Abstract
Background
We reported genome-wide significant linkage on chromosome 15q25.3–26.2 to recurrent early-onset major depressive disorder (MDD-RE). Here we present initial linkage-disequilibrium (LD) fine-mapping of this signal and sequence analysis of NTRK3 (neurotrophic receptor kinase-3), a biologically plausible candidate gene.
Methods
In 300 pedigrees informative for family-based association, 1195 individuals were genotyped for 795 SNPs. We resequenced 21 exons and seven highly conserved NTRK3 regions in 176 MDD-RE cases to test for an excess of rare functional variants, and in 176 controls for case-control analysis of common variants.
Results
LD mapping showed nominally significant association in nine genes–NTRK3, FLJ12484, RHCG, DKFZp547K1113, VPS33B, SV2B, SLCO3A1, RGMA and MCTP2–with MDD-RE. In NTRK3, five SNPs had nominally significant p-values (0.035–0.001). Sequence analysis revealed 35 variants (24 novel, including nine rare exonic); the number of rare variants did not exceed chance expectation. Case-control analysis of 13 common variants showed modest nominal association of MDD-RE with rs4887379, rs6496463 and rs3825882 (p = 0.008, 0.048, and 0.034), which were in partial LD with four of five associated SNPs from the family-based experiment.
Conclusions
Common variants in NTRK3 or one of the other genes identified might play a role in MDD-RE. However, much larger studies will be required for full evaluation of this region.
Keywords: NTRK3, TRKC, Neurotrophin, tag SNPs, Association, Major Depression
Introduction
In our Genetics of Recurrent Early-Onset Depression (GenRED) project, the 15q25.3–26.2 region produced the greatest linkage evidence to recurrent early-onset major depressive disorder MDD-RE) in preliminary (1) and final (2) genome scan analyses. Linkage fine-mapping of this region with single nucleotide polymorphisms (SNPs) in 631 European-ancestry families produced genome-wide significant evidence for linkage (3). Here we present initial linkage disequilibrium (LD) mapping of this signal--the first LD mapping study of a linkage candidate region for MDD--which suggested nine possible candidate genes, including NTRK3 (neurotrophic receptor kinase 3).
We also present here further study of NTRK3, which encodes a receptor that binds neurotrophin 3 (NT3) (4). Antidepressants are neuroprotective in the hippocampus (5) and alterations in neurotrophins, particularly BDNF and possibly also NT3, could influence MDD through loss of neuroprotective effects (6). In post-mortem MDD brain, there is evidence for upregulation of NTRK3 (7). A transgenic mouse model over-expressing ntrk3 showed increased anxiety-like behaviors (8). No MDD genetic association studies have been published for NT3 or NTRK3. We therefore carried out a resequencing experiment of NTRK3 in 176 GenRED MDD-RE cases and 176 controls.
Methods and Materials
Clinical methods have been described elsewhere (2); MDD-RE was defined as two or more episodes of DSM-IV MDD with onset before age 31 in probands or 41 in relatives, as suggested by previous family studies (9). Subjects gave written informed consent under IRB-approved protocols. Two partially overlapping samples were selected for LD mapping and resequencing, respectively, in the current study. LD mapping analyses were performed in 300 families informative for family-based association analysis by virtue of having parents and/or unaffected siblings available (813 affected/382 unaffected genotyped individuals, see Supplementary Table S1). For resequencing, from the 176 families with the greatest evidence for 15q25–26 linkage (including 98 from the LD mapping sample), we selected the case with the highest IBD sharing with affected relatives. European-American controls (N=176) selected from the NIMH repository (http://zork.wustl.edu/nimh/home/d_controls.html#) had no MDD-RE (nor bipolar disorder, schizophrenia, or other psychotic disorder) by self-report (10).
