SUMMARY
The molecular mechanisms underlying pluripotency and lineage specification from embryonic stem (ES) cells are largely unclear. Differentiation pathways may be determined by the targeted activation of lineage specific genes or by selective silencing of genome regions during differentiation. Here we show that the ES cell genome is transcriptionally globally hyperactive and undergoes global silencing as cells differentiate. Normally silent repeat regions are active in ES cells and tissue-specific genes are sporadically expressed at low levels. Whole genome tiling arrays demonstrate widespread transcription in both coding and non-coding regions in pluripotent ES cells whereas the transcriptional landscape becomes more discrete as differentiation proceeds. The transcriptional hyperactivity in ES cells is accompanied by disproportionate expression of chromatin-remodeling genes and the general transcription machinery, but not histone modifying activities. Interference with several chromatin remodeling activities in ES cells affects their proliferation and differentiation behavior. We propose that global transcriptional activity is a hallmark of pluripotent ES cells that contributes to their plasticity and that lineage specification is strongly driven by reduction of the actively transcribed portion of the genome.
INTRODUCTION
Embryonic stem (ES) cells are unique in their capacities to self-renew and to initiate differentiation into any cell type of the three germ layers. These opposing abilities are in part brought about by the presence of stem cell specific factors (Hochedlinger et al., 2005; Hough et al., 2006; Nichols et al., 1998; Pan et al., 2002; Scholer et al., 1990; Takahashi and Yamanaka, 2006). During differentiation, lineage-specific transcription factors activate the expression of specific sets of genes that are required for each specific lineage to form hierarchical transcription networks (Szutorisz and Dillon, 2005).
In addition to control by specific transcription factors, regulation via epigenetic mechanisms has recently emerged as a key mechanism in pluripotency and lineage specification (Azuara et al., 2006a; Bernstein et al., 2006; Boyer et al., 2006b; Buszczak and Spradling, 2006; Gan et al., 2007; Meshorer, 2007; Meshorer and Misteli, 2006). ES cell chromatin is characterized by several specific features, which distinguish it from that of somatic and differentiated cells (Niwa, 2007). ES cell chromatin is morphologically distinct in that heterochromatin is organized in larger and fewer domains, which become smaller, more abundant and hyper-condensed as cells differentiate (Aoto et al., 2006; Kobayakawa et al., 2007; Meshorer and Misteli, 2006; Park et al., 2004). Another characteristic feature of stem cell chromatin is the altered binding of chromatin proteins (Meshorer et al., 2006). Architectural chromatin proteins such as the heterochromatin component HP1, the linker histone H1 and core histones, display hyperdynamic and looser binding to chromatin in undifferentiated ES cells than in differentiated cells. Hyperdynamic binding is exclusively found in pluripotent cell types, but not in lineage-committed but undifferentiated cells, indicating that dynamic chromatin is associated with pluripotency rather than differentiation per se (Meshorer et al., 2006). ES cells also contain unique histone modification patterns (Spivakov and Fisher, 2007). Extensive regions of the genome are bivalently marked by transcriptionally repressive H3-triMeK27, but at the same time contain the transcription-associated histone modification H3-triMeK4 (Azuara et al., 2006b; Bernstein et al., 2006). It has been proposed that these “bivalent” domains silence developmentally-regulated genes in ES cells while keeping them poised for activation as cells enter the various differentiation pathways (Bernstein et al., 2006; Jorgensen et al., 2006). Repression of H3-triMeK27 appears to be mediated by the polycomb repression complex 2 (PRC2), which is associated with a significant number of developmental regulators (Boyer et al., 2006b; Lee et al., 2006).
A striking commonality amongst the ES cell-specific chromatin properties is that they are all indicative of transcriptionally active chromatin. We have suggested that ES cell genomes are globally transcriptionally hyperactive and express large regions of the genome, possibly indiscriminatorily and at low levels (Meshorer and Misteli, 2006). Here we have directly tested this hypothesis and demonstrate global, low-level transcriptional activity in the ES cell genome. We find elevated levels of total RNA and mRNA in pluripotent mouse ES cells, and we show that undifferentiated ES cells express repetitive sequences, mobile elements as well as lineage- and tissue-specific genes at low levels. Using whole-genome mouse tiling arrays we show that a larger fraction of the genome is active in ES cells compared to differentiating cells. The global transcriptional activity of the ES cell genome is accompanied by elevated levels of chromatin remodeling proteins and the global transcription machinery, but not histone modifying activities. Modulation of several specific chromatin remodeling activities in ES cells interferes with their proliferation and differentiation. Our results identify global, low level genome activity as a novel hallmark of ES cell genomes and they suggest that loss of pluripotency and lineage specification involves reduction of the actively transcribed portion of the genome.
RESULTS
Hallmarks of transcriptionally active chromatin in ES cells
ES cell chromatin is characterized by several distinct properties. For one, in ES cells, heterochromatin is organized in larger and fewer domains, which become smaller, more abundant and hyper-condensed as cells differentiate (Aoto et al., 2006; Kobayakawa et al., 2007; Meshorer and Misteli, 2006; Park et al., 2004). To characterize the ultrastructure of ES cell chromatin, we compared by quantitative electron microscopy undifferentiated mouse R1 ES cells with neuronal progenitor cells (NPC) derived from them by in vitro differentiation (Figure S1A, B. Consistent with global decondensation in ES cells, the majority of chromatin appears homogenous and decondensed in undifferentiated pluripotent cells (Figure S1A, left). In contrast, in R1 NPCs chromatin is heterogeneous in appearance and distinct heterochromatin domains are frequently present (Figure S1A, middle; Meshorer et al., 2006).
In addition to morphological differences, chromatin in ES cells is molecularly distinguished by a set of bivalent histone modifications of both an active and a repressive state (Azuara et al., 2006a; Bernstein et al., 2006). To extend these studies, we have compared the status of a series of histone modifications in undifferentiated R1 ES cells and NPCs derived from them (Figure S1C, D). We find enrichment of several histone marks commonly associated with active chromatin, including H3K4me3, H3K9ac, H3K14ac, H3K36me2 and H3K36me3 in ES cells compared to ES cell-derived NPCs (Figure S1C, D). The significantly elevated levels of the RNA Pol II mediated elongation-associated histone modification H3K36me2 (Bannister et al., 2005) (Figure S1C) are further suggestive of increased transcriptional activity in ES cells. The opposite behavior was observed for histone marks associated with transcriptional silencing. H4K20me2 is unchanged but the heterochromatin associated modification H3K9me3 is dramatically underrepresented in ES cells (Meshorer et al., 2006), consistent with the absence of heterochromatin domains in ES cells (Figure S1A, B; Meshorer et al., 2006). Similarly, a further hallmark of transcriptionally repressed genome regions, DNA methylation of cytidine (5meC) is also significantly reduced in ES cells compared to NPC (Bibikova et al., 2006) (Figure S1C, Figure S2).
Elevated global transcription in ES cells
Remarkably, the characteristic properties of ES cell chromatin including global decondensation, looser binding of architectural chromatin proteins, and enrichment of active histone modifications are all hallmarks of transcriptionally active chromatin. Based on these observations we hypothesized that ES cells are globally transcriptionally more active than differentiated cells (Meshorer and Misteli, 2006). To directly test this hypothesis, we measured global transcriptional activity by 3H-uridine incorporation (Kimura et al., 2002) in undifferentiated ES cells and 7d NPC derived from ES cells by in vitro differentiation (Lee et al., 2000; Meshorer et al., 2006). Strikingly, total RNA and mRNA levels (5–10% of total RNA) normalized to DNA content were almost 2-fold higher in ES cells compared to NPC (Figure 1A, p < 0.005 and 0.05, respectively, Mann-Whitney 2-tailed test). Higher levels of incorporated 3H-uridine were not due to increased RNA stability in ES cells since RNA decay rates were comparable in ES cells and NPC as demonstrated by pulse-chase labeling of newly synthesized total RNA (Figure 1B) and mRNA (Figure 1C).
Figure 1. Elevated global transcription in ES cells.
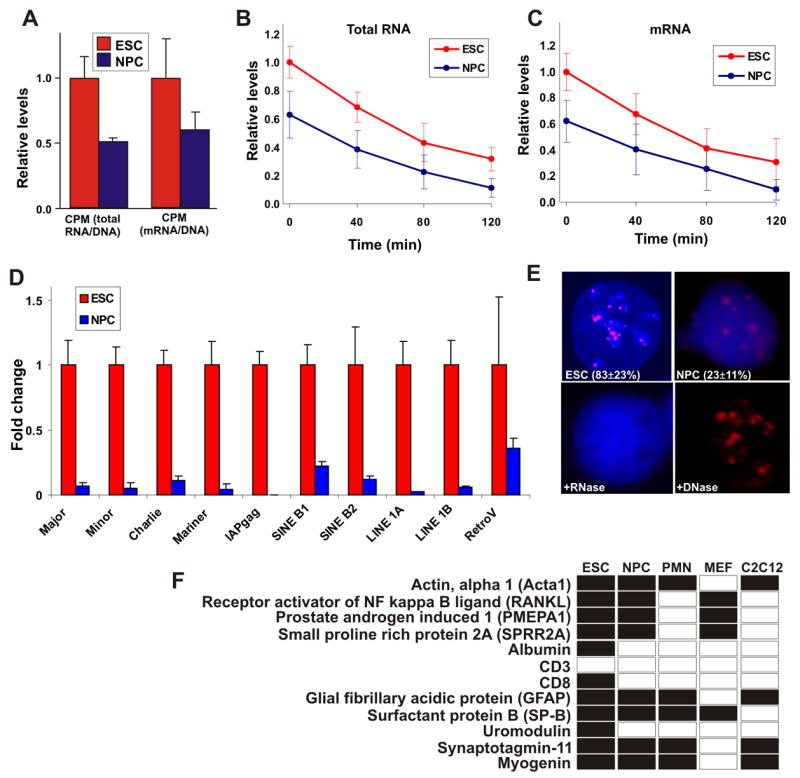
(A) Total RNA transcriptional activity (left) and mRNA transcriptional activity (right) in ES cells (red) and NPC (blue). Cells were incubated with 3H-labeled uridine for 4 hrs. Values represent averages ± SD from 3 experiments.
(B–C) As in (a) but following 2 hrs of incubation, 3H-uridine was removed and fresh medium supplemented with 0.125 μM actinomycin-D was added for additional 2 hrs. Samples were collected every 40 min and transcriptional activity of both total RNA (b) and mRNA (c) levels was determined.
