer (pH 8.5). The reaction mixture was incubated at room temperature for 6 h and left overnight at 4°C. The excess dye was removed by ethanol precipitation of the DNA. The labelled DNA was further separated from the unlabelled fraction by PAGE purification.
Holliday junction substrates were formed by first annealing strands h and r in one vial and strands b and x in another vial (10 mM Tris pH 8.0, 50 mM NaCl), and then by mixing the contents of the two vials overnight at room temperature.
For quenched-flow studies, the DNAs were purified on a 15% polyacrylamide gel/8 M urea run in TBE buffer. The major DNA band was excised and the DNA was electroeluted using an Elutrap apparatus (Schleicher & Schuell). The 5′-strand of the fork DNA was radiolabelled with 32Pi using [γ-32P]ATP (Amersham Pharmacia Biotech) and T4 polynucleotide kinase (New England Biolabs). The radiolabelled helicase substrate was prepared by mixing the 5′-strand with a 1.5-fold excess of the complementary non-radiolabelled 3′-strand. The DNA mixture was heated at 95°C for 5 min and allowed to slowly cool down to room temperature.
The sequences are as follows:
HomoJ. h: 5′ TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TCC GGT CAA CCG TAG CAG CAC GAG CGA AGG GCG AAC GCT TAT GAG CTC AT 3′.
r: 5′ ATG AGC TCA TAA GCG TTC GCC CT/iAmMC6T/CGC TCG CCT CAA CTG GGA CCG TTT CGT GAC G 3′.
b: 5′ ATG AGC TCA TAA GCG TTC GCC CT/iAmMC6T/CGC TCC TGC TGC TAC GGT TGA CCG GA 3′.
x: 5′Biotin CGT CAC GAA ACG GTC CCA GTT GAG GGG AGC GAA GGG CGA ACG CTT ATG AGC TCA T 3′.
HomoJ/het5. Strands b and x were the same as HomoJ.
h: 5′ TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TCC GGT CAA CCG TAG CAG CAC GAG GGA AGG GCG AAC GCT TAT GAG CTC AT 3′.
r: 5′ ATG AGC TCA TAA GCG TTC GCC CT/iAmMC6T/CCC TCG CCT CAA CTG GGA CCG TTT CGT GAC G 3′.
HomoJ/het16/het17. Strands b and x were the same as HomoJ.
h: 5′ TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TCC GGT CAA CCG TAG CAG CAC GAG CGA AGG GCG AAG CCT TAT GAG CTC AT 3′.
r: 5′ ATG AGC TCA TAA GGC TTC GCC CT/iAmMC6T/CGC TCG CCT CAA CTG GGA CCG TTT CGT GAC G 3′.
Bubble substrates
Substrate numbers correspond to assignment in Figure 3. Strands h and x were modified to create single base-pair mismatches (bubbles) at both sides of the junction (substrates II and III). Strand h was modified to create a single bubble in substrate IV.
Substrate II. h: 5′ TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TCC GGT CAA CCG TAG CAG TAC GAG CGA AGG GCG AAC GCT TAT GAG CTC AT 3′.
x: 5′Biotin CGT CAC GAA ACG GTC CCA GTT GAT GGG AGC GAA GGG CGA ACG CTT ATG AGC TCA T 3′.
Substrate III. h: 5′ TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TCC GGT CAA CCG TAT CAG CAC GAG CGA AGG GCG AAC GCT TAT GAG CTC AT 3′.
x: 5′Biotin CGT CAC GAA ACG GTC CCA GTT GAG GGG TGC GAA GGG CGA ACG CTT ATG AGC TCA T 3′.
Substrate IV. h: 5′ TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TCC GGT CAA CCG TAG CAG CAC TAG CGA AGG GCG AAC GCT TAT GAG CTC AT 3′.
MonoJ. The notation for the strands is different, because the dyes are on adjacent arms.
h: 5′ Cy3-CCG TAG CAG CAC GAG CGG TGG G 3′.
r: 5′ Biotin-CCC ACC GCT CGT CTC AAC TGG G 3′.
b: 5′ Cy5-CCC TAG CAA GTT GCT GCT ACG G 3′.
x: 5′ TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT CCC AGT TGA GAA CTT GCT AGG G 3′.
Supplementary Material
Supplementary Materials
Acknowledgments
This work was funded by grants from the National Institutes of Health (GM55310 (SSP) and GM065367 (TH)) and from the National Science Foundation (PHY-0134916 (TH)). SAM is a National Science Foundation graduate research fellow. TH is an investigator in the Howard Hughes Medical Institute.
