Abstract
Regulation of connective tissue growth factor (CCN2/CTGF) in gingival fibroblasts is unique and may provide therapeutic opportunities to treat oral fibrotic diseases. RhoA was previously implicated in mediating the expression of CCN2/CTGF. We now present evidence that Rho family GTPases Rac1 and Cdc42 are the principal mediators of the transforming growth factor-β1 (TGFβ1)-stimulated expression of CCN2/CTGF in primary human gingival fibroblasts. TGFβ1 does not stimulate RhoA activation in gingival fibroblasts, and the overexpression of dominant-negative RhoA does not reduce CCN2/CTGF expression in response to TGFβ1. In contrast, the overexpression of dominant-negative forms of Cdc42 or Rac1 results in a dramatic reduction of CCN2/CTGF protein levels. Lovastatin and a geranylgeranyltransferase inhibitor reduce the TGFβ1-stimulated levels of CCN2/CTGF protein by ∼75 and 100%, respectively. We previously demonstrated that JNK1 phosphorylation by TGFβ1 is also critical for TGFβ1-induced CCN2/CTGF expression, and forskolin partially reduces levels of phosphorylated JNK1. Inhibition of geranylgeranyltransferase has no effect on levels of JNK phosphorylation in response to TGFβ1 suggesting Rho-GTPases act independently of JNK1. The combination of lovastatin and forskolin results in a greater inhibitory effect than each agent alone and reduces CCN2/CTGF mRNA and protein expression by greater than 90%. This novel combination has additive inhibitory effects on the TGFβ1-stimulated expression of CCN2/CTGF in human gingival fibroblasts through the simultaneous disruption of Rho- and JNK1-mediated pathways, respectively. This combination of available therapeutic compounds may therefore be useful in designing treatment strategies for oral fibrotic conditions in which gingival CCN2/CTGF is elevated.
Connective tissue growth factor belongs to the CCN family of growth factors, designated CCN2 or CTGF (1-7). Transforming growth factor β (TGFβ)2 and lysophosphatidic acid (LPA) stimulate CCN2/CTGF expression in human fibroblastic cells (6-9). CCN2/CTGF is responsible for mediating some of the effects of TGFβ and has been implicated in the onset and progression of fibrosis in most human tissues (4, 6, 7, 10-28).
We previously reported that CCN2/CTGF is highly expressed in tissues from patients with phenytoin-induced gingival overgrowth and hereditary gingival fibromatosis (29-31). Tissues derived from these patients contain a high level of CCN2/CTGF expression and are more fibrotic compared with other forms of drug-induced gingival overgrowth, which do not overexpress CCN2/CTGF (31). We therefore consider CCN2/CTGF to be a potential therapeutic target for designing treatment alternatives against the onset and progression of phenytoin-induced and hereditary forms of gingival overgrowth. Because patients taking the anti-convulsive agent, phenytoin, may not be able to discontinue medication, the indicated treatment for gingival overgrowth tissues is the repeated, painful surgical excision of the fibrotic tissue. A less traumatic mode of intervention is needed.
It has been reported that the re-growth of fibrotic gingival tissues in these patients is rapid following surgery (32-34). The standard surgical excision of gingival tissues in patients with hereditary or phenytoin-induced gingival overgrowth results in the recruitment of platelets and normal clotting factors early in the post-surgical healing processes. A potential mechanism for the rapid re-growth of gingival tissues is the early recruitment of platelets to the wound. Platelets are rich in LPA and also carry CCN2/CTGF protein (35-39). In this study we investigate the mechanism of LPA and TGF-β1 stimulation of CCN2/CTGF in primary cultures of gingival fibroblasts and identify additive effects and unique signaling pathways that mediate their effects.