Initially, 1,056 SNPs with adequate Illumina design scores (≥ 0.6) were selected from HapMap, ABI, Celera and dbSNP (Build 34) to cover the linkage peak from rs1822237 to rs727896 (85,776,199 to 94,499,478 on build 36.2). SNP density was 5–6 kb within 78 genes and predicted genes, and 10–12 kb in intergenic regions. 795 SNPs were successfully genotyped by the Center for Inherited Disease Research using the Illumina GoldenGate assay. Of these, 11 were excluded for minor allele frequencies < 0.01, and three for deviations from Hardy-Weinberg equilibrium, computed using unrelated unaffecteds, at p ≤ 0.001, so that results for 781 SNPs are presented here. Quality control results were excellent: of 1,615,890 attempted genotypes, 99.89% were called and 0.04% were inconsistent with family structure. There were 0.011% discordances among 52,960 duplicate genotypes, and 0.011% inconsistencies in parent-child controls. For either parent-child pairs or trios, when the rate of errors or inconsistencies exceeded 2%, these were excluded. Three families surpassed this threshold.
Thirty primer sets were used to resequence ~16 kb of NTRK3 in 176 MDD cases, including all exons and flanking intronic regions, as well as highly conserved regions, on an ABI 3100 Genetic Analyzer. We covered 21 exons from three alternative transcripts (Figure 1), and the seven most conserved (LOD value: 173–292; size: 172–465 bp) non-exonic regions from the UCSC Genome Browser “most conserved region” track (11). The resequenced regions in which we identified non-synonymous variants or common (MAF > 0.05) polymorphisms in cases were also resequenced in 176 controls to determine whether these were unique to or over-represented in cases.
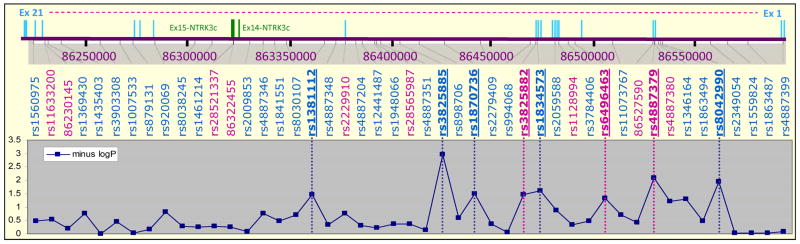
Figure. Genomic organization and allelic association results for the NTRK3 gene.
The upper panel shows the exonic structure of the NTRK3 gene while the middle panel depicts the distribution of 47 common variants (MAF > 0.05) analyzed in the family-based (blue) and case-control (pink) association studies. The lower panel shows the allelic association –log p-values obtained through TRANSMIT and Fisher’s exact test statistics. The associated SNPs (p < 0.05) are marked in bold and underlined.
Family-based association analyses were carried out using TRANSMIT (12), using the robust variance estimator to correct for prior linkage and the use of multiple siblings in the families (see Supplementary Methods). Power was evaluated by simulating 1,000 replicates of the sample under a multiplicative model (Table 1). For a threshold of p = 0.001 (in the range of the best results observed here), power was reasonable for RR = 1.5, but low for RR = 1.3. Larger samples will be required to achieve sufficient power to detect significant association after correction for multiple testing.
Table 1.
Power of the LD mapping sample to detect association using TRANSMIT under a multiplicative model
| Power for p-values of:
|
|||
|---|---|---|---|
| RRa | Freqb | 0.05 | 0.001 |
| 1.3 | 0.1 | 0.64 | 0.17 |
| 1.3 | 0.25 | 0.75 | 0.25 |
| 1.3 | 0.5 | 0.73 | 0.22 |
| 1.5 | 0.1 | 0.97 | 0.68 |
| 1.5 | 0.25 | 0.98 | 0.77 |
| 1.5 | 0.5 | 0.97 | 0.73 |
RR refers to allelic relative risk.
Freq refers to risk allele frequencies.
For the case-control resequencing experiment, we determined whether the number of variants identified in cases was significantly higher than would be expected in this length of sequence, using a population genetics-based method (13). This method compares the number of variations detected to the number expected in a particular sequence, taking into account sequence length and differing rates of variation in exons vs. introns. For more common variants, allelic case-control association was tested using Fisher’s exact test statistic.