References
- Adams DE, West SC (1996) Bypass of DNA heterologies during RuvAB-mediated three- and four-strand branch migration. J Mol Biol 263: 582–596 [DOI] [PubMed] [Google Scholar]
- Amit R, Gileadi O, Stavans J (2004) Direct observation of RuvAB-catalyzed branch migration of single Holliday junctions. Proc Natl Acad Sci USA 101: 11605–11610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyoshi M, Nishino T, Iwasaki H, Shinagawa H, Morikawa K (2000) Crystal structure of the holliday junction DNA in complex with a single RuvA tetramer. Proc Natl Acad Sci USA 97: 8257–8262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL Jr, Puglisi JD, Chu S (2004) tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA 101: 12893–12898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Mazina OM, Mazin AV (2006) Rad54 protein promotes branch migration of Holliday junctions. Nature 442: 590–593 [DOI] [PubMed] [Google Scholar]
- Byrd AK, Raney KD (2004) Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat Struct Mol Biol 11: 531–538 [DOI] [PubMed] [Google Scholar]
- Dawid A, Croquette V, Grigoriev M, Heslot F (2004) Single-molecule study of RuvAB-mediated Holliday-junction migration. Proc Natl Acad Sci USA 101: 11611–11616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis C, Fedorov A, Kas E, Salome L, Grigoriev M (2004) RuvAB-directed branch migration of individual Holliday junctions is impeded by sequence heterology. EMBO J 23: 2413–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez I, Rajagopal V, Jeong YJ, Patel SS (2007) Nucleic acid unwinding by hepatitis C virus and bacteriophage t7 helicases is sensitive to base pair stability. J Biol Chem 282: 21116–21123 [DOI] [PubMed] [Google Scholar]
- Duckett DR, Murchie AI, Diekmann S, von Kitzing E, Kemper B, Lilley DM (1988) The structure of the Holliday junction, and its resolution. Cell 55: 79–89 [DOI] [PubMed] [Google Scholar]
- Egelman EH, Yu X, Wild R, Hingorani MM, Patel SS (1995) Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc Natl Acad Sci USA 92: 3869–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S (2002) Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature 419: 638–641 [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ (2006) Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol 7: 932–943 [DOI] [PubMed] [Google Scholar]
- Hingorani MM, Patel SS (1993) Interactions of bacteriophage T7 DNA primase/helicase protein with single-stranded and double-stranded DNAs. Biochemistry 32: 12478–12487 [DOI] [PubMed] [Google Scholar]
- Hingorani MM, Patel SS (1996) Cooperative interactions of nucleotide ligands are linked to oligomerization and DNA binding in bacteriophage T7 gene 4 helicases. Biochemistry 35: 2218–2228 [DOI] [PubMed] [Google Scholar]
- Holliday R (1964) Mechanism for gene conversion in fungi. Genet Res 5: 282–304 [DOI] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM (2001) Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291: 121–125 [DOI] [PubMed] [Google Scholar]
- Jeong YJ, Levin MK, Patel SS (2004) The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc Natl Acad Sci USA 101: 7264–7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Bai L, Smith BY, Patel SS, Wang MD (2007) Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell 129: 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C, McKinney SA, Lilley DM, Ha T (2004) Exploring rare conformational species and ionic effects in DNA Holliday junctions using single-molecule spectroscopy. J Mol Biol 341: 739–751 [DOI] [PubMed] [Google Scholar]
- Kaplan DL, O'Donnell M (2002) DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol Cell 10: 647–657 [DOI] [PubMed] [Google Scholar]
- Kaplan DL, O'Donnell M (2004) Twin DNA pumps of a hexameric helicase provide power to simultaneously melt two duplexes. Mol Cell 15: 453–465 [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309 [DOI] [PubMed] [Google Scholar]
- Kreuzer KN (2000) Recombination-dependent DNA replication in phage T4. Trends Biochem Sci 25: 165–173 [DOI] [PubMed] [Google Scholar]
- Liao JC, Jeong YJ, Kim DE, Patel SS, Oster G (2005) Mechanochemistry of t7 DNA helicase. J Mol Biol 350: 452–475 [DOI] [PubMed] [Google Scholar]
- Lloyd RG, Sharples GJ (1993) Processing of recombination intermediates by the RecG and RuvAB proteins of Escherichia coli. Nucleic Acids Res 21: 1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Freeman AD, Lilley DM, Ha T (2005) Observing spontaneous branch migration of Holliday junctions one step at a time. Proc Natl Acad Sci USA 102: 5715–5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris PD, Raney KD (1999) DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry 38: 5164–5171 [DOI] [PubMed] [Google Scholar]
- Murchie AI, Clegg RM, von Kitzing E, Duckett DR, Diekmann S, Lilley DM (1989) Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature 341: 763–766 [DOI] [PubMed] [Google Scholar]
- Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T (2004) Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J 86: 2530–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Rasnik I, Joo C, Lohman TM, Ha T (2005) Repetitive shuttling of a motor protein on DNA. Nature 437: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Panyutin IG, Hsieh P (1993) Formation of a single base mismatch impedes spontaneous DNA branch migration. J Mol Biol 230: 413–424 [DOI] [PubMed] [Google Scholar]
- Panyutin IG, Hsieh P (1994) The kinetics of spontaneous DNA branch migration. Proc Natl Acad Sci USA 91: 2021–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Hingorani MM (1995) Nucleotide binding studies of bacteriophage T7 DNA helicase-primase protein. Biophys J 68: 186S–189S; discussion 189S–190S [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Rosenberg AH, Studier FW, Johnson KA (1992) Large scale purification and biochemical characterization of T7 primase/helicase proteins. Evidence for homodimer and heterodimer formation. J Biol Chem 267: 15013–15021 [PubMed] [Google Scholar]
- Picha KM, Ahnert P, Patel SS (2000) DNA binding in the central channel of bacteriophage T7 helicase-primase is a multistep process. Nucleotide hydrolysis is not required. Biochemistry 39: 6401–6409 [DOI] [PubMed] [Google Scholar]
- Picha KM, Patel SS (1998) Bacteriophage T7 DNA helicase binds dTTP, forms hexamers, and binds DNA in the absence of Mg2+. The presence of dTTP is sufficient for hexamer formation and DNA binding. J Biol Chem 273: 27315–27319 [DOI] [PubMed] [Google Scholar]
- Potter H, Dressler D (1976) Mechanism of genetic-recombination—electron-microscopic observation of recombination intermediates. Proc Natl Acad Sci USA 73: 3000–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasnik I, McKinney SA, Ha T (2006) Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods 3: 891–893 [DOI] [PubMed] [Google Scholar]
- Toth EA, Li Y, Sawaya MR, Cheng Y, Ellenberger T (2003) The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol Cell 12: 1113–1123 [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials