Abstract
Mitogen-activated protein kinase kinase 4/c-Jun NH2-terminal kinase kinase 1 (MKK4/JNKK1; hereafter referred to as MKK4) is a dual-specificity kinase with a critical role in regulating the activity of c-Jun NH2-terminal kinase and p38 kinases. We identified a novel biological function for MKK4 in the regulation of growth of ovarian and prostate cancer metastases. Clinical correlative studies showed that MKK4 protein levels were reduced in high-grade prostate cancer and prostate and ovarian cancer metastases compared with normal tissue, which prompted investigation into the mechanism(s) responsible for down-regulation of MKK4 in a panel of cancer cell lines. Initial studies found that low levels of MKK4 protein did not correlate with either exon deletion or decreased levels of MKK4 mRNA, suggesting that MKK4 protein levels were regulated posttranscriptionally by either reduced translation or reduced protein stability. Endogenous MKK4 was highly stable and not subject to altered proteolysis. Instead, MKK4 biosynthesis seemed to be regulated by altered translation. In support of this assertion, we found that cytosolic MKK4 mRNA was shifted toward active polysomes in cells with higher levels of MKK4 protein, suggesting that MKK4 mRNA was translated more efficiently in these cells. This study supports a novel mechanism for the regulation of MKK4 protein levels. Further, these findings have potential therapeutic implications for modulating the expression of a signaling kinase involved in the regulation of metastatic growth.
Introduction
Mitogen-activated protein kinase kinase 4/c-Jun NH2-terminal kinase kinase 1 (MKK4; also known as JNKK1, MEK4, and SEK1) is a widely expressed dual-specificity mitogen-activated protein kinase kinase and a critical mediator of stress-activated protein kinase signaling (1–3). In response to cytokines or damaging stimuli such as pH changes and hypoxia, activated MKK4 can phosphorylate either the c-Jun NH2-terminal kinase or p38 mitogen-activated protein kinases. Depending on the environment of the cells and the specific signal, MKK4 activation and the subsequent activation of c-Jun NH2-terminal kinase and/or p38 mitogen-activated protein kinases can lead to a variety of biological responses, including cell differentiation, growth, and death (4–6). Homozygous knockout of MKK4 in mice is embryonic lethal (7–9), and MKK4-null cells have deficiencies in c-Jun NH2-terminal kinase–dependent and p38-dependent signaling (9).
We previously identified a novel function for MKK4 in the regulation of metastasis formation in both prostate and ovarian cancer models. Clinical correlative studies suggested the involvement of MKK4 down-regulation in the acquisition of metastatic ability of certain cancers. Specifically, immunohistochemical studies showed high levels of MKK4 protein in the epithelial, but not the stromal compartment, of normal prostatic and ovarian tissues. MKK4 protein was found to be decreased in advanced prostate cancers and ovarian cancer metastases, and the gene is infrequently mutated in cancer metastases (10, 11). Similarly, in prostate and ovarian cancer cell lines, expression of MKK4 was lost or down-regulated in many of the cell lines evaluated (10, 11). Immunohistochemical analysis of pancreatic cancers revealed that decreases in MKK4 protein expression correlated with poor prognosis (12). Additionally, in a study of breast cancer metastases in the brain, Stark et al. (13) reported finding decreased MKK4 mRNA by reverse transcription-PCR and corresponding decreases in protein levels. These studies show that the MKK4 protein is frequently down-regulated in cancers and support a role for dysregulation of its signaling cascade in clinical disease.
The mechanism(s) of MKK4 inactivation in cancer is not well understood. Initial studies found that 15% of breast cancer cell lines harbored MKK4 genetic mutations (14, 15), but additional studies suggest that the percentage of mutation in breast cancers may be much lower, comparable with the 2% to 5% of mutations found in all cancer lines tested to date (16). Consistent with cell line data, one group reported that 5% of ovarian tumors had deletion of the MKK4 gene locus, whereas the remaining 95% of samples retained wild-type MKK4 sequence and expressed MKK4 mRNA (17). Chae et al. (18) reported finding no MKK4 mutations in gastric carcinoma cell lines as well as no decrease in MKK4 mRNA or protein in primary gastric carcinomas.
Taken together, findings from clinical and experimental studies prompted the question, “How is MKK4 protein down-regulated in cancer cells?” Although the kinase activity of MKK4 has been studied extensively, there is a dearth of information about the mechanisms that control endogenous protein levels of mitogen-activated protein kinase kinases. We sought to determine the mechanism by which MKK4 protein levels are modulated using high MKK4–expressing and low MKK4–expressing prostate and ovarian cancer cell lines as a model system.
To test the hypothesis that decreased wild-type MKK4 protein expression levels could result from posttranscriptional regulatory events, we studied the mechanisms controlling protein expression in high MKK4–expressing and low MKK4–expressing human cancer cell lines. Studies were conducted to determine whether the two cell types displayed differences in (a) the relative stability of MKK4 protein and (b) the relative translation of the MKK4 mRNA. Our studies found that MKK4 protein was highly stable in all cell lines tested and displayed no differential sensitivity to protease inhibitors. The compartmentalization of the MKK4 mRNA was also unchanged when comparing high- and low-expressing cells, all showing a highly stable transcript with a predominantly nuclear localization. Interestingly, however, within the cytoplasmic MKK4 mRNA subset, there was a distinct increase in the association of the MKK4 mRNA with the translational machinery in high MKK4–expressing cells compared with low MKK4–expressing cells. These findings suggest that MKK4 expression levels are influenced by the translational engagement of the MKK4 mRNA.
Results
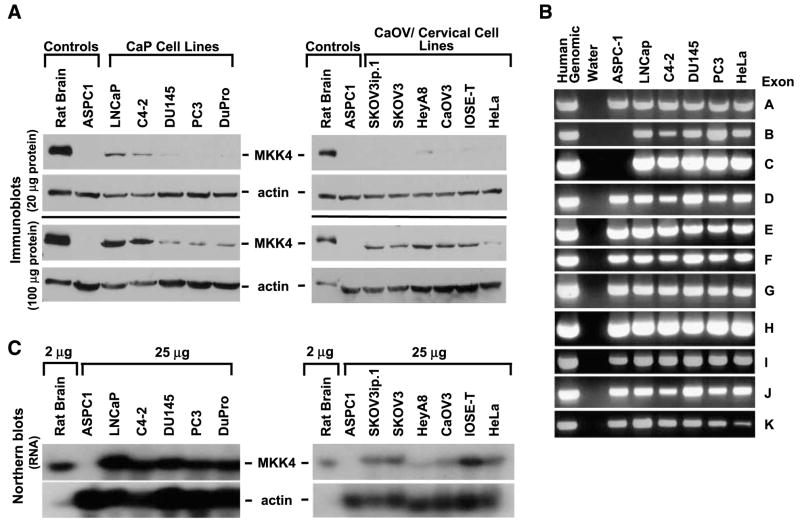
We previously observed low endogenous MKK4 levels in many human prostate and ovarian cancer cell lines (10, 11). As shown in Fig. 1A, the human prostate cancer cell lines DU145, PC3, and DuPro have low to undetectable MKK4 protein levels under standard assay conditions (20 μg). Immunoblotting with 100 μg of total protein facilitated detection of MKK4 in all cell lines examined and revealed that low MKK4–expressing cell lines have ~20-fold less MKK4 compared with the high MKK4–expressing lines LNCaP and C4-2 (Fig. 1A, left). Cell lines of ovarian and cervical origin were examined to extend this initial finding. Interestingly, all of these cell lines had low endogenous levels of MKK4 protein, which could only be detected when 100 μg of total protein were examined. To rule out the possibility that decreased levels of MKK4 protein were due to genomic deletion(s), cell lines were screened for the presence of MKK4 exons A to K. Representative data are shown in Fig. 1B. Total human genomic DNA was used as a positive control, whereas DNA prepared from the human pancreatic cancer cell line ASPC-1, which harbors homozygous deletion of exons B and C of MKK4, was used as a negative control (14). All exons of the MKK4 gene were present in each of the four prostate cancer cell lines (PC3, LNCaP, C4-2, and DU145; Fig. 1B). Similarly, three ovarian cancer cell lines (HeyA8, SKOV3, and SKOV3ip.1) were positive for all exons (data not shown). To test the possibility that decreased MKK4 levels were due to decreased levels of MKK4 RNA, the relative amount of MKK4 mRNA present in low- and high-expressing cell lines was assessed. Northern blot analysis showed that both high- and low-expressing prostate cancer cell lines had comparable levels of MKK4 mRNA (Fig. 1C, left). Similar results were obtained in ovarian cancer cell lines, as robust expression of MKK4 mRNA was detected in HeyA8, SKOV3, SKOV3ip.1, and CaOV3 (Fig. 1C, right), although expression is lower than prostate cancer cell lines. Moreover, HeLa cervical cancer cells and IOSE-T immortalized ovarian epithelial cells have similar levels of MKK4 mRNA. Actinomycin D treatment of PC3, LNCaP, and SKOV3ip.1 cells revealed that MKK4 mRNA stability is similar between cell lines (Supplementary Fig. S1). The discrepancy between the relative levels of MKK4 protein and mRNA observed in many of these cell lines led us to hypothesize that the steady-state level of MKK4 protein is regulated by a posttranscriptional mechanism.
FIGURE 1.
Low levels of MKK4 protein do not correlate with genomic deletions or low levels of MKK4 mRNA. A. MKK4 protein expression in a panel of prostate (CaP), ovarian, and cervical cancer cell lines. Protein (20 or 100 μg) was immunoblotted for MKK4. Rat brain (5 μg) and ASPC-1 (20 or 100 μg) cell protein served as positive and negative controls for MKK4 expression, respectively. β-Actin was used as a loading control. Immunoblots are representative of data from six independent experiments. B. Genomic DNA was isolated and PCRs were done with nested primers specific for each of the 11 exons (exons A-E) of the MKK4 gene. Human genomic DNA was used as a positive control for all reactions. Water and ASPC-1, a pancreatic cancer cell line harboring a homozygous deletion of MKK4 exons B and C, served as negative controls. Data are representative of two independent experiments. C. MKK4 mRNA expression in cancer cell lines. Polyadenylated mRNA was isolated from total RNA for Northern blotting with a probe complementary to the MKK4 coding region. Rat brain and ASPC-1 cells served as positive and negative controls for MKK4 expression, respectively. Representative data from three independent experiments are shown.
We next considered the possibility that low levels of MKK4 protein were due to differential MKK4 protein turnover. A prominent protein elimination system is the ubiquitin-proteasome degradation pathway, used by cells to regulate protein levels and to degrade proteins to recycle their amino acid components to form new proteins (19). PC3, LNCaP, HeLa, and SKOV3ip.1 cells were treated with the proteasome inhibitors ALLN (N-Acetyl-Leu-Leu-Norlea-al; inhibitor of the proteasome and cysteine proteases), lactacystin (specific inhibitor of proteasome), or vehicle control, with cell lysates collected at specific time intervals followed by immunoblotting for MKK4. As shown in Fig. 2A (and Supplementary Fig. S2), treatment with proteasome inhibitors did not result in the accumulation of MKK4 protein, indicating that proteasome-mediated degradation does not contribute significantly to the regulation of MKK4 abundance and suggesting that MKK4 is a stable protein. The levels of the proteasome-regulated protein MKP-1 were assessed as an internal control. As anticipated, MKP-1 protein accumulated following treatment with inhibitors (20).
FIGURE 2.
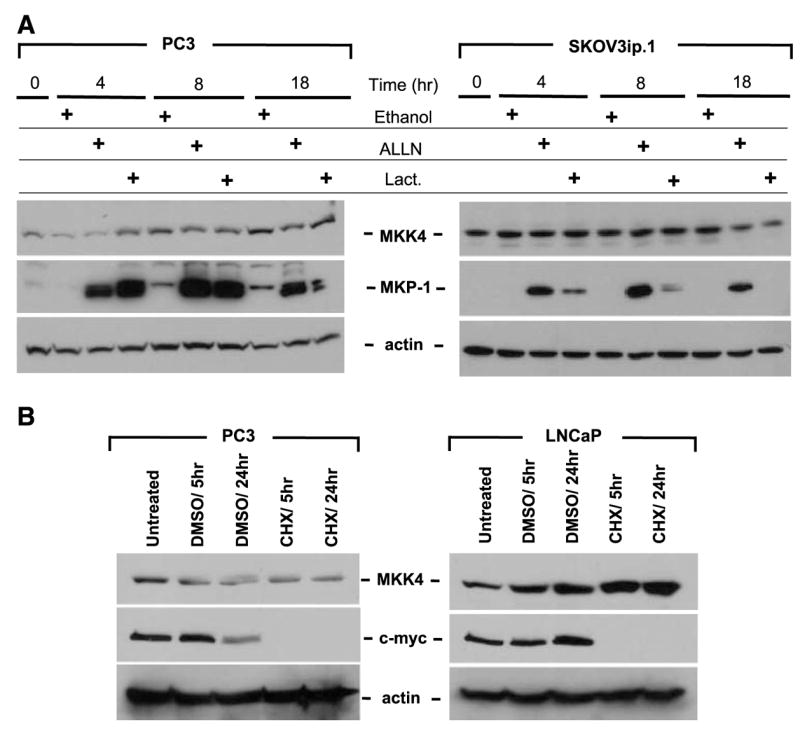
The MKK4 protein is highly stable and inhibition of the proteasome does not increase MKK4 protein levels. A. Cells were incubated with medium supplemented with proteasome inhibitors (ALLN or lactacystin) or vehicle control (ethanol) for the indicated times. Protein lysates were prepared from PC3 and SKOV3ip.1 cells. Total protein (100 μg) was immunoblotted for MKK4. Membranes were stripped and reprobed for MKP-1 as a control for inhibition of proteasome function and subsequently stripped and reprobed for β-actin as a control for protein loading. Loss of MKP-1 expression in the lactacystin 18-h time points may be due to hydrolysis of the covalent linkage between lactacystin and the proteasome (42). Representative data from at least three experiments are shown. B. Treatment of prostate cancer cells with cycloheximide (CHX) does not result in degradation of MKK4 protein. PC3 and LNCaP cells were incubated with cycloheximide or DMSO vehicle control for 5 or 24 h. Top, protein (100 μg) was immunoblotted for MKK4; middle, membranes were stripped and reprobed with an antibody against c-myc as a control for inhibition of protein synthesis; bottom, membranes were stripped and reprobed for β-actin as a loading control. Representative data from at least three experiments are shown.
To determine the relative stability of MKK4 protein in cells expressing different MKK4 levels, we treated PC3, LNCaP, SKOV3ip.1, and HeLa cells with cycloheximide to block ribosomal function. We then assessed MKK4 protein levels by immunoblotting to monitor the rate of MKK4 protein loss, which serves as a measure of its relative half-life. Figure 2B illustrates the finding that MKK4 levels were unaltered even after 24 h of cycloheximide treatment in both LNCaP (high MKK4–expressing cells) and PC3 (low MKK4–expressing cells), further indicating that the protein is quite stable. c-myc was used as a control for this assay, as it has a short half-life (21). Cycloheximide treatment of SKOV3ip.1 and HeLa cells yielded similar results(data not shown). These data suggest that MKK4 protein is highly stable and that protein degradation is unlikely to contribute to MKK4 down-regulation in low-expressing cell lines.
Next, we examined the possibility that decreased protein production is due to a mechanism involving RNA transport and/or translation. A critical, although often overlooked, step in gene regulation is the transport of mRNA from the nucleus to the cytoplasm, where it is translated into protein (22). The bulk of cellular mRNAs seems to be exported at constant rates, but mRNA transport has been shown to critically dictate the expression patterns for some genes (23). To test the possibility that MKK4 mRNA is inefficiently transported from the nucleus in low MKK4–expressing cells, PC3 and LNCaP cells were fractionated into cytosolic and nuclear components. Northern blotting of cellular fractions showed that MKK4 mRNA is primarily nuclear in both cell lines (Fig. 3) and was not preferentially elevated in the cytoplasm of LNCaP cells. Protein was also isolated from fractions and cytoplasmic and nuclear markers (α-tubulin and lamin A/C, respectively) were immunoblotted to confirm the complete and specific preparation of cytoplasmic and nuclear lysates.
FIGURE 3.
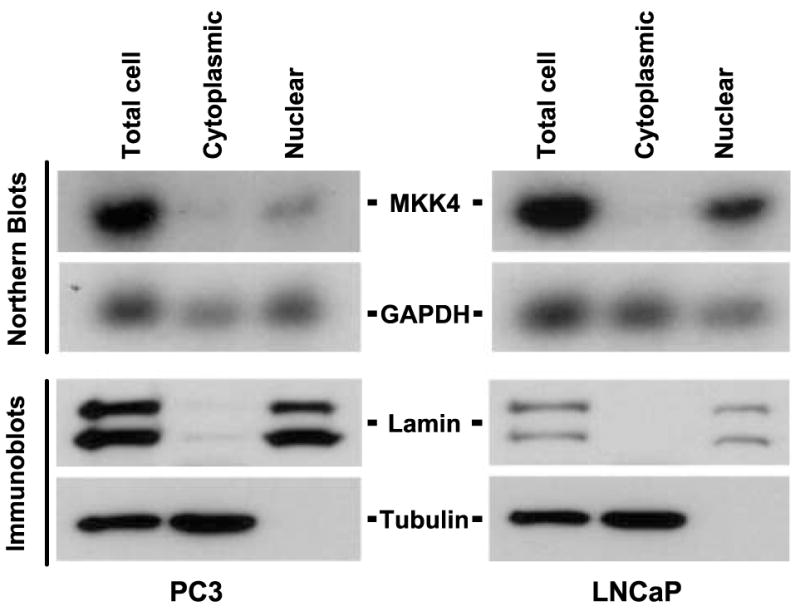
MKK4 mRNA is primarily nuclear. PC3 and LNCaP cells were fractionated into nuclear and cytoplasmic components, and RNA and protein samples were prepared. Total RNA (5 μg) was used for Northern blot analysis to detect MKK4 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs. Protein (20 μg) was used for immunoblotting for the nuclear markers lamin A and C, and the cytoplasmic marker α-tubulin. Data are representative of three independent experiments.
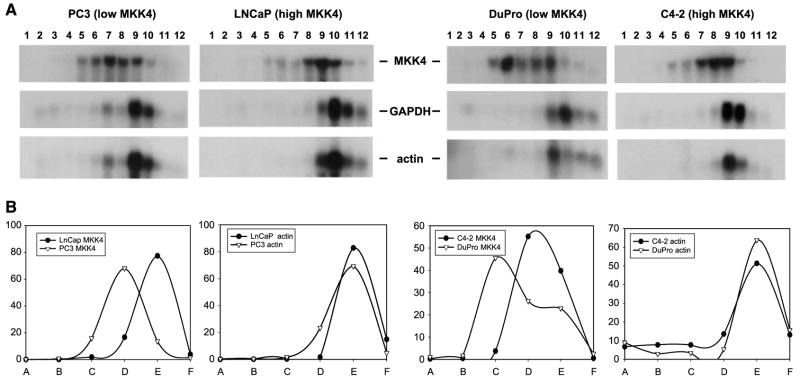
Having systematically excluded the involvement of key gene regulatory mechanisms in determining MKK4 abundance, we asked whether the relative translation rate of MKK4 mRNA may underlie the observed differences in MKK4 protein levels. To this end, we studied the association of the MKK4 mRNA with the translational machinery in cells expressing different levels of MKK4. Although there are many levels of control within the process of translation, translational initiation is often the rate-limiting step (24). The association of a given mRNA with high–molecular weight polysomes (complexes of multiple ribosomes translating a single mRNA) reveals the degree to which that mRNA has completed initiation and is engaged in active translation. Consequently, analysis of polysome-associated mRNA provides a measure of the synthesis rate of the corresponding protein (25). To ascertain the translational status of the MKK4 mRNA, we investigated its relative association with the translational apparatus. Cytoplasmic lysates (containing ribosomal subunits, individual ribosomes, and polysomes of increasing molecular weight) were size fractionated using sucrose density gradients into 12 individual fractions. Following the extraction of total RNA, the relative levels of the MKK4 and glyceraldehyde-3-phosphate dehydrogenase and β-actin housekeeping gene mRNAs in each fraction were examined by Northern blot analysis. As expected, housekeeping gene mRNAs clustered in two to three fractions, serving as a reference to indicate fractions containing actively translating, higher-order polysomes as well as a control to ensure sufficient size fractionation. A comparison of lysates from low MKK4–expressing (PC3 and DuPro) and high MKK4–expressing (LNCaP and C4-2) cells revealed that the MKK4 mRNA in LNCaP and C4-2 cells was preferentially associated with polysomes of higher molecular weight, indicating a greater engagement of MKK4 in translation in these cells (Fig. 4). Together, these observations point to the critical involvement of translational regulation as a key process governing the levels of MKK4 protein.
FIGURE 4.
Polysome analysis of cell lines expressing low (PC3 and DuPro) and high (LNCaP and C4-2) levels of MKK4 protein. A. Cytoplasmic cell lysates were separated in sucrose density gradients and fractions were collected. Fraction 1 corresponds to the lightest fraction; fraction 12, the heaviest. Total RNA was prepared from each fraction, separated by gel electrophoresis, and transferred to membranes for Northern blotting with a probe spanning the MKK4 coding region. Membranes were stripped and reprobed to monitor the expression of glyceraldehyde-3-phosphate dehydrogenase mRNA (encoding a housekeeping gene) in these fractions. Membranes were subsequently stripped and reprobed for expression of β-actin mRNA, encoding a second housekeeping gene. These two genes had strikingly similar mRNA patterns, supporting their use as a measure of actively translating polysome fractions. Representative data from three experiments are shown. B. Graphic representative of polysome analysis. The two pairs of low MKK4–expressing versus high MKK4–expressing cells were arbitrarily selected (PC3 versus LNCaP and DuPro versus C4-2). Pooled fractions are indicated as follows: fractions 1 and 2 (A), fractions 2 and 3 (B), fractions 4 and 5 (C), etc.
Discussion
A topic of increasing interest is determining how the levels and activities of metastasis suppressor proteins, as well as other cancer-related proteins, are regulated. Our data support a model in which MKK4 protein levels are controlled by a translational regulatory mechanism via differential recruitment of the MKK4 mRNA to polysomes. Initial observations indicated that some cell lines express abundant MKK4 mRNA yet very low levels of MKK4 protein. A series of studies was conducted to rule out gross DNA alterations and differential message stability, proteasomal degradation, turnover, or RNA transport as mechanisms responsible for differences in MKK4 protein levels. At the same time, we conducted complementary studies showing that there is increased engagement of MKK4 mRNA on higher-order polysomes in cells with high levels of protein compared with cells with low levels of MKK4 protein. Translational efficiency is determined by sequence (“cis”) elements within the mRNA and by “trans” factors that interact with these sequences. In addition to ubiquitous constituents of virtually all mammalian mRNAs (the 5′ cap structure and 3′-polyadenylic acid tail), several structural features and regulatory sequences within the mRNA can influence translational efficiency. Theses include internal ribosomal entry sites, which mediate cap-independent translation initiation; upstream open reading frames, which normally reduce translation from the main open reading frame; secondary or tertiary RNA structures, such as hairpins; and regulatory sites within untranslated regions (26). At this time, there is no information about structural elements within the MKK4 mRNA.
Cancer metastasis is a complex process that begins with the formation of a primary tumor and culminates in the formation of overt metastases at a discontinuous secondary site(s). We are in the midst of a revolution in the way we view metastasis, and the study of metastasis suppressor proteins is revealing new facets of the molecular regulation of this complex process. The existence of a translational mechanism for control of MKK4 protein levels in prostate cancer cells, and possibly other cancer types, fits well into the overall view of metastasis suppressor regulation. Metastatic cells may need to quickly adapt to a new environment by altering levels of metastasis-related proteins. It may not be favorable for cancer cells to permanently alter metastasis genes (through mutation), as their function may be required at one or more steps of metastasis formation. Therapies based on “reexpression” of metastasis suppressors may be feasible if posttranscriptional mechanisms, such as translational suppression, control their expression. More than one mechanism may contribute to the posttranscriptional regulation of MKK4. A recent study of ovarian cancer is consistent with this view. Spillman et al. (27) found no evidence of MKK4 promoter methylation, only small variations in MKK4 mRNA levels, and low levels of MKK4 protein (compared with normal epithelium) in a set of primary ovarian cancers. However, the majority of MKK4 protein seemed to be in the inactivated (unphosphorylated) form. Thus, it is possible that two mechanisms exist for cancer cells to rapidly change levels of MKK4 kinase activity (translation and phosphorylation). Although several studies have shown the contribution of transcriptional regulation in down-regulation of metastasis suppressors (28, 29), this is the first investigation to reveal the translational regulation of a metastasis suppressor. There is strong precedent for the involvement of alterations in protein translation in tumor initiation and progression on both the global and individual transcript level (30–34). As we expand our understanding of MKK4-mediated metastasis suppression, elucidating the specific mechanisms governing endogenous MKK4 expression will be essential for identifying novel targets for therapeutic intervention.
The MKK4 mRNA untranslated regions may contain sequences responsible for regulating translation of the transcript through interactions with regulatory molecules, such as RNA-binding proteins (RBP) and microRNAs (miR; ref. 35). RBPs are a large group of proteins, and although many of these are required for the normal biogenesis of mRNAs, others directly regulate the amount of protein synthesized from a given transcript. miRs function as posttranscriptional regulators of gene expression through several different mechanisms, including promoting RNA degradation, repressing translation initiation, and blocking protein elongation, and possibly additional mechanisms, such as engaging in complexes of sequestered mRNAs (26, 36). Normal cells may require a certain level of RBP(s) and/or miR(s) to maintain appropriate translation of MKK4 mRNA and thus wild-type protein levels. If the levels or function of positive regulatory molecules are decreased, less mRNA is translated. Alternatively, cancer cells with less efficient translation could have up-regulated a RBP or miR that binds to the untranslated region to prevent translation. In other words, there could be a mechanism required for maintaining MKK4 protein levels in normal cells, which is altered in low MKK4–expressing cancer cells, or there could be a cancer-specific mechanism that is not involved in the normal regulation of MKK4 translation.
An important goal will be to identify the regulatory elements of the MKK4 mRNA sequence, presumably within the 5′- and/or 3′-untranslated regions. Heterologous reporter constructs may be helpful in this regard. An additional goal is to identify the translation-regulatory molecules that function by interacting with MKK4 mRNA, possibly RBPs and/or miRs. Several RBPs that regulate translation and have potential roles in cancer are candidates: HuR is a member of the ELAV family of RBPs (37) and may contribute to malignancy by controlling the expression of specific mRNAs (38). T-cell–restricted intracellular antigen-1 and T-cell–restricted intracellular antigen-1–related protein are two translational repressors that have roles in development and stress responses (39). The influence of these and other RBPs on MKK4 translation is a current topic of interest. In addition, examination of functional interactions between miRs with MKK4 mRNA awaits direct investigation.
Materials and Methods
Cell Lines and Culture Conditions
PC3, LNCaP, C4-2, DuPro, and DU145 human prostate carcinoma cell lines were cultured in RPMI 1640 (Mediatech) supplemented with 10% FCS (Atlanta Biologicals) and 1% penicillin (100 units/mL)/streptomycin (100 μg/mL; BioWhittaker/Cambrex). SKOV3ip.1, SKOV3, CaOV3, HeyA8 human ovarian carcinoma cells, IOSE-T transformed ovarian epithelial cells, and HeLa cervical carcinoma cells were cultured in DMEM with L-glutamine and high glucose (4.5 g/L; Mediatech) supplemented with 5% FCS, 1% penicillin/streptomycin, 10 mmol/L sodium pyruvate (Mediatech), 1× nonessential amino acids (Mediatech), and 2× MEM vitamin solution (Mediatech). ASPC-1 cells were grown in DMEM supplemented with 20% FCS and 1% penicillin/streptomycin (Mediatech).
PCR
Nested PCR was done using standard methods (New England Biolabs). Products were run on 2% agarose/Tris-borate EDTA gels containing 0.2 μg/mL ethidium bromide for visualization and photographed. Primer sequences used for amplification of the 11 exons of MKK4 were as previously described (14).
Protein Lysate Preparation and Immunoblotting
Total protein lysates were prepared using radioimmunoprecipitation assay buffer supplemented with protease inhibitors and quantified as described (40). Total protein (20–100 μg) was resolved by SDS-PAGE on 10% gels and transferred to nitrocellulose membranes, and immunoblotting was conducted as described (41). The antibodies and dilutions were as follows: MKK4 (1:5,000; Santa Cruz Biotechnology), c-myc (1:500; Cell Signaling), MKP-1 (1:500; Santa Cruz Biotechnology), tubulin (1:1,000; Santa Cruz Biotechnology), lamin A/C (1:2,000; Upstate), β-actin (1:10,000; Calbiochem), anti-rabbit horseradish peroxidase (1:10,000; Cell Signaling), and anti-mouse IgG-horseradish peroxidase (1:10,000; Sigma). Densitometric analysis was done using Un-Scan-It software (Silk Scientific). MKK4 protein expression was normalized against actin protein expression from the corresponding lysates. The ratios LNCaP/PC3 and C4-2/DuPro were calculated to yield fold increases of 23.81 and 16.15, respectively.
Northern Blotting
Total RNA was extracted with Trizol reagent as directed (Invitrogen). For polyadenylated mRNA isolation, Oligotex oligo(dT) bead technology was used (Qiagen). RNA was resolved on 1.2% agarose/MOPS gels (Cambrex), imaged, and photographed. Capillary transfer of RNA onto a Zeta-Probe membrane (Bio-Rad) was done overnight in 10× SSC and subjected to UV cross-linking. Random primer labeling of probes with [γ-32P]dCTP was done using the Megaprime DNA Labeling System (Amersham). Unincorporated nucleotides were removed using Chroma Spin-100 columns (Clontech). Membranes were prehybridized for 30 min in ExpressHyb solution (Clontech) and hybridized with probe overnight (both at 65°C), and membranes were washed and exposed to film for 30 min to 24 h. cDNA probes were as follows: MKK4 is a 1-kb fragment of MKK4 cDNA isolated by BglI/SphI digestion of pLNCX2-jnkk1 plasmid, glyceraldehyde-3-phosphate dehydrogenase is a 0.9-kb fragment of human glyceraldehyde-3-phosphate dehydrogenase amplified by PCR (primer sequences: 5′-GAGTCAACGGATTTGGTCGT-3′ and 5′-TGAGCTTGACAAAGTGGTCG-3′), and β-actin was a 838-bp cDNA (BioChain).
Drug Treatments
Drug solutions and vehicle controls were prepared to final concentrations in the appropriate medium and cells were incubated for the indicated times. Concentrations were as follows: 200 μg/mL cycloheximide (Sigma), 10 μmol/L ALLN (Sigma), 10 μmol/L lactacystin (Sigma), 1 μL/mL ethanol, 10μg/mL actinomycin D (Sigma), and 2 μL/mL DMSO.
Polysome Isolation
Cells were incubated with cycloheximide (0.1 mg/mL) for 20 min, washed with 1× PBS [137 mmol/L NaCl, 10 mmol/L phosphate, 2.7 mmol/L KCl (pH 7.4)], lysed with polysome extraction buffer [0.3 mol/L NaCl, 15 mmol/L MgCl2, 15 mmol/L Tris-Cl (pH 7.6), 1% Triton X-100, 0.1 mg/mL cycloheximide, 1 mg/mL heparin], incubated on ice for 10 min, and centrifuged at 14,000 rpm for 10 min to remove nuclear and membrane material. The soluble protein concentration was measured as above to normalize the samples. Lysates (2 mg) were brought to a final volume of 1 mL with polysome extraction buffer and layered onto sucrose gradients. Sucrose gradients were prepared with 2.2 mL of 10% to 50% sucrose layers in 0.3 mol/L NaCl, 15 mmol/L MgCl2, 15 mmol/L Tris-Cl (pH 7.6), 0.1 mg/mL cycloheximide, and 1 mg/mL heparin and allowed to settle overnight at 4°C. Gradients were centrifuged in a Beckman Ultracentrifuge SW-41 rotor at 39,000 rpm for 90 min. Fractions (1 mL) were collected and vortexed with 1.5 mL of guanidine HCl, and RNA was precipitated with ethanol. Northern blotting was done as above.
Nuclear Fractionation
To prepare fractions, the BioVision Nuclear/Cytosol Fractionation kit was used according to the manufacturer’s instructions with the following addition: the RNase inhibitor Superasin (Ambion) was added to buffers for a final concentration of 0.1 unit/μL to preserve RNA integrity. Cells (1 × 106) were fractionated, and nuclear and cytoplasmic fractions were pooled and then divided in half for RNA and protein preparation as described above. Northern and immunoblotting analysis was done as described above.
Acknowledgments
We thank Dr. James O’Keefe for his assistance with data analysis.
Grant support: U.S. Army Medical Research and Command grants W81XWH-04-1-0852 (V.L. Robinson, O. Shalhav, and C.W. Rinker-Schaeffer) and W81-XWH-06-1-0041 (K. Otto and C.W. Rinker-Schaeffer); RO1 CA-89569 (V.L. Robinson, K. Otto, O. Shalhav, and C.W. Rinker-Schaeffer); and The University of Chicago Research, Cure, and Education fund.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
References
- 1.Sanchez I, Hughes RT, Mayer BJ, et al. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature. 1994;372:794 – 8. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 2.Derijard B, Raingeaud J, Barrett T, et al. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682 – 5. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 3.Lin A, Minden A, Martinetto H, et al. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science. 1995;268:286 – 90. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 4.Cuenda A. Mitogen-activated protein kinase kinase 4 (MKK4) Int J Biochem Cell Biol. 2000;32:581 – 7. doi: 10.1016/s1357-2725(00)00003-0. [DOI] [PubMed] [Google Scholar]
- 5.Robinson VL, Hickson JA, Vander Griend DJ, Dubauskas Z, Rinker-Schaeffer CW. MKK4 and metastasis suppression: a marriage of signal transduction and metastasis research. Clin Exp Metastasis. 2003;20:25 – 30. doi: 10.1023/a:1022586318678. [DOI] [PubMed] [Google Scholar]
- 6.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239 – 52. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 7.Nishina H, Vaz C, Billia P, et al. Defective liver formation and liver cell apoptosis in mice lacking the stress signaling kinase SEK1/MKK4. Development. 1999;126:505 – 16. doi: 10.1242/dev.126.3.505. [DOI] [PubMed] [Google Scholar]
- 8.Nishina H, Bachmann M, Oliveira-dos-Santos AJ, et al. Impaired CD28-mediated interleukin 2 production and proliferation in stress kinase SAPK/ERK1 kinase (SEK1)/mitogen-activated protein kinase kinase 4 (MKK4)-deficient T lymphocytes. J Exp Med. 1997;186:941 – 53. doi: 10.1084/jem.186.6.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganiatsas S, Kwee L, Fujiwara Y, et al. SEK1 deficiency reveals mitogen-activated protein kinase cascade crossregulation and leads to abnormal hepatogenesis. Proc Natl Acad Sci U S A. 1998;95:6881 – 6. doi: 10.1073/pnas.95.12.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HL, Vander Griend DJ, Yang X, et al. Mitogen-activated protein kinase kinase 4 metastasis suppressor gene expression is inversely related to histological pattern in advancing human prostatic cancers. Cancer Res. 2001;61:2833 – 7. [PubMed] [Google Scholar]
- 11.Yamada SD, Hickson JA, Hrobowski Y, et al. Mitogen-activated protein kinase kinase 4 (MKK4) acts as a metastasis suppressor gene in human ovarian carcinoma. Cancer Res. 2002;62:6717 – 23. [PubMed] [Google Scholar]
- 12.Xin W, Yun KJ, Ricci F, et al. MAP2K4/MKK4 expression in pancreatic cancer: genetic validation of immunohistochemistry and relationship to disease course. Clin Cancer Res. 2004;10:8516 – 20. doi: 10.1158/1078-0432.CCR-04-0885. [DOI] [PubMed] [Google Scholar]
- 13.Stark AM, Tongers K, Maass N, Mehdorn HM, Held-Feindt J. Reduced metastasis-suppressor gene mRNA-expression in breast cancer brain metastases. J Cancer Res Clin Oncol. 2005;131:191 – 8. doi: 10.1007/s00432-004-0629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teng DH, Perry WL, Hogan JK, et al. Human mitogen-activated protein kinase kinase 4 as a candidate tumor suppressor. Cancer Res. 1997;57:4177 – 82. [PubMed] [Google Scholar]