Results
Results of association tests for all 781 SNPs are shown in Supplementary Table S2. The 43 tests with nominal p < 0.05 are shown in Table 2. There were not more such tests than would be expected by chance, and no p-value would be considered significant after correction for multiple testing. Nominally positive results were observed for SNPs in nine known genes–NTRK3, FLJ12484, RHCG, DKFZp547K1113, VPS33B, SV2B, SLCO3A1, RGMA and MCTP2.
Table 2.
LD mapping results: SNPs with nominally significant p-values
| rs ID | Loc (Build 36.2) | MAF | Gene | Region | Chi-sq | p-value |
|---|---|---|---|---|---|---|
| rs1381112 | 86 366 855 | 0.125 | NTRK3 | Intron | 4.44 | 0.0351 |
| rs3825885 | 86 403 845 | 0.328 | NTRK3 | Intron | 10.70 | 0.0011 |
| rs1870736 | 86 425 625 | 0.425 | NTRK3 | Intron | 4.56 | 0.0327 |
| rs1834573 | 86 471 117 | 0.298 | NTRK3 | Intron | 5.11 | 0.0239 |
| rs8042990 | 86 532 457 | 0.318 | NTRK3 | Intron | 6.46 | 0.0110 |
| rs1530309 | 86 640 008 | 0.043 | 5.37 | 0.0204 | ||
| rs1561806 | 86 657 188 | 0.495 | 5.79 | 0.0161 | ||
| rs2028389 | 86 967 765 | 0.143 | FLJ12484 | Intron | 5.33 | 0.0209 |
| rs2072693 | 87 815 949 | 0.481 | RHCG | Exon synon | 4.45 | 0.0349 |
| rs1256840 | 88 329 016 | 0.122 | 5.71 | 0.0169 | ||
| rs1256841 | 88 349 309 | 0.121 | DKFZp547K1113 | Intron | 4.77 | 0.0290 |
| rs1867225 | 89 364 833 | 0.401 | VPS33B | Intron | 4.61 | 0.0317 |
| rs1005060 | 89 389 551 | 0.425 | 4.53 | 0.0334 | ||
| rs2597909 | 89 462 723 | 0.082 | SV2B | Intron | 4.75 | 0.0293 |
| rs4525467 | 89 840 777 | 0.398 | 8.02 | 0.0046 | ||
| rs4300626 | 89 896 689 | 0.198 | 5.65 | 0.0175 | ||
| rs4632107 | 89 941 183 | 0.334 | 7.22 | 0.0072 | ||
| rs7165398 | 90 195 099 | 0.255 | SLCO3A1 | Promoter | 6.86 | 0.0088 |
| rs1568209 | 90 382 294 | 0.467 | SLCO3A1 | Intron | 6.21 | 0.0127 |
| rs2892291 | 90 430 815 | 0.337 | SLCO3A1 | Intron | 10.71 | 0.0011 |
| rs871167 | 90 435 137 | 0.335 | SLCO3A1 | Intron | 11.13 | 0.0008 |
| rs2286355 | 90 439 197 | 0.374 | SLCO3A1 | Exon synon | 4.79 | 0.0287 |
| rs207974 | 90 439 656 | 0.256 | SLCO3A1 | Intron | 8.62 | 0.0033 |
| rs2048945 | 90 443 405 | 0.381 | SLCO3A1 | Intron | 4.90 | 0.0269 |
| rs1517620 | 90 451 267 | 0.292 | SLCO3A1 | Intron | 9.63 | 0.0019 |
| rs207964 | 90 453 177 | 0.149 | SLCO3A1 | Intron | 6.33 | 0.0119 |
| rs2238360 | 90 457 120 | 0.442 | SLCO3A1 | Intron | 4.03 | 0.0447 |
| rs732546 | 90 491 552 | 0.358 | SLCO3A1 | Intron | 4.96 | 0.0259 |
| rs2132616 | 91 039 384 | 0.407 | 4.36 | 0.0368 | ||
| rs1534780 | 91 426 217 | 0.422 | RGMA | Intron | 7.06 | 0.0079 |
| rs7180175 | 92 080 839 | 0.485 | 6.49 | 0.0109 | ||
| rs1351306 | 92 388 421 | 0.434 | 5.33 | 0.0210 | ||
| rs11853883 | 92 520 920 | 0.288 | 4.72 | 0.0298 | ||