(D) Real-time quantitative PCR of the indicated repeat sequences and transposable and retroviral elements in ES cells (red) and NPC (blue) normalized against Cyclophilin B. Values represent averages ± SD from 3 independent experiments.
(E) RNA-FISH for the major satellite repeat using Cy3-labled locked nucleic acid (LNA) probes in embryonic stem cell (ESC) and ES cells-derived neuronal progenitor cells (NPC). When ES cells were pretreated with RNase A signal was abolished (+ RNase), while DNase I treatment retained the signal (DNase). Values represent averages ± SD from 3 experiments. At least 50 cells were scored per experiment.
(F) Lineage-specific transcription in undifferentiated ES cells. Shown is a detection table (black, detected; white, undetected) of a selection of lineage-specific genes detected by RT-PCR in undifferentiated ES cells, NPC, ES cells-derived post-mitotic neurons (PMN), MEFs or differentiated C2C12 cells. Genes were considered not expressed when undetected in 2 independent experiments. Several genes required re-amplification for detection (Figure S3). All samples were treated similarly. For copy number determination see Supplementary Methods.
The elevated transcriptional activity in undifferentiated ES cells could either be due to the activity of a specific set of genes or might alternatively reflect global activation of the genome in ES cells. To begin to distinguish between these possibilities, we analyzed the activity status of satellite repeat sequences, LINEs, SINEs and several retrotransposons (Martens et al., 2005), which are normally repressed in differentiated cells. Transcription of all elements including major and minor satellite repeats, LINEs and SINEs was significantly higher in ES cells than in ES cells-derived NPC (Figure 1D; p < 0.05; normalized to the constantly expressed cyclophilin-B mRNA). While these elements were detected at very low levels in NPC, their expression was increased by 2–10 fold in ES cells (Figure 1D). We obtained similar results when undifferentiated ES cells were compared to other cell types, such as MEFs or differentiated C2C12 muscle cells (data not shown). To exclude the trivial possibility that the detection of these transcripts by RT-PCR was due to their expression in only a small subpopulation of cells in the ES cell population, we visualized major satellite repeat expression in undifferentiated (Figure 1E, ESC) and differentiated (Figure 1E, NPC) ES cells by RNA fluorescence in situ hybridization. Expression of major satellite repeats was detected in 83 ± 23% of the undifferentiated ES cells but only in 23 ± 11% of NPC (Figure 1E). When ES cells were pre-treated with RNAse, the signal was abolished (Figure 1E, + RNase), whereas DNase treatment left the signal intact (Figure 1E, + DNase). These results show that undifferentiated ES cells express regions of the genome that are normally silenced in differentiated cells.
Low level expression of tissue-specific genes in ES cells
To ask whether transcription of silent genome regions was limited to repeat sequences or was a general property of the ES cell genome, we probed the transcription status of specific genes using RT-PCR. We selected several tissue-specific genes and genes associated with terminal differentiation, which are not expected to be expressed in undifferentiated ES cells (Table S1). In order to avoid false positives originating from DNA contamination, all PCR primers were designed to flank long introns, so that the genomic products would be significantly longer than the cDNA products (Table S1). Transcripts for 11 out of 12 lineage-restricted genes were detected in undifferentiated ES cells (Figure 1F). The transcription level of these genes was very low with an estimated 0.25–20 copies per cell as determined by direct comparison with known quantities of plasmid cDNA (Supplemental Methods), suggesting their transcription likely occurs stochastically within the population. The low abundance prevented accurate quantification by real-time PCR in most cases, and some required re-amplification (Figure S3A). However, in the cases for which we were able to quantitatively compare transcription levels in ESC, 7d NPC and ES cells-derived fully differentiated post-mitotic neurons (PMN) by quantitative real-time PCR, transcription levels decreased during differentiation (Figure S3B). In 7-day NPC, 8 of 12 genes were still detected, but this number dropped to 5 in PMN (Figure 1F). As a control, we tested several differentiated cell lines. Most of the genes analyzed were silenced in MEFs and C2C12 muscle cells, except the ones specific to that particular lineage such as acta1 and myogenin in C2C12 cells and SPRR2A in MEFs. Interestingly, comparison with publicly available chromatin immunoprecipitation followed by genomic sequencing (ChIP-seq) data (Mikkelsen et al., 2007) revealed that all detected genes (except SPRR2A, for which ChIP data were not available) were marked with higher levels of both H3K4m3 and H3K27m3 in undifferentiated ES cells than in NPC, and the H3K27m3:H3K4m3 ratio increased following differentiation (Figure S4A–B), supporting increased silencing of non-related lineage-specific genes in the differentiated state. Attempts to detect protein products from these transcripts using Western blotting yielded negative results (data now shown).
Genome-wide transcriptional activity in ES cells
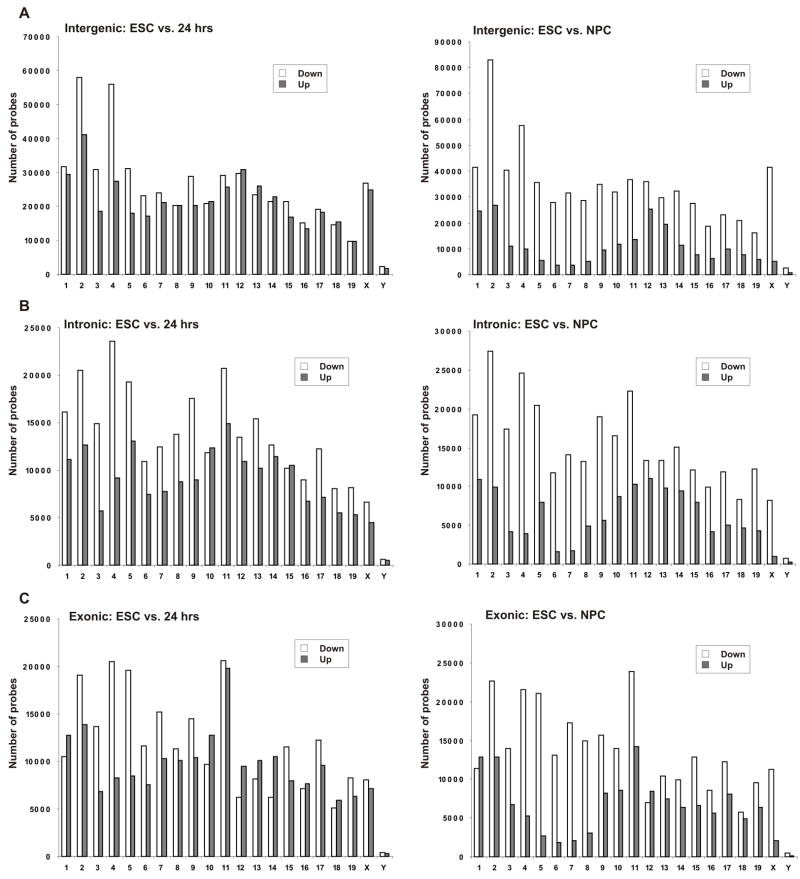
To systematically assess the transcriptional status of the entire genome and to extend our analysis to non-coding genome regions, we used an Affymetrix whole-genome mouse tiling array at 30 bp resolution to compare genome wide transcription profiles in pluripotent ES cells and in ES cells-derived NPC (see Methods for details). Microarray analysis was validated by the detection of downregulation of several stem cell specific genes including Oct4 and Sox15 during differentiation, whereas expression of neuronal genes including Sox4 and Sox11 (Bergsland et al., 2006) was increased as confirmed by qPCR (Figure S5A–B). The tissue-specific transcripts detected in ES cells by RT-PCR showed low-level or no transcription on the tiling array indicating the sensitivity limits of the microarray (Figure S5C). A detailed list of gene and transcript lengths, untranslated regions (UTRs) length and their averages are given in Table S2. To ask whether the genome of undifferentiated ES cells is globally transcriptionally more active, we first compared the number of positive probes on microarrays hybridized with poly(A)+ RNA from ES cells or from ES cells-derived 7d NPC. We used the number of single positive probes as a general surrogate for transcriptional activity. In support of increased global transcriptional activity in ES cells, the number of positive probes was significantly elevated in ES cells compared to NPC. While in ES cells 1,041,879 probes displayed intensity values above threshold (defined as 90% noise level, calculated using the mismatch probes data. See Materials and Methods for details), this number was reduced to 838,787 positive probes in NPC, corresponding to ~ 20% decrease (p < 10−5, 2-tailed Student’s t-test). The number of reduced probes is an underestimate since equal amounts of RNA rather than RNA from equal number of cells were hybridized and only RNAs of more than 200 nt transcripts in length were purified for use in hybridization. In addition, a stringent threshold was used to avoid measurements of false positive probes.
The reduction of detected probes occurred across all regions of the genome, including intergenic (Figure 2A), intronic (Figure 2B) and exonic (Figure 2C) domains. Importantly, the reduction was evident as early as 24 hrs after the withdrawal of LIF, demonstrating that the reduction in transcriptional activity is not restricted to a particular lineage and was not due to more rapid proliferation of undifferentiated ES cells, since ES cells after 24h withdrawal of LIF are still highly proliferative. The degree of reduction varied amongst chromosomes but was typically between 20–50% in NPC (Figure 2). Global transcriptional reduction was most prominent in intergenic and intronic regions. In these regions, probe counts were significantly reduced in 9–15 of the 21 chromosomes by 24 hrs and in 12–17 of the 21 chromosomes by 7 days. The remaining chromosomes did not show significant changes (Figure 2A–B). In exonic regions, 5 chromosomes displayed reductions at the 24 hrs time point and 11 chromosomes showed reduced counts in NPC (Figure 2C). Interestingly, in NPC two chromosomes (1 and 12) showed significantly elevated counts in exonic regions only. These are likely due to a disproportionately high number of differentiation-induced genes on these chromosomes, including Sox11, Sox17 and many others (Table S3). The overall reduction of active probes was not due to elevated levels of ribosomal genes as the rDNA bearing chromosomal regions are not represented on the array. To verify that the elevated number of probes in ES cells was not due to increased background noise, we analyzed the distribution of the probes that were exclusively present in ES cells but not in NPC. Slightly more than 50% of probes were clustered in groups of 3 or longer, corresponding to 100 bp or longer transcripts (Figure S6). Comparison of the size distribution of the positive probes with a random distribution using computer-assisted statistical simulations confirmed that they were contiguous transcripts (p < 10−6).
Figure 2. Whole-genome mouse tiling array analysis.
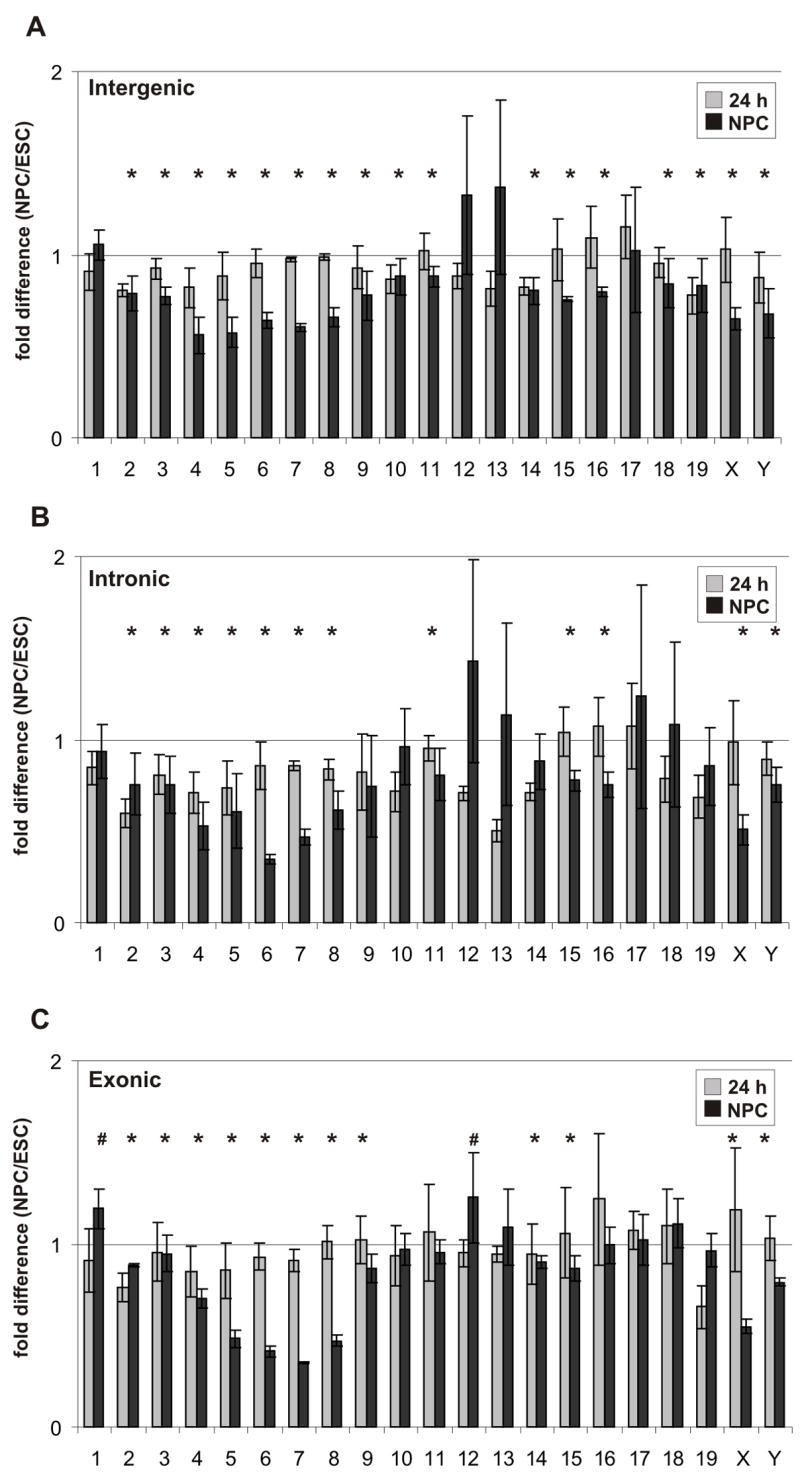
(A–C) Comparison of average fold-difference (± standard deviations) for positive probes from each chromosome between undifferentiated ES cells, cells 24 hrs after LIF withdrawal (gray columns) and NPC (black columns). The fold-difference is depicted relative to the 1.0 fold change shown as a straight line for intergenic regions (A), intronic regions (B) and exonic regions (C). Data are from 3 independent experiments. Asterisks denote significant reduction and number sign (#) denote significant increase between ESC and NPC (p < 0.017). P-values were estimated by one side hypothesis testing, adjusted with Bonferroni correction for multiple comparisons.
The more global transcriptional activity in ES cells was also evident from inspection of selected intergenic and intronic regions (Figure 3). Some intergenic regions in ES cells exhibited ‘transcription bursts’ displaying intermittent active and silent regions, which were reduced to near threshold levels in NPC (Figure 3A). In other cases, intergenic regions were active over large contiguous stretches in ES cells and activity was dramatically reduced over the entire region in NPC (Figure 3B). Some regions were characterized by ‘transcriptional islands’ in which parts of a region were active in both ES cells and NPC (Figure 3C). Significant transcription occurred in many cases along entire introns in ES cells (Figure 3D) or was often confined to a limited region of varying extent within the intron in ES cells, but was significantly reduced in NPC (Figure 3E, F). These reductions were specific, since numerous intronic regions were more highly expressed in NPC (Figure 3F). These analyses of intronic and intergenic regions point to a wide-spread elevated genomic transcriptional activity in undifferentiated ES cells.
Figure 3. Elevated intergenic and intronic transcription patterns in ES cells.
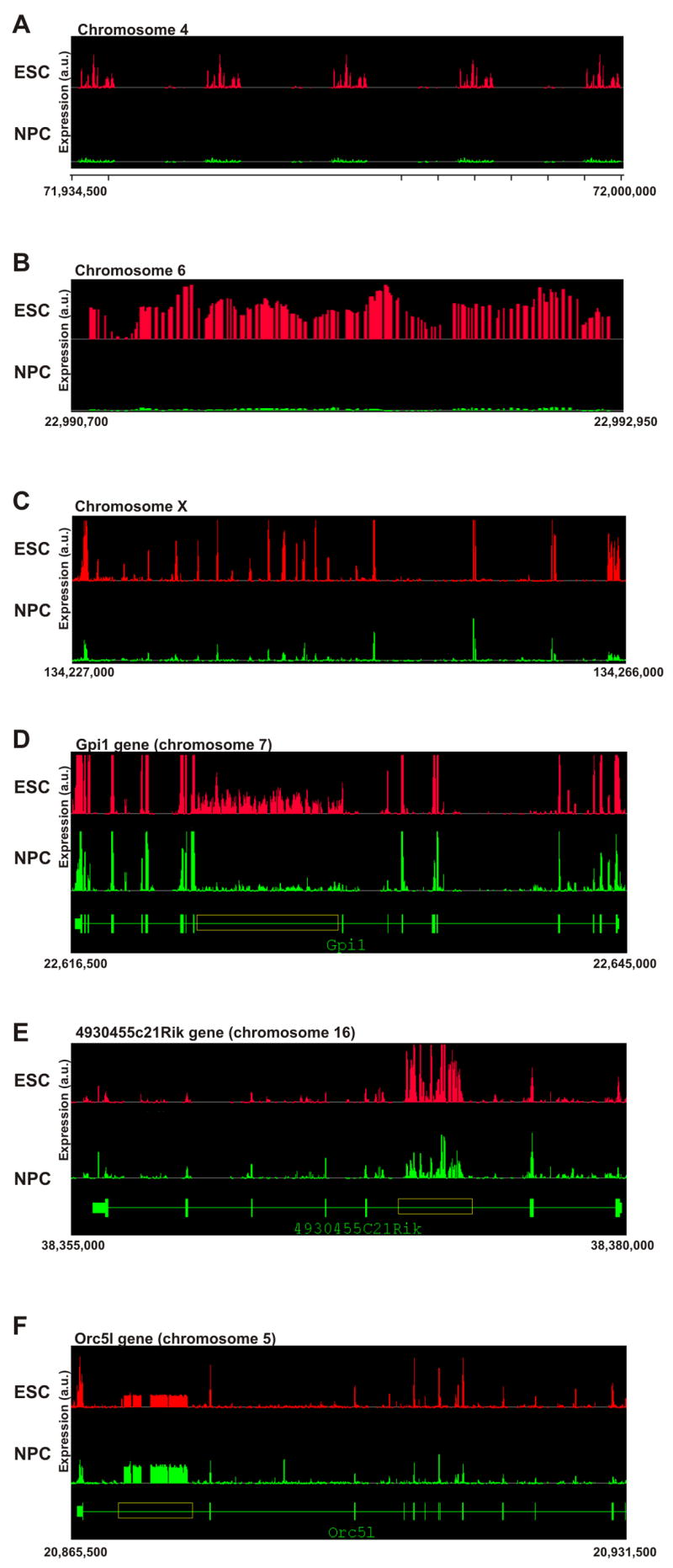
Composite graphs depicting signal intensity form the independent biological replicas represent probe intensity per genomic coordinates. All represented coordinates are in the mm.NCBIv33 version of the mouse genome and are indicated below each panel. Y-axis denoted arbitrary units of expression.
(A–C) Intergenic transcription.
(A) A ~65 kb intergenic region on chromosome 4 displaying repetitive bursts of transcription in ES cells (red, top), but not in neuronal progenitor cells (green, bottom).
(B) A 2,250 bp intergenic region on chromosome 6, which is active in ES cells (red, top) but not in NPC (green, bottom).
(C) A 29 kb intergenic region on chromosome X, where parts are active in both ES cells and NPC and parts are active in ES cells only.
(D–F) Intronic transcription.
(D) The annotated region of the Gpi1 gene (~28 kb) on chromosome 7 (green, bottom) shows intronic transcription (yellow box, > 7.5 kb) in both ES cells (red, top) and NPC (green, middle) but transcription level is considerably higher in ES cells.
(E) The annotated region of the 4930455C21Rik gene (~25 kb) on chromosome 16 (green, bottom) shows a burst of transcription inside the fifth intron in both ES cells (red, top) and NPC (green, middle). Despite higher expression of the 4930455C21Rik gene in NPC, intronic transcription is higher in ES cells. Note that unlike Gpi1, exons in this case are active at lower levels than the intronic transcription.
(F) The annotated region of the Orc5l gene (~66 kb) on chromosome 5 (green, bottom). A long intronic region (yellow box, > 8 kb) inside the Orc5l gene is active. The Orc5l gene itself is also active and the intronic transcription is lower than the exonic transcription. In this example, intronic transcription is higher in NPC (green, middle) than in undifferentiated ES cells (red, top).
Elevated transcription levels in active genome regions of ES cells
Inspection of regions which were active both in ES cells and in NPC often indicated higher transcription levels in ES cells (Figure 3E), suggesting that the increased overall transcription level might not only be due to additional active regions, but also generally higher levels of transcription in constitutively active regions. To test this possibility, we analyzed the expression level of all probes which were active in both undifferentiated ES cells and cells differentiated for either 24 hrs or 7 days (NPC). By 24 hrs, between 14 and 19 chromosomes displayed a higher number of down-regulated than up-regulated probes in all regions (Figure 4A–C, left). After 7 days, both intergenic and intronic regions displayed a higher number of down-regulated than up-regulated probes on all chromosomes (Figure 4A–B, right). Exonic regions displayed the same general trend of a higher number of down-regulated probes in 19 of the 21 chromosomes (Figure 4C, right). The two remaining chromosomes (1 and 12) showed a higher number of up-regulated probes, in agreement with the number of active probes in these chromosomes (see Figure 2C) and the higher number of differentiation-induced genes. The same trend was observed when the 24 h time point was compared with NPC (Figure S7). These data indicate that the activity of genomic regions which are both active in undifferentiated and in differentiating ES cells is higher in the undifferentiated state. In sum, based on the genome-wide analysis of active probe number, probe distribution and signal level, we conclude that the genome of undifferentiated ES cells exhibits global transcriptional activity, which becomes restricted during differentiation.
Figure 4. Global expression changes during ES cell differentiation.
(A–C) Comparison of positive probes between ES cells and cells 24 hrs after LIF withdrawal (left) or between ES cells and neuronal progenitor cells (NPC, right). Total number of down-regulated and up-regulated probes is depicted as white and gray bars respectively for intergenic regions (A), intronic regions (B) and exonic regions (C) for all mouse chromosomes. Only probes which were positive in both time points were used for this analysis. Data represents the average of three independent experiments.
Elevated transcription of chromatin remodeling factors and general transcription factors in undifferentiated ES cells
We hypothesized that the global changes in chromatin structure and the low-level transcription of large regions of the genome may be brought about by differences in the levels of chromatin proteins. To determine the basis of global transcription in ES cells, we performed a genome-wide comparison of the transcript levels of known general transcription factors (GTFs), chromatin remodeling factors and several types of histone modifying activities including histone acetyl transferases (HATs), histone deacetylases (HDACs) and histone methyl transferases (HMTs) in ES cells and NPC (Figure 5, Figure S8). To compare changes in the transcription levels of these groups of genes to the entire transcriptome, we first examined the complete set of annotated genes on the tiling arrays. Based on lower intensities of constitutive probes, 54% of all genes were reduced during differentiation into NPC, 38% were elevated and 8% of the genes were unchanged or undetected at both time points. We then compared these numbers to the expression patterns of the various groups of chromatin proteins. While all histone modifiers including HATs (Figure 5A), HDACs (Figure 5B) and HMTs (Figure 5C) showed a similar reduction in their transcription levels as the complete transcriptome (p = 0.34, 0.66 and 0.59, respectively), GTFs (Figure 5D) and chromatin-remodeling genes (Figure 5E) displayed a statistically significant more pronounced reduction in their transcription level (p = 0.0005 and 0.009 compared to all other genes, respectively), suggesting a disproportionately high level of expression of general transcription factors and chromatin remodeling proteins in ES cells. Out of 25 detectable chromatin-remodeling genes, 20 were significantly down-regulated in NPC and five slightly elevated. Amongst the 21 detectable general transcription factors, 19 were down-regulated in NPC and only 2 were slightly elevated (Figure 5E). The expression patterns for the chromatin remodeling factors were confirmed by qRT-PCR (Table S4, Figure S9). Consistent with the transcriptional downregulation of GTFs and chromatin remodelers, their protein levels were reduced in NPCs compared with ES cells (Figure S10).
Figure 5. Disproportionate over-representation of general transcription factors and chromatin remodeling genes in undifferentiated ES cells.
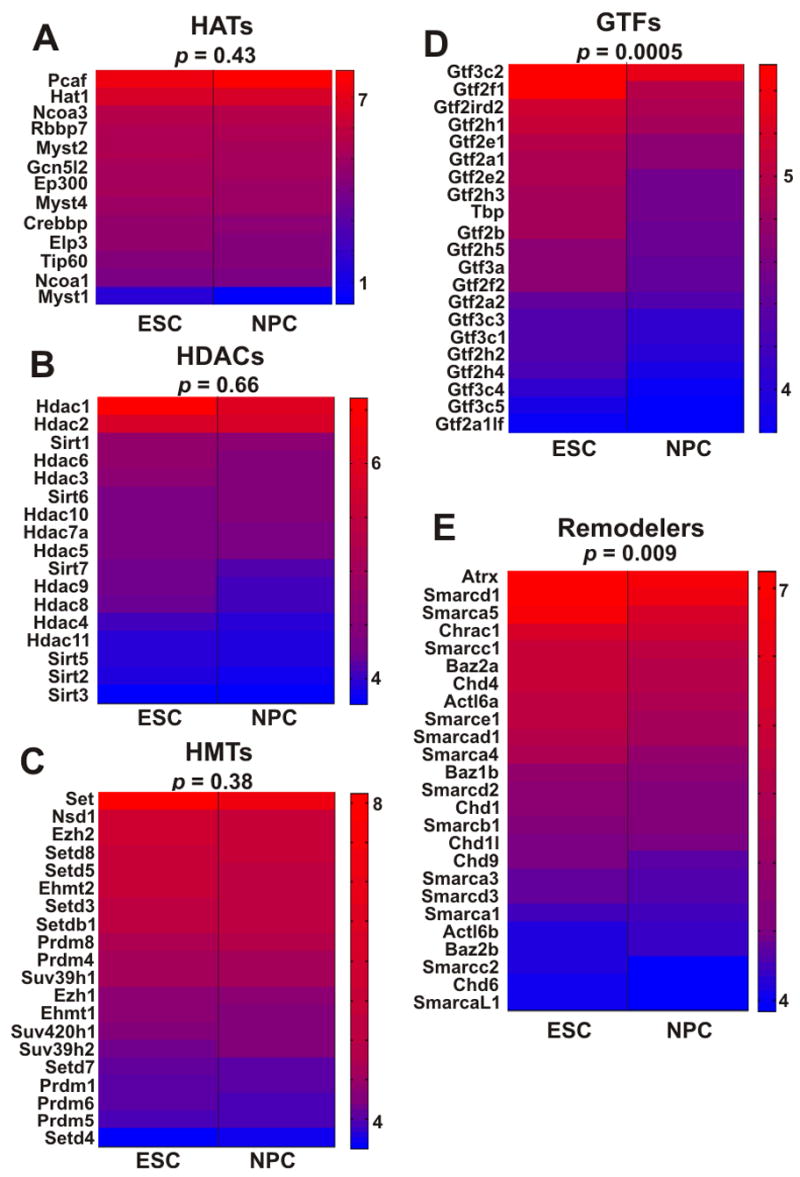
(A–E) Transcription level heat-maps of different groups of genes that are associated with transcription and regulation of chromatin, including histone acetyltransferases (A), histone deacetylases (B), histone methyltransferases (C), general transcription factors (D) and chromatin remodeling proteins (E). Gene names are given on the left of each map, p values (binomial hypothesis testing) are indicated on top. Chromatin remodeling factors and general transcription factors are disproportionately expressed in ES cells. Heat-maps were generated using microarray signal levels displayed as arbitrary units. Red-to-blue corresponds to high-to-low signal intensity.
Reduction of chromatin remodeling activity impairs ES cell proliferation and differentiation
To test whether the overrepresentation of chromatin remodeling factors was functionally relevant for ES cells and their differentiation we selectively tested the effect of knockdown of the SWI/SNF remodeling component Brg1 (Smarca4), the SWI/SNF component Smarcd2 and the ISWI-related chromodomain helicase DNA binding protein 1-like (Chd1l) by RNAi. Knockdown for Brg1 factor was confirmed by Western blotting (Figure 6A) and for Smarcd2 and Chd1l by quantitative real-time PCR due to the absence of antibodies (Figure 6B). RNAi against luciferase was used as a negative control (Figure 6). ES cells treated with Brg1 siRNAs displayed marked reduction in both their proliferation and differentiation capacities (Figure 6A, C–D). After 96 hrs of Brg1 siRNA treatment, the proliferation rate was roughly 60% of that of luciferase RNAi treated cells (Figure 6C, top left). In addition, while luciferase siRNA treated cells generated nestin-positive neuronal progenitor cells at a rate of 74%, this number dropped to 15% in the Brg1 siRNA treated cells. Knockdown of Chd1l, which displayed the second most pronounced up-regulation in ES cells (by 8.9 ± 3.9-fold) resulted in an ES cell proliferation defect but did not appear to affect differentiation (Figure 6B–C), while knockdown of Smarcd2, which displayed the most pronounced up-regulation in ES cells (by 9.6 ± 2.8-fold) resulted in no apparent phenotype (Figure 6B–C). Treatment with both Smarcd2 and Chd1l RNAi appeared similar to Chd1l RNAi treatment alone (Figure 6C, bottom right). These results suggest that while some chromatin remodelers play important roles in ES cell proliferation and differentiation, partial depletion of single factors may have subtle or no effects, supporting the notion that the group of chromatin remodeling proteins, rather than individual factors, helps to support stem cell maintenance and pluripotency.
Figure 6. Knockdown of specific chromatin remodeling factors inhibits ES cell differentiation.
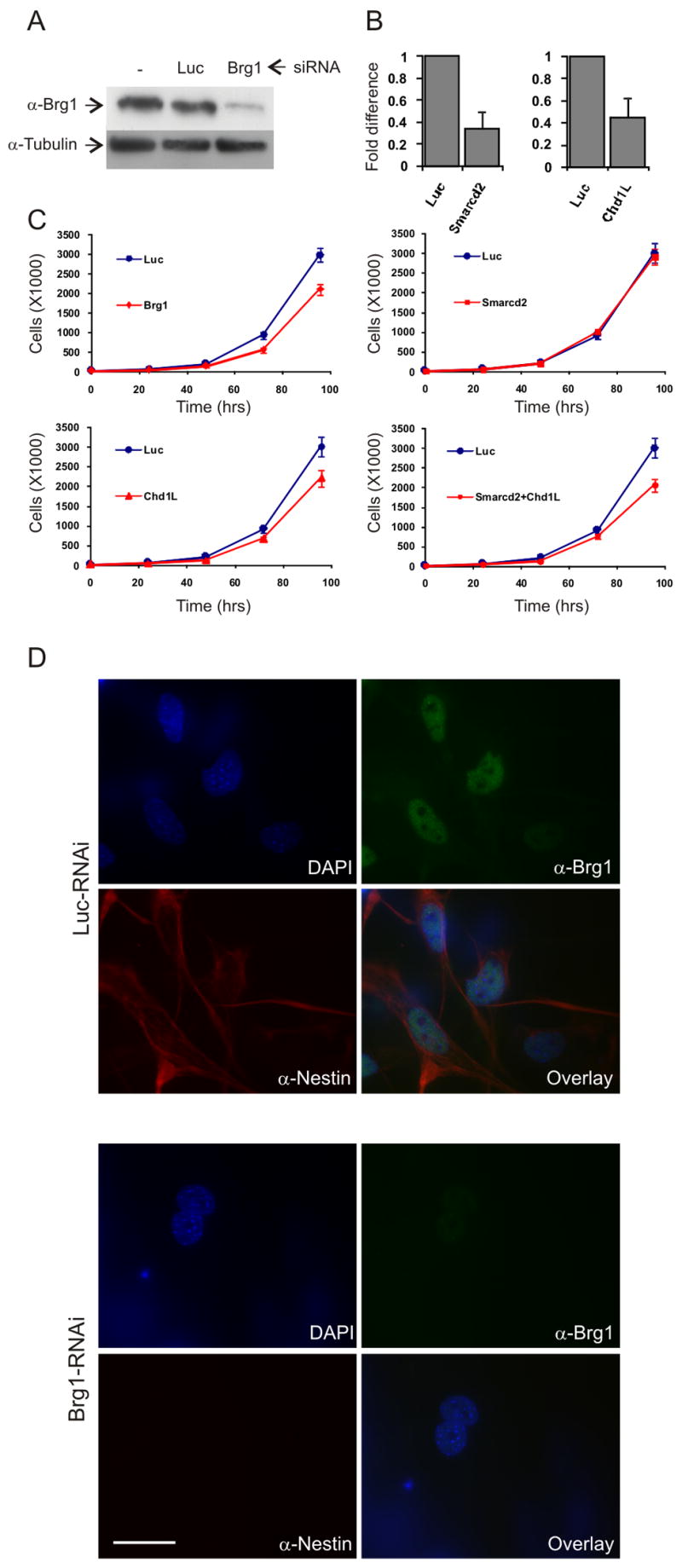
(A) Knockdown of Smarca4 (Brg1) using siRNAs (SmartPool, Dharmacon). Western Blot showing levels of Brg1 protein in ES cells in the absence of siRNA (left), with siRNA against Luciferase (middle) and with siRNA specific to Brg1 (left). Levels of tubulin is used as control (bottom).
(B) Real-time RT-PCR of RNA levels after siRNA treatment to Smarcd2 and Chd1L.
(C) Proliferation rate of luciferase siRNA treated cells (Luc, blue lines) and of ES cells treated with siRNA against the three chromatin remodeling factors indicated.
(D) Top: ES cells-derived NPC treated with luciferase siRNA oligos. Brg1 is shown in green, Nestin in red and DAPI in blue. Lower right panel shows overlay image. Bottom: ES cells treated with siRNA against Brg1 fail to differentiate into NPC. Brg1 is absent in these cells (upper right) and so is Nestin (lower left).
DISCUSSION
Our results suggest that pluripotent ES cells are characterized by elevated global transcriptional activity and that loss of pluripotency and lineage specification involves reduction of the actively transcribed portion of the genome. The increased global transcriptional activity observed here is consistent with the unique properties of chromatin in ES cells including a globally open structure, a specific set of histone modifications and looser binding of architectural proteins (Arney and Fisher, 2004; Boyer et al., 2006a; Boyer et al., 2006b; Buszczak and Spradling, 2006; Gan et al., 2007; Lee et al., 2006; Meshorer, 2007; Meshorer and Misteli, 2006; Meshorer et al., 2006; Szutorisz and Dillon, 2005; Szutorisz et al., 2006). Global low-level transcription in ES cells is also in line with the presence of bivalent chromatin marks of both active and repressive histone modifications on silent lineage-specific genes (Azuara et al., 2006a; Bernstein et al., 2006; Mikkelsen et al., 2007). Global, possibly stochastic, transcription in ES cells is also suggested by the identification in mouse ES cells of over 40,000 different transcripts using High-Coverage Gene Expression Profiling (HiCEP) (Araki et al., 2006) as well as detection of transcription initiation at most genes in human ES cells (Guenther et al., 2007). Global genome transcriptional activity likely also occurs in human ES cells since an increased number of expressed genes has been demonstrated in human ES cells using microarray analysis (Golan-Mashiach et al., 2005). Elevated transcriptional activity and permissive expression of lineage-restricted genes has also been observed in the hematopoietic system, where expression of genes of multiple lineages was detected prior to commitment (Hu et al., 1997) and where a larger fraction of the genes is active in the undifferentiated state (Eckfeldt et al., 2005; Terskikh et al., 2003; Zipori, 2004).
The finding that general transcription factors and chromatin-remodeling proteins are disproportionately over-expressed in ES cells suggests that they are critical in maintaining chromatin in an open state and contribute to global transcriptional activity. Indeed we find that loss of chromatin remodeling factors affects ES cell proliferation and differentiation in a factor-specific fashion. A critical role for chromatin remodeling complexes in ES cell differentiation has been hinted at by the observation that disruption of several of these proteins, including Brg1 (Bultman et al., 2000; Bultman et al., 2006), Snf5 (Klochendler-Yeivin et al., 2000), SSRP1 (Cao et al., 2003) and Snf2h (Stopka and Skoultchi, 2003), results in embryonic death at the blastocyst stage before implantation, during the period when the inner cell mass (ICM), the source of all ES cells, is being formed. In Drosophila, chromatin remodeling is involved in germline stem cell self-renewal and differentiation (Xi and Xie, 2005). In mice, the NuRD chromatin remodeling complex is essential for ES cell differentiation (Kaji et al., 2006) and we now show here that loss of Brg1 leads to ES cell differentiation defects. Furthermore, reduction of Chd1l impairs ES cell proliferation. These observations are in line with the finding that the chromatin assembly factor CAF-1 is essential for heterochromatin formation in mouse ES cells and depletion of CAF-1 in ES cells results in heterochromatin reorganization and deformation and subsequent lethality (Houlard et al., 2006). In differentiated MEFs, however, CAF-1 depletion had little effect. Since we performed group analysis we do not rule out important contributions of individual genes inside groups that did not display significant differences between ES cells and NPC, i.e. chromatin modifying enzymes. For example, the polycomb group gene Suz12 (an H3K27 HMT) was down-regulated following differentiation and was shown to play a role in ES cell maintenance (Pasini et al., 2007). In another more recent example, the H3K9m2 and H3K9m3 demethylase genes, Jmjd1a and Jmjd2c, were shown to be positively regulated in ES cells by Oct4 and their depletion results in ES cell differentiation (Loh et al., 2007). The changes in individual histone modifier genes may well be responsible for the changes we observed in histone modifications during differentiation. Taken together observations strongly point towards an active role of chromatin-remodeling factors in the maintenance of stem cell identity and the initial stages of stem cell differentiation and they are consistent with their disproportionate upregulation in ES cells.
We propose that the higher abundance of chromatin remodeling factors in ES cells maintains the ES cell genome in a preferentially open state allowing freer access of the general transcription machinery and facilitating the stochastic formation of pre-initiation complexes (PICs) even on silenced genes. In support of this view, RNA polymerase II complexes are found at promoters of most protein coding genes in ES cells (Guenther et al., 2007). The formation of these PICs might be actively counterbalanced as the 26S subunit of the proteasome has recently been demonstrated to remove forming PICs from promoters of pluripotent ES cells (Szutorisz et al., 2006). Importantly, no such role for the 26S proteasome was found in differentiated cells, suggesting that the higher propensity of PIC formation is a property of undifferentiated ES cells (Szutorisz et al., 2006). The involvement of the 26S proteasome in removal of the PIC from ES genes implies that the transcriptional hyperactivity of the ES genome is under regulatory control. It is unclear at present whether global transcription is merely a bi-product of the chromatin properties in ES cells or whether it is essential for pluripotency and control of differentiation, particularly since it is unclear whether the permissive transcripts generated in ES cells are full length and whether they lead to production of functional protein. The possibility that global transcription is functionally important for.differentiation is attractive in the light of the observation that in fission yeast heterochromatin silencing is mediated by the RNAi pathway (Volpe et al., 2002) and requires RNA pol II (Kato et al., 2005). Similar types of mechanisms might be operating in mammalian cells, especially during ES cell differentiation, when heterochromatin domains are formed (Meshorer and Misteli, 2006). In support, loss of Dicer, one of the key factors in the RNAi pathway, leads to a significant reduction in heterochromatin silencing in ES cells and to severe defects in ES cell differentiation in vitro and in vivo (Kanellopoulou et al., 2005). Similarly, production of non-coding RNAs in ES cells may serve as precursors for regulatory small RNAs (Kapranov et al., 2007). Default global transcription in ES cells may thus be a key mechanism in the maintenance of the pluripotent state and in the silencing of specific genome regions during differentiation.
METHODS
Cells
Mouse R1 male ES cells (from A. Nagy, Toronto, Canada) were grown and differentiated into 7 days ES cells-derived neuronal progenitor cells (NPC). The R1 ES cell differentiation system has previously been extensively characterized (Lee et al., 2000; Meshorer et al., 2006).
Electron spectroscopic Imaging
Following immunolabelling, cells were prepared by standard fixation, embedding and thin sectioning methods (Dellaire et al., 2004). Electron micrographs were taken at 200 kV on a transmission electron microscope (Tecnai 20, FEI). Energy filtered images were collected using a post-column imaging filter (Gatan) as described elsewhere (Dellaire et al., 2004).
Antibodies, Western blots and immunofluorescence (IF)
Oct4 (goat polyclonal, Santa Cruz Biotechnologies, Santa Cruz, CA, cs-8628); Nestin (rabbit polyclonal, R. McKay); TUJ1 (mouse monoclonal, Chemicon, Temecula, CA, MAB1637), H3K4me3 (rabbit monoclonal, Upstate 05-745); H3K9ac (06-942), H3K14ac (06-911), H3K36me2 (07-274), H3K36me3 (07-549), H3S28p (07-145), H4K20me2 (06-031) (all rabbit polyclonal, Upstate), and 5-meC (mouse monoclonal, Eurogentec, BI-MECY-0100) antibodies were used. Blots were performed on purified nuclei (Meshorer et al., 2006). Detection was with anti-rabbit or anti-mouse antibodies conjugated to HRP for Western blots and either Texas Red or FITC (Jackson ImmunoResearch, West Grove, PA) for IF. IF was performed as described (Misteli et al., 2000).
Transcription assay
3H-uridine was added to the culture media at a final concentration of 3.7 Mbq/ml for 4 hrs. Cells were harvested and RNA and DNA were simultaneously purified using the RNA/DNA Mini Kit (Qiagen). Messenger RNA was purified using the Oligotex Mini mRNA isolation kit (Qiagen). Optical density was measured using the ND-1000 spectrophotometer (NanoDrop) and radiation was measured using an LS 6000IC scintillation counter (Beckman).
RNA FISH
Cells grown on gelatin coated (for ES cells) or poly-L-lysine/fibronectin coated (for NPC and PMNs) glass cover-slips were treated with CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES pH 6.8) supplemented with 0.5% Triton X-100 and 200 mM vanadyl ribonucleoside complex (VRC) (Ambion), fixed in 4% paraformaldehyde PBS for 15 min, washed 3 X PBS for 5 min each and treated in an ascending EtOH series (70%, 80%, 90%, 100%, 5 min each). A locked nucleic acid (LNA) Cy3-labeled 36-mer probe (1 μg/ul) (5′-Cy3-CtCgCcAtAtTtCaCgTcCtAaAgTgTgTaTtTcTc-3′; LNA bases are capitalized) was mixed with unlabeled 18S and 28S rDNA probes (1 μg/ul) (Gift from M. Dundr) and denatured for 5 min at 80ºC followed by 30 min at 37ºC. Probe was applied overnight in a hybridization solution (50% formamide, 2 × SSC, 10% dextran sulfate, 1 mg/ml BSA) at 37ºC in a humidified chamber. Cells were washed 3 × in 50% formamide, 2 × SSC, then 3 × in 2 × SSC at 39ºC, 5 min each and in 1 × SSC for 5 min at room temp. Cells were DAPI stained and mounted.
RT-PCR
BioRad MyiQ real-time PCR machine in a 96-well format with IQ SYBR Green Supermix (BioRad) was used for all experiments. Reverse transcription was with High Capacity cDNA Archive Kit (Applied biosystems, Foster City, CA) and StrataScript RT-PCR System (Stratagene, Cedar Creek, TX) using 250–1000 ng of total RNA (RNeasy kit supplemented with RNase-free DNase set, Qiagen, Valencia, CA) with a mix of random hexamers and poly dT primers. For quantification, standard curves were generated for each primer pair by serial dilution of the starting template. Cyclophilin B was used for normalization. Primers for repetitive sequences and transposable elements were as described elsewhere (Martens et al., 2005). Primers for lineage-specific genes are given in Table S1. Primers for chromatin remodeling factors are given in Table S4.
Microarray design and hybridization
We prepared total poly(A)+ RNA from undifferentiated ES cells, differentiating ES cells at 12 and 24 hrs and 7 days NPC. Three biological replicates were generated for each time point. Samples were prepared and labeled as described (Kapranov et al., 2002). Briefly, total RNA was enriched for poly(A)+ species using the Oligotex protocol (Qiagen). Double stranded cDNA was prepared from poly(A)+ RNA and 2 μg of double-stranded cDNA was labeled and hybridized to the GeneChip® Mouse Tiling Array 1.0R Array set (Affymetrix) containing the entire mouse non-repetitive genome on 16 chips. Sequences used in the design were selected from NCBI mouse genome assembly (Build 32, mm4). Repetitive elements were removed by RepeatMasker. All probes on the chip are tiled at an average resolution of 30 bp, as measured from the central position of adjacent 25-mer oligos, leaving a gap of approximately 5 bp between probes. All Graphs were generated in Mouse genome Assembly 33 (mm5) using probe coordinates that were remapped to mm5. A total of 192 chips were used for the whole experiment and composite graphs combining the three biological replica were generated for each time point using standard Affymetrix pipeline (Kampa et al., 2004; Kapranov et al., 2002). All graph files have been submitted to GEO. We first examined whether the signal corresponds to annotated regions and found a perfect correlation between the two. With 5 exons and 4 introns, the stem cell marker Oct4 serves as an example (Figure S5A, top left). For analysis purpose only the undifferentiated ES cells and differentiated 7 days NPC samples were used.
Microarray data analysis
To generate the number of positive probes in each chromosome (Figure 2), we used the intensity data from the entire set of the mismatch probes (MM) to determine the threshold (X). We assumed a Gamma distribution of the MM intensity values and calculated the mean (μ) and variance (σ). The threshold (X) was then defined as the intensity level under which 90% of the MM signal is contained. We then counted the number of probes above X. In order to determine if the level of expression decreases or increases for the same set of probes between ES cells and NPC (Figure 4), we compared the expression of all probes in ES cells to all probes in NPC, filtering out probes below the detection threshold, defined as the threshold that generates a false-positive rate of 2.9% from the bacterial controls on all arrays (Kampa et al., 2004). For a probe to be included in this analysis, it must be present above threshold in both time points. Normalization was done essentially as described (Kampa et al., 2004). Positive probes were determined by the difference (PM-MM), and are thus insensitive to the normalization method, as both PM values and MM values are scaled together.
Supplementary Material
Figure S1. Decondensed and actively marked chromatin in ES cells
(A) Phosphorus (yellow) and nitrogen-phosphorus (N-P) (blue) maps in undifferentiated ES cells (ESC), ES cells-derived 7 days neuronal progenitor cells (NPC) and mouse embryonic fibroblasts (MEF). ES cells exhibit homogeneous chromatin texture and are devoid of detectable heterochromatin foci. Bar = 500 nm.
(B) Line scans of mass distribution (nitrogen maps) in ES cells (left), NPC (middle) and MEFs (right). CoV = Coefficient of Variation (% standard deviation/mean). At least 10 scans of each of 10 cells were analyzed for ESC and NPC and 4 cells for MEFs. Representative images are shown.
(C) Western blots for histone modifications as well as methylated DNA (5-meC) in undifferentiated ES cells (left), cells 24 hrs after withdrawal of LIF (middle) and NPC (right). Active histone marks are enriched in ES cells. Equal amounts of protein were loaded and histone H3 serves as a loading control.
(D) Immunofluorescence analysis for H3K4m3 in undifferentiated ES cells (ESC) and ES cell-derived neuronal progenitor cells (NPC). Fluorescence intensity quantification (n>30 cells) is shown on the right.
Figure S2. Detection of DNA methylation by Western blot
(A) Purified nuclei from three preparations of undifferentiated ES cells (lanes 1–3) and three preparations of NPC (lanes 4–6) were denatured in Laemmli buffer and electrophoresed on a 4–20% gradient SDS-PAGE gel (BioRad). Gel was blotted using a polyvinylidene fluoride (PVDF) transfer membrane (Immobilon-P, Millipore, Billerica, MA), which readily blots both proteins and nucleic acids, blocked, and incubated 2 hrs at room temperature or over-night at 4°C with a mouse monoclonal 5-methylcitidine antibody (Eurogentec) in 3% skim milk and 2% BSA (1:1000). Following standard washes and blocking, the blot was incubated with a secondary anti-mouse antibody (GE Healthcare, Buckinghamshire, UK) for 2 hrs at room temperature. Detection was performed with ECL-plus kit (GE). Note that one ‘heavy’ band is visible, likely corresponding to bulk DNA.
(B) Specificity of the detected band. Purified nuclei from undifferentiated ES cells were pre-incubated in DNase I buffer (10 mM Tris-HCl; 2.5 mM MgCl2; 0.5 mM CaCl2) in the absence (lane 1, 40 min) or presence (lane 2, 20 min and lane 3, 40 min) of DNase I enzyme (5 μg/ml). Pre-incubation with DNAse I was performed to ensure that DNA and only DNA is detected using Western blots. Note complete disappearance of detectable bands following DNase I treatment, attesting to the specificity of the reaction. Also note appearance of an additional band in lane 1, due to pre-incubation.
Figure S3. Detection of lineage-specific genes in ES cells
(A) RT-PCR detection. Top: RT-PCR products for 12 lineage-specific genes. Five genes (Acta1, PMEPA1, SPRR2A, Albumin and Synaptotagmin-11) were detected in the first RT-PCR reaction. Bottom: second PCR was performed for the seven genes that failed to yield a product in the first RT-PCT reaction. Of these, the six noted genes were detected in the second reaction, suggesting low-level transcription.
(B) Real-time RT-PCR for three lineage-specific genes, the transcription level of which is reduced following differentiation. ESC: undifferentiated ES cells; NPC: ES cells-derived neuronal progenitor cells; PMN: ES cells-derived pot-mitotic neurons.
Figures S4. ChIP-seq data for the selected lineage-specific genes
(A–B) Chromatin immunoprecipitation (ChIP) followed by single-molecule-based sequencing (ChIP-seq) using Illumina/Solexa technology was performed and described by Mikkelsen et al (2007). In this approach, DNA molecules that are pulled-down by ChIP are spotted on an array and amplified locally. Then, successive rounds of primer-mediated single-base extension are applied, using fluorescently labeled reversible terminators, and imaged after every round. ChIP was performed with the noted antibodies: H3K4m3 is shown in green and H3K27m3 in red for both ES cells (ESC) and ES cell-derived neuronal progenitor cells (NPC). Gene names and their genomic locations appear at the top of each panel and complete gene annotations at the bottom. While genes are bivalently marked in ESC, the H3K4m3/H3K27m3 ratio is always higher in ESC except for Synaptotagmin which is expressed in NPC. These results support low-level transcription of these transcripts in ES cells.
(C) ChIP-seq data for classic markers of the three germ layers are shown. FGF5 is a marker for ectoderm, BMP4 for mesoderm and Sox17 for endoderm. In all cases, the H3K4m3/H3K27m3 ratio is higher in ESC supporting transcriptional activation at low levels. All data in this figure is from Mikkelsen et al, 2007.
Figure S5. Microarray data validation
(A) Tiling microarray expression pattern of the stem cell markers Oct4 (Pou5f1) (top left) and Sox15 (top, right) as well as the neuronal progenitor markers Sox4 (bottom, left) and Sox11 (bottom, right) in ES cells (ESC, white) and in ES cells-derived neuronal progenitor cells (NPC, green). Each panel represent composite graphs describing signal intensity form the three independent biological replicas to represent probe intensity per genomic coordinates. All coordinates are in the mm.NCBIv33 version of the mouse genome.
(B) Semi-quantitative real-time RT-PCR for Oct4, Sox15, Sox4 and Sox11. The following primers were used: Oct4-F: 5-AGCTGCTGAAGCAGAAGAGG-3; Oct4-R: 5-CCTGGGAAAGGTGTCCTGTA-3 Sox4-F: 5-CGCCTTGGTGATTTCTTGTT-3; Sox4-R: 5-ATCACCCCAGAGCCTTCTTT-3 Sox11-F: 5-CAGGACCTGTTGCTGTCAGA-3; Sox11-R: 5-GGTCACGAAGCTGGTTTGAT-3 Sox15-F: 5-CGGCGTAAGAGCAAAAACTC-5; Sox15-R: 5-TGGGATCACTCTGAGGGAAG-3
(C) Microarray detection. Shown are the annotated regions of albumin (top) and CD8 (bottom). Both genes display only background levels of transcription on the microarray and most exons do not generate transfrags.
Figure S6. Cluster size distribution maps of the probes present in ESC and absent in NPC
In order to determine the contiguity of the detected transcription in ESC cells, probes selected to be positive in ES cells but absent in NPC were clustered into bins of varying sizes. The frequency of each cluster is represented in the ordinate and the size (# of adjacent probes) in the abscissa. For example, a frequency of 100,000 clusters of size 3 that fit the criteria. This means 100,000 instances of three adjacent probes that were both present in ES cells and absent in NPC across the whole genome (both intragenic and intergenic regions). Since each probe encompasses 25 bp of genomic DNA with a gap of 5 bp between two adjacent probes, a cluster of 3 probes represent an approximate genomic size of 100 bp. Cluster size of 200 corresponds to 6 kb.
Figure S7. Global expression changes during ES cell differentiation – 24 hrs Vs. NPC
(A–C) Comparison of positive probes between cells 24 hrs after LIF withdrawal and between ES cells-derived neuronal progenitor cells (NPC). Total numbers of down-regulated and up-regulated probes are depicted as white and gray bars respectively for intergenic regions (A), intronic regions (B) and exonic regions (C) for all mouse chromosomes. Only probes which were positive in both time points were used for this analysis. Data represents the average of three independent experiments.
Figure S8. Comparison of expression levels of specific classes of genes in differentiating ES cells
Shown are relative expression levels of microarray data in ES cells (ESC, blue) and ES cells-derived neuronal progenitor cells (NPC, red) of histone acetyl transferases (HATs), histone deacetylases (HDACs), histone methyl transferases (HMTs), general transcription factors (GTFs) and chromatin remodeling factors (Remodelers). Data is the same as presented in Figure 5 but displayed as bar graphs rather than heat-maps.
Figure S9. Real-time qRT-PCR analysis of chromatin remodeling genes
Validation of microarray expression data for chromatin remodelers by quantitative real-time RT-PCR. From a total of 21 genes for which signal could be obtained from both ESC and NPC states, 17 displayed reduced expression in NPC, most of which correlated with the array data. Of the four genes that displayed either slight elevation or no change (Chd1, Chd4, Chd6, Smarcc2) three (Chd1, Chd6, Smarcc2) were also in agreement with the microarray data (p < 0.05). An additional 4 genes (Smarca1, Smarca5, Smarcad1 and Baz2b) which were detected only in the ES cells qRT-PCR measurements were excluded. All measurements were normalized against Cyclophilin B. Values shown are fold difference representing averages ± SD from 3 independent experiments. These results strongly corroborate the microarray analysis and reconfirm the abundance of chromatin remodeling factors and general transcription factors in undifferentiated ES cells.
Figure S10. Protein levels for various chromatin related factors during ES cell differentiation
Western blots for a variety of chromatin proteins from nuclear extracts of undifferentiated ES cells (ESC), 24 hrs following LIF withdrawal (24 h) or 7 d after induction of differentiation (NPC) were probed for the indicated proteins. Total histones served as a loading control.
Acknowledgments
We thank Drs. Liran Carmel (NIH) and Robert Clifford (NIH) for help with statistical analysis, Dr. Mirek Dundr (Boston) for providing rDNA plasmids and Dr. Bradley Bernstein for the ChIP-seq data shown in Figure S4. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research; the Israel Science Foundation (215/07) (to EM); the European Union (IRG-206872) (to EM); Alon Fellowship (to EM); and an operating grant (to DPB-J) from the Canadian Institutes of Health Research. DPB-J holds a Canada Research Chair in Molecular and Cellular Imaging. Design and hybridization of all tiling arrays utilized in this study were supported by Affymetrix. Inc.
Footnotes
Accession numbers. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) (Barrett et al., 2007; Edgar et al., 2002) and are accessible through GEO Series accession number GSE10834.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoto T, Saitoh N, Ichimura T, Niwa H, Nakao M. Nuclear and chromatin reorganization in the MHC-Oct3/4 locus at developmental phases of embryonic stem cell differentiation. Dev Biol. 2006;298:354–367. doi: 10.1016/j.ydbio.2006.04.450. [DOI] [PubMed] [Google Scholar]
- Araki R, Fukumura R, Sasaki N, Kasama Y, Suzuki N, Takahashi H, Tabata Y, Saito T, Abe M. More than 40,000 transcripts, including novel and noncoding transcripts, in mouse embryonic stem cells. Stem Cells. 2006;24:2522–2528. doi: 10.1634/stemcells.2006-0005. [DOI] [PubMed] [Google Scholar]
- Arney KL, Fisher AG. Epigenetic aspects of differentiation. J Cell Sci. 2004;117:4355–4363. doi: 10.1242/jcs.01390. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006a;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006b doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic Acids Res. 2007;35:D760–765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen T, Xie X, Kamal M, Huebert D, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Chudin E, Wu B, Zhou L, Garcia EW, Liu Y, Shin S, Plaia TW, Auerbach JM, Arking DE, et al. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006a;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006b;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Spradling AC. Searching chromatin for stem cell identity. Cell. 2006;125:233–236. doi: 10.1016/j.cell.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Cao S, Bendall H, Hicks GG, Nashabi A, Sakano H, Shinkai Y, Gariglio M, Oltz EM, Ruley HE. The high-mobility-group box protein SSRP1/T160 is essential for cell viability in day 3.5 mouse embryos. Mol Cell Biol. 2003;23:5301–5307. doi: 10.1128/MCB.23.15.5301-5307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaire G, Nisman R, Bazett-Jones DP. Correlative light and electron spectroscopic imaging of chromatin in situ. Methods Enzymol. 2004;375:456–478. doi: 10.1016/s0076-6879(03)75028-9. [DOI] [PubMed] [Google Scholar]
- Eckfeldt CE, Mendenhall EM, Verfaillie CM. The Molecular Repertoire of the ‘Almighty’ Stem Cell. Nat Rev Mol Cell Biol. 2005:2–13. doi: 10.1038/nrm1713. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Q, Yoshida T, McDonald OG, Owens GK. Concise review: epigenetic mechanisms contribute to pluripotency and cell lineage determination of embryonic stem cells. Stem Cells. 2007;25:2–9. doi: 10.1634/stemcells.2006-0383. [DOI] [PubMed] [Google Scholar]
- Golan-Mashiach M, Dazard JE, Gerecht-Nir S, Amariglio N, Fisher T, Jacob-Hirsch J, Bielorai B, Osenberg S, Barad O, Getz G, et al. Design principle of gene expression used by human stem cells: implication for pluripotency. Faseb J. 2005;19:147–149. doi: 10.1096/fj.04-2417fje. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Hough SR, Clements I, Welch PJ, Wiederholt KA. Differentiation of mouse embryonic stem cells after RNA interference-mediated silencing of OCT4 and Nanog. Stem Cells. 2006;24:1467–1475. doi: 10.1634/stemcells.2005-0475. [DOI] [PubMed] [Google Scholar]
- Houlard M, Berlivet S, Probst AV, Quivy JP, Hery P, Almouzni G, Gerard M. CAF-1 Is Essential for Heterochromatin Organization in Pluripotent Embryonic Cells. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- Jorgensen HF, Giadrossi S, Casanova M, Endoh M, Koseki H, Brockdorff N, Fisher AG. Stem cells primed for action: polycomb repressive complexes restrain the expression of lineage-specific regulators in embryonic stem cells. Cell Cycle. 2006;5:1411–1414. doi: 10.4161/cc.5.13.2927. [DOI] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, Macleod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006 doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, Drenkow J, Piccolboni A, Bekiranov S, Helt G, et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermueller J, Hofacker IL, et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science. 2007 doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- Kimura H, Sugaya K, Cook PR. The transcription cycle of RNA polymerase II in living cells. J Cell Biol. 2002;159:777–782. doi: 10.1083/jcb.200206019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayakawa S, Miike K, Nakao M, Abe K. Dynamic changes in the epigenomic state and nuclear organization of differentiating mouse embryonic stem cells. Genes Cells. 2007;12:447–460. doi: 10.1111/j.1365-2443.2007.01063.x. [DOI] [PubMed] [Google Scholar]
- Lee SH, Lumelsky N, Studer L, Auerbach JM, McKay RD. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. Embo J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E. Chromatin in embryonic stem cell neuronal differentiation. Histol Histopathol. 2007;22:311–319. doi: 10.14670/HH-22.311. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H. Open conformation chromatin and pluripotency. Genes Dev. 2007;21:2671–2676. doi: 10.1101/gad.1615707. [DOI] [PubMed] [Google Scholar]
- Pan GJ, Chang ZY, Scholer HR, Pei D. Stem cell pluripotency and transcription factor Oct4. Cell Res. 2002;12:321–329. doi: 10.1038/sj.cr.7290134. [DOI] [PubMed] [Google Scholar]
- Park SH, Kook MC, Kim EY, Park S, Lim JH. Ultrastructure of human embryonic stem cells and spontaneous and retinoic acid-induced differentiating cells. Ultrastruct Pathol. 2004;28:229–238. doi: 10.1080/01913120490515595. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholer HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- Stopka T, Skoultchi AI. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci U S A. 2003;100:14097–14102. doi: 10.1073/pnas.2336105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szutorisz H, Dillon N. The epigenetic basis for embryonic stem cell pluripotency. Bioessays. 2005;27:1286–1293. doi: 10.1002/bies.20330. [DOI] [PubMed] [Google Scholar]
- Szutorisz H, Georgiou A, Tora L, Dillon N. The proteasome restricts permissive transcription at tissue-specific gene Loci in embryonic stem cells. Cell. 2006;127:1375–1388. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Terskikh AV, Miyamoto T, Chang C, Diatchenko L, Weissman IL. Gene expression analysis of purified hematopoietic stem cells and committed progenitors. Blood. 2003;102:94–101. doi: 10.1182/blood-2002-08-2509. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Xi R, Xie T. Stem cell self-renewal controlled by chromatin remodeling factors. Science. 2005;310:1487–1489. doi: 10.1126/science.1120140. [DOI] [PubMed] [Google Scholar]
- Zipori D. The nature of stem cells: state rather than entity. Nat Rev Genet. 2004;5:873–878. doi: 10.1038/nrg1475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Decondensed and actively marked chromatin in ES cells
(A) Phosphorus (yellow) and nitrogen-phosphorus (N-P) (blue) maps in undifferentiated ES cells (ESC), ES cells-derived 7 days neuronal progenitor cells (NPC) and mouse embryonic fibroblasts (MEF). ES cells exhibit homogeneous chromatin texture and are devoid of detectable heterochromatin foci. Bar = 500 nm.