Primary human gingival fibroblasts are unique in their resistance to the inhibition of the TGFβ1-induced expression of CCN2/CTGF by prostaglandin E2 (PGE2), whereas in primary human renal mesangial cells and IMR90 human lung fibroblasts PGE2 potently blocks CCN2/CTGF expression stimulated by TGFβ1 (40). Although it has been demonstrated that renal fibroblasts require activated RhoA to promote CCN2/CTGF expression in response to LPA and TGFβ (8, 9), the role of the Rho family GTPases in mediating the TGFβ1-stimulated expression of CCN2/CTGF has yet to be determined in gingival cells. Here we demonstrate that the Rho family GTPases Rac1 and Cdc42, and not RhoA, are essential mediators in the TGFβ1-stimulated expression of CCN2/CTGF in primary human gingival fibroblasts. These findings demonstrate another novel aspect of the cell/tissue-specific differences between human fibroblastic cell types in regulating CCN2/CTGF expression. Data presented provide potentially useful information that may lead to the development of novel therapeutic strategies designed to minimize fibrosis caused by the overexpression of CCN2/CTGF in forms of oral tissue overgrowth.
EXPERIMENTAL PROCEDURES
Reagents—Forskolin, H89, and mevalonolactone were obtained from Sigma. PGE2, mouse anti-actin monoclonal antibody, Y27632, geranylgeranyltransferase inhibitor (GGTI), farnesyltransferase inhibitor (FTI), lovastatin-sodium and simvastatin-sodium were obtained from Calbiochem; recombinant adenovirus, adenoviral titer kit, and β-galactosidase staining kit were from CellBiolabs; LPA was from Avanti Polar Lipids; the exotoxin C3-ADP-ribosyltransferase of Clostridium botulinum was from Cytoskeleton, Inc.; Chariot protein delivery reagent was acquired from Active Motif; RNeasy mini-RNA purification kits were purchased from Qiagen; TGFβ1 was from PeproTech; rabbit anti-human CCN2/CTGF polyclonal antibody was kindly provided by FibroGen Corp., South San Francisco, CA; LumiGLO chemiluminescent detection system, rabbit anti-mitogen-activated protein kinase (MAPK) antibodies against phospho-stress-activated protein kinase/JNK (Thr-183, Tyr-184; catalog number 9251), total stress-activated protein kinase/JNK (catalog number 9252), and secondary anti-rabbit horseradish peroxidase-conjugated antibodies were obtained from Cell Signaling Inc.; Dulbecco's modified Eagle's medium, phosphate-buffered saline, trypsin, nonessential amino acids, and antibiotics (penicillin/streptomycin) were from Invitrogen; all solutions, Taqman probes, and instruments for real time qPCR were obtained from Applied Biosystems; 100-mm and 6-well cell culture plates were from Fisher; Restore Western blot Stripping Solution and nuclear extraction kits were from Pierce.
Cell Culture—Primary human gingival fibroblasts were obtained from donors from the Clinical Research Center, Boston University Goldman School of Dental Medicine, as described previously (41). Gingival fibroblasts from two different adult donors were used, and data obtained were reproducible. Data shown are from one donor (HCT-11). Fibroblasts from all tissues were grown in Dulbecco's modified Eagle's medium supplemented with 0.1 mm nonessential amino acids, 10% fetal bovine serum, and 100 units/ml penicillin and 0.1 mg/ml streptomycin at 37 °C and 5% CO2 in a fully humidified incubator in 100-mm cell culture dishes. Cells were grown to 80% confluence and then placed in serum-free medium containing 0.1% bovine serum albumin for a minimum of 12 h prior to treatment. Specified inhibitors were next added in serum-free bovine serum albumin medium for 30 or 60 min prior to the addition of TGF-β1 or vehicle. TGF-β1 treatments were generally for either 4 h for RNA analyses or 6 h for protein analyses, based on previous studies. Cells were not passed beyond four passages.
Cultures of gingival fibroblasts were treated with simvastatin, lovastatin, GGTI, or FTI for 12 h prior to treatment with TGFβ1. Cultures were then incubated for an additional 6 h prior to harvest of total cell layer protein to assess CCN2/CTGF protein expression via Western blot. Mevalonolactone (Sigma) hydrolysis to mevalonate salt was accomplished according to protocols described previously (41). Briefly, mevalonolactone was incubated in 0.1 n NaOH, pH 7.4, for 2 h at 50 °C. Aliquots of 0.1 m mevalonate salt solution were then stored at -20 °C until required. In experiments involving the C3 exotoxin of C. botulinum, the C3-mediated inhibition of Rho-GTPases, gingival fibroblast cultures were transfected with 5 μg/ml C3 using the Chariot protein transfection reagent. Transfection complexes were added to cultures and incubated for 12 h prior to the introduction of TGFβ1. Cultures were incubated for an additional 6 h prior to harvest of total cell layer protein for Western blot analysis.
Real Time Quantitative PCR (qPCR)—Total RNA was obtained using RNeasy Midi columns (Qiagen) following the manufacturer's protocol. Total RNA was isolated from fibroblast cell cultures and purified using RNeasy mini-RNA purification kits (Qiagen). RNA obtained was run on a 1% agarose gel to ensure RNA quality by analyzing for the presence of 18 S and 28 S rRNA in proper relative proportions. RNA was quantified via spectrophotometry, and 1 μg of RNA per treatment condition was added to 30 μl of reverse transcription reactions using random primers and the Applied Biosystems reverse transcription kit. Thermal cycler conditions for reverse transcription were 25 °C for 10 min, 37 °C for 60 min, and 95 °C for 5 min, and cDNA was stored at -20 °C prior to use in real time PCR. Four microliters of each reverse transcription reaction was used for 50-μl real time PCRs using the traditional 96-well format. Taqman probes for human CCN2/CTGF (catalog number Hs00170014_m1) and glyceraldehyde-3-phosphate dehydrogenase (catalog number Hs99999905_m1) were used in real time PCR analyses. Real time PCRs were run in an Applied Biosystems GeneAmp Prism 7700 system at thermal cycler conditions of 50 °C for 2 min, 95 °C for 10 min, and 60 °C for 1 min for 40 cycles. Data were analyzed using the 2-ΔΔct method, and CCN2/CTGF mRNA levels were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA and no treatment controls.
Western Blot Analysis—Fibroblast cell cultures were grown as described above, and total cell layer protein was harvested by dissolving cell layers in 500 μl of sample buffer containing 62.5 mm Tris, 10% glycerol, 2% SDS, and 5% β-mercaptoethanol per 100-mm culture dish. Samples were then boiled for 5 min and stored at -80 °C. Samples were then subjected to 10% SDS-PAGE and Western blotting with antibodies specific for protein of interest. Proteins were transferred to polyvinylidene difluoride membrane overnight at 4 °C in blotting buffer containing 0.025 m Tris, 0.192 m glycine, and 20% methanol. Membranes were blocked with 5% dry milk in TBST (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.1% Tween) for 1 h followed by the addition of specific primary rabbit antibody in blocking solution overnight at 4 °C with mild shaking. Membranes were then washed with TBST 5 times for 10 min each. Membranes were next incubated with secondary anti-rabbit horseradish peroxidase-conjugated antibodies in 5% milk and TBST for 2 h at room temperature with mild shaking. Membranes were washed with TBST five times for 5 min each. Chemiluminescent detection of bound horseradish peroxidase-conjugated secondary antibodies was determined using the LumiGlo/peroxide (Cell Signaling) and exposed to film. Membranes were subsequently stripped using Restore Western Stripping Solution and re-probed for loading controls as required. Densitometric analysis was performed using a Bio-Rad VersaDoc 3000 Imaging System.
Extraction of Nuclear Proteins—Nuclei were collected, and nuclear lysates were purified using the nuclear extract kit from Active Motif with strict adherence to the manufacturer's guidelines. Briefly, cultures of primary human gingival fibroblasts were grown to near confluence and serum-starved for 12 h and then treated with 10 μm LPA and/or 5 ng/ml TGFβ1 for the times indicated. Cells were washed twice with cold phosphate-buffered saline containing the phosphatase mixture provided and then scraped and collected in the same buffer. Detached cells were spun at 500 rpm at 4 °C, and the supernatant was discarded. Hypotonic buffer (1×) was added to the pellets and incubated on ice for 15 min. Detergent was added, and the cells were vortexed for 10 s and then centrifuged for 30 s at 14,000 × g at 4 °C. Supernatant (cytosolic fraction) was saved, and Complete Lysis Buffer was added to each pellet and vortexed for 10 s. Samples were incubated at 4 °C for 30 min while rocking, vortexed for 30 s, and then centrifuged at 14,000 × g at 4 °C for 10 min. Supernatant (nuclear fraction) was saved and stored at -80 °C.