| rs2117215 | 92 680 688 | 0.461 | MCTP2 | Intron | 5.67 | 0.0172 |
| rs2388779 | 93 058 282 | 0.288 | 3.84 | 0.0499 | ||
| rs2388881 | 93 084 371 | 0.344 | 6.48 | 0.0109 | ||
| rs2388883 | 93 122 138 | 0.193 | 6.64 | 0.0100 | ||
| rs1026453 | 93 377 084 | 0.241 | 4.48 | 0.0343 | ||
| rs4984553 | 93 444 400 | 0.275 | 5.57 | 0.0182 | ||
| rs1471169 | 93 463 523 | 0.306 | 6.56 | 0.0104 | ||
| rs1834212 | 93 876 557 | 0.403 | 4.27 | 0.0388 | ||
| rs2397813 | 93 929 511 | 0.393 | 5.75 | 0.0165 | ||
| rs766716 | 94 489 374 | 0.208 | 4.52 | 0.0336 |
Two long genes contained multiple nominally significant SNPs: NTRK3 and SLCO3A1. Resequencing of SLCO3A1 is ongoing. Resequencing of NTRK3 revealed 35 sequence variations. The 24 novel variants we identified did not exceed the number expected by chance (N = 40). Rarer variants (frequency < 5%) are listed in Table 3 along with the number of cases who were carriers. There were nine rare variants in exons including two novel rare missense mutations (Arg306His in exon 9 and Asn714Ser in exon 17), each observed in one case, but absent in controls. The remaining rare variants were observed in other regions as shown in the table.
Table 3.
Frequencies of 22 uncommon variants identified through resequencing of NTRK3 in 176 MDD cases
| Loc (Build 36.2) | Regionb,c | Variant | # of Heterozygotes (MAF) |
|---|---|---|---|
| 86 600 697 | 5′ of gene | G/T | 2 (0.006) |
| 86 600 687 | 5′ of gene | C/A | 1 (0.003) |
| 86 600 681 | 5′ of gene | G/C | 1 (0.003) |
| 86 600 601 | Exon 1 (5′UTR) | CGG repeatd | 2 (0.006) |
| 86 600 250 | Exon 2 | CGG→AGG (synonymous Arg) | 4 (0.011) |
| 86 577 488 | Intron 2 (cons 1) | A/G | 1 (0.003) |
| 86 528 357 | Intron 3 | C/T | 1 (0.003) |
| 86 489 484 | Intron 5 (cons 4) | G/A | 9 (0.026) |
| 86 489 409 | Intron 5 (cons 4) | T/C | 1 (0.003) |
| 86 480 234 | Exon 8 | ACG→ACA (synonymous Thr) | 1 (0.003) |
| 86 479 623 | Exon 9 | CGT→CAT (Arg→His)e | 1 (0.003) |
| 86 323 519 | Intron 13 (Exon 15 of NTRK3c-3′UTR) | T/C | 1 (0.003) |
| 86 323 054 | Intron 13 (Exon 15 of NTRK3c-3′UTR) | G/A | 1 (0.003) |
| 86 322 802 | Intron 13 (Exon 15 of NTRK3c-3′UTR) | C/A | 2 (0.006) |
| 86 314 079 | Intron 13 (cons 7) | C/T | 1 (0.003) |
| 86 313 972 | Intron 13 (cons 7) | C/T | 2 (0.006) |
| 86 313 902 | Intron 13 (cons 7) | C/T | 3 (0.009) |
| 86 284 909 | Exon 14 | GCC→GCT (synonymous Ala) | 1 (0.003) |
| 86 277 456 | Intron 14 | G/A | 8 (0.023) |
| 86 277 203 | Intron 15 | C/T | 2 (0.006) |
| 86 277 170a | Intron 15 | T/G | 16 (0.045) |
| 86 229 963 | Exon 17 | AAT→AGT (Asn→Ser)e | 1 (0.003) |
dbSNP ID rs1006046 (Genotype data from Hap Map project available at www.hapmap.org). All other variants are novel.