(B) Line scans of mass distribution (nitrogen maps) in ES cells (left), NPC (middle) and MEFs (right). CoV = Coefficient of Variation (% standard deviation/mean). At least 10 scans of each of 10 cells were analyzed for ESC and NPC and 4 cells for MEFs. Representative images are shown.
(C) Western blots for histone modifications as well as methylated DNA (5-meC) in undifferentiated ES cells (left), cells 24 hrs after withdrawal of LIF (middle) and NPC (right). Active histone marks are enriched in ES cells. Equal amounts of protein were loaded and histone H3 serves as a loading control.
(D) Immunofluorescence analysis for H3K4m3 in undifferentiated ES cells (ESC) and ES cell-derived neuronal progenitor cells (NPC). Fluorescence intensity quantification (n>30 cells) is shown on the right.
Figure S2. Detection of DNA methylation by Western blot
(A) Purified nuclei from three preparations of undifferentiated ES cells (lanes 1–3) and three preparations of NPC (lanes 4–6) were denatured in Laemmli buffer and electrophoresed on a 4–20% gradient SDS-PAGE gel (BioRad). Gel was blotted using a polyvinylidene fluoride (PVDF) transfer membrane (Immobilon-P, Millipore, Billerica, MA), which readily blots both proteins and nucleic acids, blocked, and incubated 2 hrs at room temperature or over-night at 4°C with a mouse monoclonal 5-methylcitidine antibody (Eurogentec) in 3% skim milk and 2% BSA (1:1000). Following standard washes and blocking, the blot was incubated with a secondary anti-mouse antibody (GE Healthcare, Buckinghamshire, UK) for 2 hrs at room temperature. Detection was performed with ECL-plus kit (GE). Note that one ‘heavy’ band is visible, likely corresponding to bulk DNA.
(B) Specificity of the detected band. Purified nuclei from undifferentiated ES cells were pre-incubated in DNase I buffer (10 mM Tris-HCl; 2.5 mM MgCl2; 0.5 mM CaCl2) in the absence (lane 1, 40 min) or presence (lane 2, 20 min and lane 3, 40 min) of DNase I enzyme (5 μg/ml). Pre-incubation with DNAse I was performed to ensure that DNA and only DNA is detected using Western blots. Note complete disappearance of detectable bands following DNase I treatment, attesting to the specificity of the reaction. Also note appearance of an additional band in lane 1, due to pre-incubation.
Figure S3. Detection of lineage-specific genes in ES cells
(A) RT-PCR detection. Top: RT-PCR products for 12 lineage-specific genes. Five genes (Acta1, PMEPA1, SPRR2A, Albumin and Synaptotagmin-11) were detected in the first RT-PCR reaction. Bottom: second PCR was performed for the seven genes that failed to yield a product in the first RT-PCT reaction. Of these, the six noted genes were detected in the second reaction, suggesting low-level transcription.
(B) Real-time RT-PCR for three lineage-specific genes, the transcription level of which is reduced following differentiation. ESC: undifferentiated ES cells; NPC: ES cells-derived neuronal progenitor cells; PMN: ES cells-derived pot-mitotic neurons.
Figures S4. ChIP-seq data for the selected lineage-specific genes
(A–B) Chromatin immunoprecipitation (ChIP) followed by single-molecule-based sequencing (ChIP-seq) using Illumina/Solexa technology was performed and described by Mikkelsen et al (2007). In this approach, DNA molecules that are pulled-down by ChIP are spotted on an array and amplified locally. Then, successive rounds of primer-mediated single-base extension are applied, using fluorescently labeled reversible terminators, and imaged after every round. ChIP was performed with the noted antibodies: H3K4m3 is shown in green and H3K27m3 in red for both ES cells (ESC) and ES cell-derived neuronal progenitor cells (NPC). Gene names and their genomic locations appear at the top of each panel and complete gene annotations at the bottom. While genes are bivalently marked in ESC, the H3K4m3/H3K27m3 ratio is always higher in ESC except for Synaptotagmin which is expressed in NPC. These results support low-level transcription of these transcripts in ES cells.
(C) ChIP-seq data for classic markers of the three germ layers are shown. FGF5 is a marker for ectoderm, BMP4 for mesoderm and Sox17 for endoderm. In all cases, the H3K4m3/H3K27m3 ratio is higher in ESC supporting transcriptional activation at low levels. All data in this figure is from Mikkelsen et al, 2007.
Figure S5. Microarray data validation
(A) Tiling microarray expression pattern of the stem cell markers Oct4 (Pou5f1) (top left) and Sox15 (top, right) as well as the neuronal progenitor markers Sox4 (bottom, left) and Sox11 (bottom, right) in ES cells (ESC, white) and in ES cells-derived neuronal progenitor cells (NPC, green). Each panel represent composite graphs describing signal intensity form the three independent biological replicas to represent probe intensity per genomic coordinates. All coordinates are in the mm.NCBIv33 version of the mouse genome.
(B) Semi-quantitative real-time RT-PCR for Oct4, Sox15, Sox4 and Sox11. The following primers were used: Oct4-F: 5-AGCTGCTGAAGCAGAAGAGG-3; Oct4-R: 5-CCTGGGAAAGGTGTCCTGTA-3 Sox4-F: 5-CGCCTTGGTGATTTCTTGTT-3; Sox4-R: 5-ATCACCCCAGAGCCTTCTTT-3 Sox11-F: 5-CAGGACCTGTTGCTGTCAGA-3; Sox11-R: 5-GGTCACGAAGCTGGTTTGAT-3 Sox15-F: 5-CGGCGTAAGAGCAAAAACTC-5; Sox15-R: 5-TGGGATCACTCTGAGGGAAG-3
(C) Microarray detection. Shown are the annotated regions of albumin (top) and CD8 (bottom). Both genes display only background levels of transcription on the microarray and most exons do not generate transfrags.
Figure S6. Cluster size distribution maps of the probes present in ESC and absent in NPC
In order to determine the contiguity of the detected transcription in ESC cells, probes selected to be positive in ES cells but absent in NPC were clustered into bins of varying sizes. The frequency of each cluster is represented in the ordinate and the size (# of adjacent probes) in the abscissa. For example, a frequency of 100,000 clusters of size 3 that fit the criteria. This means 100,000 instances of three adjacent probes that were both present in ES cells and absent in NPC across the whole genome (both intragenic and intergenic regions). Since each probe encompasses 25 bp of genomic DNA with a gap of 5 bp between two adjacent probes, a cluster of 3 probes represent an approximate genomic size of 100 bp. Cluster size of 200 corresponds to 6 kb.
Figure S7. Global expression changes during ES cell differentiation – 24 hrs Vs. NPC
(A–C) Comparison of positive probes between cells 24 hrs after LIF withdrawal and between ES cells-derived neuronal progenitor cells (NPC). Total numbers of down-regulated and up-regulated probes are depicted as white and gray bars respectively for intergenic regions (A), intronic regions (B) and exonic regions (C) for all mouse chromosomes. Only probes which were positive in both time points were used for this analysis. Data represents the average of three independent experiments.
Figure S8. Comparison of expression levels of specific classes of genes in differentiating ES cells
Shown are relative expression levels of microarray data in ES cells (ESC, blue) and ES cells-derived neuronal progenitor cells (NPC, red) of histone acetyl transferases (HATs), histone deacetylases (HDACs), histone methyl transferases (HMTs), general transcription factors (GTFs) and chromatin remodeling factors (Remodelers). Data is the same as presented in Figure 5 but displayed as bar graphs rather than heat-maps.
Figure S9. Real-time qRT-PCR analysis of chromatin remodeling genes
Validation of microarray expression data for chromatin remodelers by quantitative real-time RT-PCR. From a total of 21 genes for which signal could be obtained from both ESC and NPC states, 17 displayed reduced expression in NPC, most of which correlated with the array data. Of the four genes that displayed either slight elevation or no change (Chd1, Chd4, Chd6, Smarcc2) three (Chd1, Chd6, Smarcc2) were also in agreement with the microarray data (p < 0.05). An additional 4 genes (Smarca1, Smarca5, Smarcad1 and Baz2b) which were detected only in the ES cells qRT-PCR measurements were excluded. All measurements were normalized against Cyclophilin B. Values shown are fold difference representing averages ± SD from 3 independent experiments. These results strongly corroborate the microarray analysis and reconfirm the abundance of chromatin remodeling factors and general transcription factors in undifferentiated ES cells.
Figure S10. Protein levels for various chromatin related factors during ES cell differentiation
Western blots for a variety of chromatin proteins from nuclear extracts of undifferentiated ES cells (ESC), 24 hrs following LIF withdrawal (24 h) or 7 d after induction of differentiation (NPC) were probed for the indicated proteins. Total histones served as a loading control.