Recombinant Adenovirus Dominant-negative Studies—Gingival fibroblasts were grown in 12-well plates to 80% confluence. Adenovirus expressing β-galactosidase was used as the control for assessment of the level of infection and gene expression in our cells. Different dominant-negative adenoviral constructs (1010 infectious units/ml) were introduced, and cultures were grown for 48 h to permit expression of virus-encoded genes. At this concentration we achieved 100% infection of gingival fibroblast cultures as determined by β-galactosidase staining. Forty eight hours post-infection, TGFβ1 was added, and cultures were incubated for times indicated prior to harvest of total cell layer protein for Western blot analysis. β-Galactosidase staining and estimated viral titer was conducted according to the manufacturer's guidelines. Recombinant adenovirus expressing the dominant-negative forms of the following were used in our experiments and were purchased from Cell Biolabs, Inc.; Cdc42 (N17, catalog number ADV-153), RhoA (N19, catalog number ADV-156), Ras (N17, catalog number ADV-145), and Rac1 (N17, catalog number ADV-150).
RhoA Activation Assay—The presence of activated RhoA was determined in response to various stimuli using the EZ-detect RhoA activation kit from Pierce. Briefly, 100-mm plates of gingival fibroblasts were grown to 80% confluence and serum-starved for 12 h. Two culture plates were then treated with the indicated concentrations of TGFβ1, LPA, PGE2, or forskolin for 3 min. Cells were then lysed with 500 μl of lysis/binding/wash buffer, and lysates were transferred to a labeled microcentrifuge tube and briefly vortexed. Lysates were incubated on ice for 5-min then microcentrifuged at 16,000 × g for 15 min at 4 °C. Supernatants were stored on ice in a fresh microcentrifuge tube. 400 μg of GST-Rhotekin-RBD was added to individual spin columns, and 700 μl of sample lysate was added to each column and briefly vortexed. Binding was permitted to continue for 1 h at 4 °C. Spin columns were then microcentrifuged at 7,200 × g for 30 s, and resin was washed three times with 400 μl of kit wash buffer. Fifty microliters of sample buffer (see under “Western Blot Analysis”) was then added to each column, and samples were boiled for 5 min. Finally, columns were microcentrifuged at 7,200 × g for 2 min, and flow-through was stored at -20 °C and then subjected to 2% SDS-PAGE for analysis of activated/bound RhoA. Lysates treated with GTPγSor GDP were used as positive and negative controls of RhoA activation, respectively.
C3/Chariot Transfection Study—Cell cultures of primary gingival fibroblasts were grown to 80% confluence. Each culture was treated with the Chariot small protein transfection reagent as described in the manufacturer's guidelines (Active Motif) combined with 5 μg of purified C3 exotransferase from C. botulinum (Cytoskeleton, Inc.). Negative and positive (TGFβ1 alone) control cultures were mock-transfected with Chariot without the addition of C3. Cultures were transfected using Chariot/C3 for 12 h in the presence of serum-free medium. Culture media were then removed and replaced with media supplemented with 5 ng/ml TGFβ1 and incubated for an additional 6 h prior to harvest of total cellular protein to be used for Western blot analysis of CCN2/CTGF protein expression.
RESULTS
Kinetics of CCN2/CTGF mRNA and Protein Expression Differ in Response to TGFβ1 and LPA in Human Gingival Fibroblasts—In human renal fibroblasts and mesangial cells, 10 μm LPA induces CCN2/CTGF mRNA expression more than TGFβ1 (8, 9, 42). Because the regulation of CCN2/CTGF differs between human renal and gingival fibroblasts (40), we wished to determine and compare the kinetics of CCN2/CTGF expression in response to TGFβ1 and LPA in primary gingival fibroblasts. Primary human gingival fibroblast cultures were grown to 80% confluence and serum-starved for 12 h. Cultures were then treated with either 5 ng/ml TGFβ1 or 10 μm LPA for 4 h prior to harvest of total RNA or 6 h prior to harvest of total cell layer protein.
Data indicate that the expression of both CCN2/CTGF mRNA and protein increases gradually after treatment with TGFβ1 (Fig. 1, A and B). In contrast, stimulation with LPA produced a rapid increase in CCN2/CTGF mRNA 2 h after treatment, which rapidly returned to nearly unstimulated levels by 6 h (Fig. 1A). The LPA-induced expression of CCN2/CTGF protein remained elevated 6 h after treatment (Fig. 1B). Interestingly, the effect of the combined treatment of TGFβ1 and LPA appears to increase CCN2/CTGF protein levels above LPA or TGFβ1 alone (Fig. 1B).
FIGURE 1.
The kinetics of the TGFβ1- and LPA-stimulated expression of CCN2/CTGF mRNA and protein levels differ in primary human gingival fibroblasts. Near confluent cultures of primary gingival fibroblasts were serum-starved for 12 h and then treated with 10 μm LPA, 5 ng/ml TGFβ1, or both for 2 or 6 h prior to harvest of total RNA or protein. A, real time qPCR analysis of the fold change in CCN2/CTGF mRNA expression versus no treatment control cultures. B, Western blot analysis of CCN2/CTGF protein expression. Each experiment was conducted at least twice with identical results. Data are represented as the mean ± S.D. and statistical evaluation per each experimental condition determined versus control, no treatment, cultures. *, p < 0.001; **, p < 0.05; #, p < 0.01.
TGFβ1 and LPA Stimulate Similar Levels of JNK1 Phosphorylation in Human Gingival Fibroblasts—We previously demonstrated that JNK1 phosphorylation induced by TGFβ1 is essential for maximal CCN2/CTGF expression (40). We hypothesize that, like TGFβ1, LPA might stimulate JNK1 activation. To test this, cultures of primary gingival fibroblasts were grown to near confluence and serum-starved for 12 h. Cells were then stimulated with either 5 ng/ml TGFβ1 and/or 10 μm LPA for 0-15 min. Western blot analysis of JNK1 phosphorylation indicates that LPA stimulates JNK1 (Fig. 2A), and densitometric analyses demonstrate that the LPA-induced stimulation of JNK1 activation is similar in intensity as that observed following stimulation with TGFβ1 (Fig. 2B). Because we had observed an apparent additive effect of TGFβ1 in combination with LPA on CCN2/CTGF expression (Fig. 1B), we wished to determine whether the levels of activated JNK1 were similarly increased in response to co-stimulation. Data show no increase in activated JNK1 beyond that observed in response to TGFβ1 alone (Fig. 2C). This finding suggests that LPA stimulation of CCN2/CTGF in gingival fibroblasts may be mediated by signal transduction pathways in addition to JNK1 activation.
FIGURE 2.
TGFβ1 and LPA stimulate comparable levels of phospho-JNK1 in human gingival fibroblasts. Gingival fibroblast cultures serum-starved for 12 h were treated with 10 μm LPA, 5 ng/ml TGFβ1, or both for times indicated prior to harvest of total cell layer protein. A, representative Western blot of phosphorylated JNK1 in response to LPA or TGFβ1. B, densitometric analysis from three separate experiments showing no significant difference between the TGFβ1- and LPA-stimulated inductions of phosphorylated JNK1. Data are represented as the mean ± S.D. and statistical evaluation per each experimental condition determined versus control, no treatment, cultures. C, Western blot of JNK1 protein phosphorylation in response to TGFβ1 alone or in combination with LPA.
LPA Is a Weak Stimulator of Smad3 Nuclear Localization—Activated Smad3 has been described previously as being required for CCN2/CTGF expression (43, 44), consistent with the finding that the CCN2/CTGF promoter contains Smad-binding and TGFβ1-response elements (6, 7, 45). The ability of LPA to stimulate CCN2/CTGF expression led us to ask whether or not LPA was capable of stimulating the activation and nuclear localization of active Smad3. To test this, we first wished to determine a time course for nuclear localization of phosphorylated Smad3 in response to TGFβ1 in gingival fibroblasts. Cultures of primary gingival fibroblasts were grown and serum-starved overnight. Cells were then treated with 5 ng/ml TGFβ1 for the times indicated prior to collection and extraction of nuclei (Active Motif). The purity of the nuclear fraction was determined by the lack of a detectable cytoplasmic marker (α-tubulin) and the presence of a nuclear marker (lamin B) in these preparations (Fig. 3A). Data indicate that the TGFβ1-stimulated nuclear localization of phospho-Smad3 progressively increased with time of treatment (Fig. 3B). Interestingly, data also show that LPA alone was also capable of stimulating the nuclear localization of phospho-Smad3 (Fig. 3B). However, levels of nuclear phospho-Smad3 observed in response to LPA, at 60 min after treatment, were substantially lower than levels observed in response to TGFβ1 (Fig. 3C). These data suggest that LPA stimulation of Smad3 nuclear localization could contribute to the observed additive increase in CCN2/CTGF levels seen in Fig. 1B.