Regions are in reference to the NTRK3a transcript except where the NTRK3c transcript is specified.
“cons” refers to highly conserved regions that are numbered in accordance with their location beginning from the 5′ end of NTRK3.
Two cases were heterozygous for ‘CGG’ repeat alleles of length 4 and 8; all other subjects were homozygous for the 8-repeat allele.
These two nsSNP were also sequenced in controls, and no carriers were observed.
The experiment was designed to test the hypothesis of an excess of rare non-synonymous mutations in cases. Because only two such mutations were observed in cases, i.e., not enough for a significant excess to be observed, the test of the hypothesis was conclusively negative even without sequencing controls. However, we went on to sequence a limited number of exons in controls: those containing non-synonymous variants or common (MAF > 0.05) polymorphisms.
More common variants (frequency > 5%) are listed in Table 4, and results of case-control association tests are also shown. All were in Hardy-Weinberg equilibrium in both cases and controls. Allele frequencies of three of these SNPs differed in cases vs. controls at the p < 0.05 level without correcting for multiple tests. Two of these three SNPs were in almost complete LD with each other, and all were in modest, but significant LD with four of the five nominally associated SNPs in the family-based LD mapping experiment (Supplementary Figure 1). Thus the association observed in the family-based analysis was also observed in the case-control analysis although the results would not be significant after correcting for multiple tests. All association results for NTRK3 are depicted in Figure 1.
Table 4.
Frequency distribution and case-control association analysis of common variants in the NTRK3 gene
| Loc (Build 36.2) | Regiona,b | Variant | rs ID | Group | Genotype Counts | Allele Counts (Frequencies) | p-valued | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Maj/Maj | Het | Min/Min | Major | Minor | ||||||
| 86 528 755 | Intron 2 | T/A | rs4887380 | Cases | 107 | 62 | 7 | 276 (0.784) | 76 (0.216) | 0.063 |
| Controls | 120 | 53 | 3 | 293 (0.832) | 59 (0.168) | |||||
| 86 528 340 | Intron3 | G/C | rs4887379 | Cases | 100 | 66 | 10 | 266 (0.756) | 86 (0.244) | 0.008 |
| Controls | 120 | 53 | 3 | 293 (0.832) | 59 (0.168) | |||||
| 86 527 590 | Intron 4 | C/T | Novel | Cases | 111 | 59 | 6 | 281 (0.798) | 71 (0.202) | 0.386 |
| Controls | 108 | 61 | 2 | 277 (0.810) | 65 (0.190) | |||||
| 86 489 658 | Intron 5 | A/G | rs6496463 | Cases | 36 | 92 | 45 | 164 (0.474) | 182 (0.526) | 0.048 |
| Controls | 55 | 79 | 41 | 189 (0.540) | 161 (0.460) | |||||
| 86 481 688 | Exon6 | AAC→AAT (synonymous Asn) | rs1128994 | Cases | 94 | 70 | 12 | 258 (0.733) | 94 (0.267) | 0.466 |
| Controls | 92 | 76 | 8 | 260 (0.739) | 92 (0.261) | |||||
| 86 470 386 | Intron 12 | C/G | rs3825882c | Cases | 38 | 93 | 44 | 169 (0.483) | 181 (0.517) | 0.034 |
| Controls | 60 | 73 | 41 | 193 (0.555) | 155 (0.445) | |||||
| 86 392 534 | Intron 12 (cons 6) | G/T | rs28565987 | Cases | 138 | 34 | 2 | 310 (0.891) | 38 (0.109) | 0.431 |
| Controls | 141 | 34 | 1 | 316 (0.898) | 36 (0.102) | |||||
| 86 377 189 | Exon13 | CGC→CGG (synonymous Arg) | rs2229910 | Cases | 67 | 85 | 24 | 219 (0.622) | 133 (0.378) | 0.173 |
| Controls | 79 | 74 | 23 | 232 (0.659) | 120 (0.341) | |||||
| 86 322 455 | Intron 13 (Exon 15 of NTRK3c-3′UTR) | G/A | Novel | Cases | 156 | 17 | 2 | 329 (0.940) | 21 (0.060) | 0.556 |
| Controls | 156 | 19 | 1 | 331 (0.940) | 21 (0.060) | |||||
| 86 322 284 | Intron 13 (Exon 15 of NTRK3c-3′UTR) | G/C | rs28521337 | Cases | 37 | 92 | 47 | 166 (0.472) | 186 (0.528) | 0.530 |
| Controls | 41 | 84 | 51 | 166 (0.472) | 186 (0.528) | |||||
| 86 230 145 | Intron 16 | T/C | Novel | Cases | 152 | 24 | 0 | 328 (0.932) | 24 (0.068) | 0.616 |
| Controls | 153 | 21 | 2 | 327 (0.929) | 25 (0.071) | |||||
| 86 230 080 | Intron 16 | T/C | rs11633200 | Cases | 17 | 86 | 73 | 120 (0.341) | 232 (0.659) | 0.290 |
| Controls | 26 | 76 | 74 | 128 (0.364) | 224 (0.636) | |||||
| 86 224 467 | Intron 18 | G/A | rs1560975c | Cases | 23 | 88 | 65 | 134 (0.381) | 218 (0.619) | 0.630 |
| Controls | 23 | 81 | 67 | 127 (0.371) | 215 (0.629) | |||||
Regions are in reference to the NTRK3a transcript except where the NTRK1c transcript is specified.
“cons” refers to highly conserved regions that are numbered in accordance with their location beginning from the 5′ end of NTRK3.
Genotype data from Hap Map project available at www.hapmap.org.
Uncorrected p-values < 0.05 are marked in bold.
Discussion
An initial LD mapping study of the 15q25–26 MDD-RE linkage region produced nominally significant evidence for association of MDD-RE to common variants in nine genes, but none of these findings can be considered statistically significant given the multiple tests performed. It is possible that common variants in one or more of these genes play a role in susceptibility to MDD-RE. However, much larger studies and denser SNP maps will be required for full evaluation of this region.
We began our investigations of these genes by carrying out a resequencing study of NTRK3, with the primary goal of identifying any rare functional susceptibility variants. Although we identified 21 rare novel variants in NTRK3, we did not observe an excess of rare functional variants in cases. Further, the total number of variants identified did not exceed that expected by chance. Among the two novel missense mutations we did identify, Arg306His in exon 9 is located in the neurotrophin binding domain, within which mutations could significantly reduce the affinity of the NTRK3 receptor for neurotrophin 3 (14).
Our case-control analysis revealed modest association of three common NTRK3 variants (rs4887379, rs6496463, rs3825882). Interestingly, these SNPs are located quite close to and are in modest LD with the ones that showed family-based association. The finding that similar associations were seen both with family-based and case-control methods suggests that the signal is unlikely to be the result of genotyping or other artifact.
Several limitations of this study should be considered. We could have missed true case-control differences in the frequency of rare variants, as: a) rare variation was studied neither in the introns nor the flanking regions; and, b) there could be case-control differences in the frequency of very rare variants requiring a larger sample to detect. Further, our case-control findings could be false positives due to population stratification, though the similar self-reported ancestries of our cases and controls (Supplementary Table S3) make this less likely, as do our positive family-based association findings. We are currently conducting a genome-wide association study in a much larger group of cases and controls, which should help clarify the role of NTRK3 and other chromosome 15q25–26 genes in MDD-RE susceptibility.
Supplementary Material
Acknowledgments
This work was supported by NIMH grants 5R01MH059542 (Crowe), 5R01MH059552 (DePaulo/Potash), 5R01MH061686 and 1K24MH64197 (Levinson), 5R01MH059541 (Scheftner) and 5R01MH060912 (Weissman). The GenRED cell and data collections used in this study included contributions from Dr. George S. Zubenko and Dr. Wendy N. Zubenko, Department of Psychiatry, University of Pittsburgh School of Medicine, that were supported by R01 grant MH60866 from the National Institute of Mental Health (GSZ, PI). Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number N01-HG-65403. The NIMH Cell Repository at Rutgers University and the NIMH Center for Collaborative Genetic Studies on Mental Disorders made essential contributions to this project. Thanks to Barbara Schweizer, Brandie Craighead, and Jennifer Toolan for their assistance. The authors express their profound appreciation to the families who participated in this project, and to the many clinicians who facilitated the referral of participants to the study. Data and biomaterials were collected in six projects that participated in the National Institute of Mental Health (NIMH) Genetics of Recurrent Early-Onset Depression (GenRED) project. From 1999–2003, the Principal Investigators and Co-Investigators were: New York State Psychiatric Institute, New York, NY, R01 MH060912, Myrna M. Weissman, Ph.D. and James K. Knowles, M.D., Ph.D.; University of Pittsburgh, Pittsburgh, PA, R01 MH060866, George S. Zubenko, M.D., Ph.D. and Wendy N. Zubenko, Ed.D., R.N., C.S.; Johns Hopkins University, Baltimore, R01 MH059552, J. Raymond DePaulo, M.D., Melvin G. McInnis, M.D. and Dean MacKinnon, M.D.; University of Pennsylvania, Philadelphia, PA, RO1 MH61686, Douglas F. Levinson, M.D. (GenRED coordinator), Madeleine M. Gladis, Ph.D., Kathleen Murphy-Eberenz, Ph.D. and Peter Holmans, Ph.D. (University of Wales College of Medicine); University of Iowa, Iowa City, IW, R01 MH059542, Raymond R. Crowe, M.D. and William H. Coryell, M.D.; Rush University Medical Center, Chicago, IL, R01 MH059541-05, William A. Scheftner, M.D. Rush-Presbyterian. Control subjects from the National Institute of Mental Health Schizophrenia Genetics Initiative (NIMH-GI), data and biomaterials are being collected by the “Molecular Genetics of Schizophrenia II” (MGS-2) collaboration. The investigators and coinvestigators are: ENH/Northwestern University, Evanston, IL, MH059571, Pablo V. Gejman, M.D. (Collaboration Coordinator; PI), Alan R. Sanders, M.D.; Emory University School of Medicine, Atlanta, GA,MH59587, Farooq Amin, M.D. (PI); Louisiana State University Health Sciences Center; New Orleans, Louisiana, MH067257, Nancy Buccola APRN, BC, MSN (PI); University of California-Irvine, Irvine, CA, MH60870, William Byerley, M.D. (PI); Washington University, St. Louis, MO, U01, MH060879, C. Robert Cloninger, M.D. (PI); University of Iowa, Iowa, IA,MH59566, Raymond Crowe, M.D. (PI), Donald Black, M.D.; University of Colorado, Denver, CO, MH059565, Robert Freedman, M.D. (PI); University of Pennsylvania, Philadelphia, PA, MH061675, Douglas Levinson M.D. (PI); University of Queensland, Queensland, Australia, MH059588, Bryan Mowry, M.D. (PI); Mt. Sinai School of Medicine, New York, NY,MH59586, Jeremy Silverman, Ph.D. (PI).
Footnotes
Financial Disclosures
Dr. Weissman received investigator initiated grants from Lilly and GlaxoSmithkline. Drs. Verma, Grover, Holmans, Knowles, Grover, Evgrafov, Crowe, Scheftner, DePaulo, and Potash reported no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Holmans P, Zubenko GS, Crowe RR, DePaulo JR, Jr, Scheftner WA, Weissman MM, et al. Genomewide significant linkage to recurrent, early-onset major depressive disorder on chromosome 15q. Am J Hum Genet. 2004;74:1154–1167. doi: 10.1086/421333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmans P, Weissman MM, Zubenko GS, Scheftner WA, Crowe RR, DePaulo JR, Jr, et al. Genetics of recurrent early-onset major depression (GenRED): final genome scan report. Am J Psychiatry. 2007;164:248–258. doi: 10.1176/ajp.2007.164.2.248. [DOI] [PubMed] [Google Scholar]
- 3.Levinson DF, Evgrafov OV, Knowles JA, Potash JB, Weissman MM, Scheftner WS, et al. Genetics of recurrent early-onset major depression (GenRED): significant linkage on chromosome 15q25-q26 after fine-mapping with SNP markers. Am J Psychiatry. 2007;164:259–264. doi: 10.1176/ajp.2007.164.2.259. [DOI] [PubMed] [Google Scholar]
- 4.Lamballe F, Klein R, Barbacid M. trkC, a new member of the trk family of tyrosine protein kinases, is a receptor for neurotrophin-3. Cell. 1991;66:967–979. doi: 10.1016/0092-8674(91)90442-2. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe Y, Gould E, Daniels DC, Cameron H, McEwen BS. Tianeptine attenuates stress-induced morphological changes in the hippocampus. Eur J Pharmacol. 1992;222:157–162. doi: 10.1016/0014-2999(92)90830-w. [DOI] [PubMed] [Google Scholar]
- 6.Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. Biol Psychiatry. 2006;59:1116–1127. doi: 10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Higgs BW, Elashoff M, Richman S, Barci B. An online database for brain disease research. BMC Genomics. 2006;7:70. doi: 10.1186/1471-2164-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dierssen M, Gratacos M, Sahun I, Martin M, Gallego X, Amador-Arjona A, et al. Transgenic mice overexpressing the full-length neurotrophin receptor TrkC exhibit increased catecholaminergic neuron density in specific brain areas and increased anxiety-like behavior and panic reaction. Neurobiol Dis. 2006;24:403–418. doi: 10.1016/j.nbd.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 9.Weissman MM, Gershon ES, Kidd KK, Prusoff BA, Leckman JF, Dibble E, Hamovit J, Thompson WD, Pauls DL, Guroff JJ. Psychiatric disorders in the relatives of probands with affective disorders: The Yale NIMH collaborative family study. Arch Gen Psychiatry. 1984;41:13–21. doi: 10.1001/archpsyc.1984.01790120015003. [DOI] [PubMed] [Google Scholar]
- 10.Sanders AR, Duan J, Levinson DF, Shi J, He D, Hou C, Burrell GJ, Rice JP, Nertney DA, Olincy A, Rozic P, Vinogradov S, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Crowe RR, Cloninger CR, Martinez M, Gejman PV. Association of 14 candidate genes to schizophrenia in a large European ancestry sample: Implications for psychiatric genetics. Am J Psychiatry. doi: 10.1176/appi.ajp.2007.07101573. in press. [DOI] [PubMed] [Google Scholar]
- 11.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clayton D. A generalization of the transmission/disequilibrium test for uncertain-haplotype transmission. Am J Hum Genet. 1999;65:1170–1177. doi: 10.1086/302577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell AA, Chakravarti A, Cutler DJ. On the probability that a novel variant is a disease-causing mutation. Genome Res. 2005;15:960–966. doi: 10.1101/gr.3761405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urfer R, Tsoulfas P, O’Connell L, Hongo JA, Zhao W, Presta LG. High resolution mapping of the binding site of TrkA for nerve growth factor and TrkC for neurotrophin-3 on the second immunoglobulin-like domain of the Trk receptors. J Biol Chem. 1998;273:5829–5840. doi: 10.1074/jbc.273.10.5829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.