- 15.Su GH, Hilgers W, Shekher MC, et al. Alterations in pancreatic, biliary, and breast carcinomas support MKK4 as a genetically targeted tumor suppressor gene. Cancer Res. 1998;58:2339 – 42. [PubMed] [Google Scholar]
- 16.Su GH, Song JJ, Repasky EA, Schutte M, Kern SE. Mutation rate of MAP2K4/MKK4 in breast carcinoma. Hum Mutat. 2002;19:81. doi: 10.1002/humu.9002. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama K, Nakayama N, Davidson B, et al. Homozygous deletion of MKK4 in ovarian serous carcinoma. Cancer Biol Ther. 2006;5:630 – 4. doi: 10.4161/cbt.5.6.2675. [DOI] [PubMed] [Google Scholar]
- 18.Chae KS, Ryu BK, Lee MG, Byun DS, Chi SG. Expression and mutation analyses of MKK4, a candidate tumour suppressor gene encoded by chromosome 17p, in human gastric adenocarcinoma. Eur J Cancer. 2002;38:2048 – 57. doi: 10.1016/s0959-8049(02)00147-8. [DOI] [PubMed] [Google Scholar]
- 19.Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22:442 – 51. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 20.Lin YW, Chuang SM, Yang JL. ERK1/2 achieves sustained activation by stimulating MAPK phosphatase-1 degradation via the ubiquitin-proteasome pathway. J Biol Chem. 2003;278:21534 – 41. doi: 10.1074/jbc.M301854200. [DOI] [PubMed] [Google Scholar]
- 21.Luscher B, Eisenman RN. c-myc and c-myb protein degradation: effect of metabolic inhibitors and heat shock. Mol Cell Biol. 1988;8:2504 – 12. doi: 10.1128/mcb.8.6.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cullen BR. Nuclear RNA export. J Cell Sci. 2003;116:587 – 97. doi: 10.1242/jcs.00268. [DOI] [PubMed] [Google Scholar]
- 23.Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci U S A. 1996;93:1065 – 70. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sachs AB, Varani G. Eukaryotic translation initiation: there are (at least) two sides to every story. Nat Struct Biol. 2000;7:356 – 61. doi: 10.1038/75120. [DOI] [PubMed] [Google Scholar]
- 25.Pradet-Balade B, Boulme F, Beug H, Mullner EW, Garcia-Sanz JA. Translation control: bridging the gap between genomics and proteomics? Trends Biochem Sci. 2001;26:225 – 9. doi: 10.1016/s0968-0004(00)01776-x. [DOI] [PubMed] [Google Scholar]
- 26.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827 – 35. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spillman MA, Lacy J, Murphy SK, et al. Regulation of the metastasis suppressor gene MKK4 in ovarian cancer. Gynecol Oncol. 2007;105:312 – 20. doi: 10.1016/j.ygyno.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JH, Kim B, Cai L, et al. Transcriptional regulation of a metastasis suppressor gene by Tip60 and β-catenin complexes. Nature. 2005;434:921 – 6. doi: 10.1038/nature03452. [DOI] [PubMed] [Google Scholar]
- 29.Ouatas T, Halverson D, Steeg P. Dexamethasone and medroxyprogesterone acetate elevate Nm23-H1 metastasis suppressor expression in metastatic human breast carcinoma cells: new uses for old compounds. Clin Cancer Res. 2003;9:3763 – 72. [PubMed] [Google Scholar]
- 30.Rajasekhar VK, Holland EC. Postgenomic global analysis of translational control induced by oncogenic signaling. Oncogene. 2004;23:3248 – 64. doi: 10.1038/sj.onc.1207546. [DOI] [PubMed] [Google Scholar]
- 31.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179 – 92. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 32.Clemens MJ, Bommer UA. Translational control: the cancer connection. Int J Biochem Cell Biol. 1999;31:1 – 23. doi: 10.1016/s1357-2725(98)00127-7. [DOI] [PubMed] [Google Scholar]
- 33.Holland EC, Sonenberg N, Pandolfi PP, Thomas G. Signaling control of mRNA translation in cancer pathogenesis. Oncogene. 2004;23:3138 – 44. doi: 10.1038/sj.onc.1207590. [DOI] [PubMed] [Google Scholar]
- 34.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189 – 99. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 35.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390 – 4. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 36.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653 – 62. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 37.Gorospe M. HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle. 2003;2:412 – 4. [PubMed] [Google Scholar]
- 38.Lopez de Silanes I, Olmo N, Turnay J, et al. Acquisition of resistance to butyrate enhances survival after stress and induces malignancy of human colon carcinoma cells. Cancer Res. 2004;64:4593 – 600. doi: 10.1158/0008-5472.CAN-04-0711. [DOI] [PubMed] [Google Scholar]
- 39.Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963 – 9. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 40.Vander Griend DJ, Kocherginsky M, Hickson JA, Stadler WM, Lin A, Rinker-Schaeffer CW. Suppression of metastatic colonization by the context-dependent activation of the c-Jun NH2-terminal kinase kinases JNKK1/MKK4 and MKK7. Cancer Res. 2005;65:10984 – 91. doi: 10.1158/0008-5472.CAN-05-2382. [DOI] [PubMed] [Google Scholar]
- 41.Berger JC, Vander Griend DJ, Robinson VL, Hickson JA, Rinker-Schaeffer CW. Metastasis suppressor genes: from gene identification to protein function and regulation. Cancer Biol Ther. 2005;4:805 – 12. doi: 10.4161/cbt.4.8.1865. [DOI] [PubMed] [Google Scholar]
- 42.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397 – 403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]