FIGURE 3.
LPA is significantly less effective than TGFβ1 in stimulating the nuclear localization of phosphorylated Smad3. Gingival fibroblasts were serum-starved for 12 h and then treated with 5 ng/ml TGFβ1 and/or 10 μm LPA for times indicated. Nuclei were harvested and purified, and total nuclear protein was retained for Western blot analysis of nuclear phospho-Smad3. A, Western blot for α-tubulin (cytoplasmic marker) and lamin B (nuclear marker) demonstrating that purified nuclear extracts used in these experiments were free of cytoplasmic proteins. B, Western blot demonstrating nuclear levels of phospho-Smad3 following treatment with TGFβ1 for the times indicated. C, Western blot of nuclear phospho-Smad3 in response to TGFβ1 and LPA or both. Each experiment was conducted at least twice with the same outcomes. B and C, lamin B, a nuclear protein, was used as the loading control.
Lovastatin, Geranylgeranyltransferase Inhibition, and Forskolin Reduce CCN2/CTGF mRNA and Protein Levels and May Suggest a Novel Potential Therapeutic Strategy—LPA is a potent inducer of Rho family small GTPases (36, 37, 46). Because LPA is capable of stimulating CCN2/CTGF expression, we considered whether Rho-GTPases could be essential in the TGFβ1-induced expression of CCN2/CTGF in gingival fibroblasts. The cholesterol biosynthesis pathway plays a major role in providing key isoprene units required by Rho family and Ras GTPases. Transfer of these small lipid molecules to cytosolic forms of small GTPases is accomplished by either geranylgeranyltransferase (geranylgeranylation for Rho-GTPases) or farnesyltransferase (for Ras). This lipid modification of the cytosolic GTPase is a critical step in the GTPase localization to the plasma cell membrane where they can be available for activation by external stimuli (47-51).
Similarly, HMG-CoA reductase inhibitors, or statins, block the synthesis of cholesterol and its precursors by inhibiting an initial and rate-limiting step in this pathway and the conversion of HMG-CoA to mevalonate that is required for the post-translational modification of Rho family and Ras GTPases (47-51). If Rho or Ras GTPases were involved in mediating the induction of CCN2/CTGF expression with TGFβ1, then the inhibition of the lipid modification required for their activation at the cell membrane should reduce CCN2/CTGF levels stimulated by TGFβ1.
Primary human gingival fibroblasts were grown to near confluence and serum-starved, in the presence of 20 μm lovastatin, for 12 h. Cultures were then stimulated with 5 ng/ml TGFβ1 for 6 h prior to harvest of total cell protein for Western blot analyses. Data in Fig. 4A indicate that lovastatin is more potent at inhibiting the TGFβ1-induced expression of CCN2/CTGF than simvastatin in human gingival fibroblasts. To demonstrate that the inhibition of CCN2/CTGF by lovastatin was specific to the cholesterol pathway, we wished to determine whether the addition of exogenous mevalonate would rescue CCN2/CTGF protein expression inhibited by lovastatin. Gingival fibroblast cultures were again grown and serum-starved in the presence of lovastatin or lovastatin plus mevalonate for 12 h. Cells were then stimulated for 6 h with TGFβ1, and CCN2/CTGF protein levels were assessed via Western blot. Fig. 4B demonstrates that the addition of exogenous mevalonate completely rescues CCN2/CTGF protein levels inhibited by lovastatin.
FIGURE 4.
Lovastatin and forskolin work in concert to completely block the TGFβ1-induced expression of CCN2/CTGF in human gingival fibroblasts. A, Western blot of CCN2/CTGF protein levels in response to treatment with 20 μm of either lovastatin or simvastatin. Cell cultures were pretreated with lovastatin or simvastatin for 12 h. Media were then aspirated and replaced with media containing lovastatin or simvastatin combined with 5 ng/ml TGFβ1 and incubated for 6 h prior to harvest of total cell layer protein for Western blot analysis of CCN2/CTGF. B, Western blot of CCN2/CTGF protein in response to 20 μm lovastatin and 20 μm mevalonate. Cultures were pretreated with lovastatin for 12 h. Media were then aspirated and replaced with media containing lovastatin plus 20 μm mevalonate and 5 ng/ml TGFβ1. Cultures were incubated for an additional 6 h prior to harvest of total cell layer protein for Western blot analysis of CCN2/CTGF. C, Western blot of the TGFβ1-stimulated expression of CCN2/CTGF and the effects of 20 μm GGTI or FTI inhibitors. Cultures were pretreated with 20 μm GGTI or FTI for 12 h. Media were then aspirated, and GGTI or FTI was combined with 5 ng/ml TGFβ1. Cultures were then incubated for 6 h prior to harvest of total cell layer protein for Western blot analysis of CCN2/CTGF. D, Western blot of phosphorylated JNK1 levels in response to 5 ng/ml TGFβ1 and 20 μm GGTI. Cell cultures were pretreated with GGTI-supplemented media for 12 h. Media were then aspirated, and 20 μm GGTI was combined with 5 ng/ml TGFβ1, and cells were harvested at intervals, as indicated. E, real time qPCR analysis of CCN2/CTGF mRNA expression in response to challenge with 10 μm forskolin, 20 μm lovastatin, or both. Cell cultures were pretreated with 20 μm lovastatin (12 h) or 10 μm forskolin (1 h). Media were then aspirated and either lovastatin or forskolin was combined with 5 ng/ml TGFβ1 and incubated for an additional 4 h prior to harvest of total mRNA for real time qPCR of CCN2/CTGF mRNA. In experiments combining lovastatin and forskolin, cultures were pretreated with 20 μm lovastatin for 11 h. Medium was aspirated and replaced with medium containing 20 μm lovastatin and 10 μm forskolin and incubated for 1 h. Culture medium was again replaced with lovastatin and forskolin containing 5 ng/ml TGFβ1. Cultures were incubated for an additional 4 h prior to isolation of total RNA for real time qPCR analysis; *, p < 0.0001; **, p < 0.000005 compared with TGF-β1-stimulated cells. F, Western blot analysis demonstrating the strong combined inhibitory effect of lovastatin and forskolin on the TGFβ1-induced expression of CCN2/CTGF protein. Cultures were treated as described in E, but the final incubation time was 6 h prior to the harvest of total cell layer protein for Western blot analysis of CCN2/CTGF. Each experiment was repeated at least twice with the same outcomes. Representative blots are shown. Real time qPCR experiments were conducted twice in triplicate, and data are represented as the mean ± S.D. and statistical evaluation per each experimental condition determined versus control, no treatment, cultures.
To confirm our hypothesis that Rho family GTPases are required in the TGFβ1-induced expression of CCN2/CTGF, we utilized specific inhibitors of the GGTI and FTI and followed CCN2/CTGF protein levels stimulated by TGFβ1 by Western blot. Gingival fibroblasts were grown and serum-starved in the presence of either 20 μm GGTI or FTI. Data indicate that complete inhibition of the TGFβ1-induced expression of CCN2/CTGF protein could be achieved by inhibiting geranylgeranyltransferase, the pathway required for the subsequent activation of Rho family GTPases (RhoA, Rac, and Cdc42) (Fig. 4C). Inhibition of farnesyltransferase (Ras) resulted in ∼25% reduction in CCN2/CTGF protein levels (Fig. 4C).
Activation of the c-Jun N-terminal kinase 1 (JNK1) is essential for maximal expression of CCN2/CTGF in response to TGFβ1 in gingival fibroblasts (40). We hypothesized that Rho family GTPases were required in mediating the TGFβ1-induced activation of JNK. Thus, we wished to determine whether JNK phosphorylation by TGFβ1 was a consequence of stimulation by Rho-GTPases. Because GGTI was effective at completely blocking CCN2/CTGF expression, we proposed that GGTI would also strongly inhibit JNK phosphorylation induced by TGFβ1. Primary gingival fibroblast cultures were grown and serum-starved in the presence or absence of 20 μm GGTI for 12 h. Cultures were then stimulated with TGFβ1 for 5 or 10 min prior to harvest of total cell layer protein for Western blot analysis of JNK1 phosphorylation. Surprisingly, data indicate that treatment with GGTI had no effect on the TGFβ1-stimulated phosphorylation of JNK1 (Fig. 4D) suggesting that Rho-GTPase activity is not required for the phosphorylation of JNK1. Data indicate that multiple TGF-β1-stimulated signaling pathways work in parallel to stimulate CCN2/CTGF expression in gingival fibroblasts.
As noted, geranylgeranyltransferase inhibition blocks CCN2/CTGF expression but has no effect on JNK1 activation (Fig. 4, C and D). The diminished CCN2/CTGF levels in response to inhibition of the cholesterol biosynthesis pathway are therefore because of effects other than inhibiting JNK activation. We hypothesized that the combination of partial JNK inhibition with forskolin (40), and the independent inhibitory effects of lovastatin directed at Rho family GTPases, could together more fully reduce CCN2/CTGF protein levels in response to TGFβ1 in gingival fibroblasts. To test this, gingival cell cultures were treated with 10 μm forskolin alone or in combination with 20 μm lovastatin for 12 h in serum-free media. Media were then removed and replaced with serum-free media containing TGFβ1 in combination with equivalent concentrations of forskolin, lovastatin, or both. Cultures were incubated an additional 4 h for real time PCR analysis of CCN2/CTGF mRNA expression or 6 h prior to harvest of total cellular protein for Western blot analysis of CCN2/CTGF protein. We found that each forskolin and lovastatin produced a similar reduction in CCN2/CTGF mRNA expression, whereas the combination of forskolin and lovastatin reduced CCN2/CTGF mRNA levels to that observed in untreated cultures (Fig. 4E). Likewise, the combination of forskolin and lovastatin reduced CCN2/CTGF protein expression induced by TGFβ1 to basal levels (Fig. 4F). Because both lovastatin and forskolin can be employed clinically, these findings may suggest a novel therapeutic strategy to address oral fibrotic conditions.
Similar Reductions of CCN2/CTGF Levels Can Be Achieved with Lower Concentrations of Lovastatin—According to previously published data in renal and lung fibroblasts (9), the IC50 values of HMG-CoA reductase inhibitors simvastatin, lovastatin, and pravastatin were found to be ∼1, 3, and 500 μm, respectively. In experiments to this point, we have used 20 μm lovastatin to observe the effects of this inhibitor on CCN2/CTGF expression and a short incubation period (12 h) prior to stimulation with TGFβ1. We wished to determine whether lower concentrations of lovastatin would yield a similar reduction in CCN2/CTGF protein levels. We also theorized that a longer incubation time (24 versus 12 h) might result in an enhanced inhibition of CCN2/CTGF levels because it provided more time to deplete cellular levels of lipid isoprene units required for GTPase activation. To test these hypotheses, gingival cell cultures were grown and serum-starved for 12 h in the presence of 1, 5, 10, or 20 μm lovastatin. Media were then replaced with serum-free media containing equivalent levels of lovastatin with TGFβ1, and cultures were incubated for an additional 6 h. Total cellular protein was harvested for Western blot analysis of CCN2/CTGF protein expression. Data indicate that 10 μm lovastatin produced a nearly identical level of inhibition of CCN2/CTGF protein induced by TGFβ1 as 20 μm lovastatin (Fig. 5A). Concentrations of lovastatin less than 10 μm were less effective at reducing CCN2/CTGF protein levels (Fig. 5A). Moreover, cultures challenged with 10 or 20 μm lovastatin for 12 or 24 h demonstrate that the inhibitory effects of these concentrations of lovastatin were not enhanced with longer treatment times (Fig. 5B). Although the longer incubation may reduce CCN2/CTGF protein levels further, the difference between the 12- and 24-h incubation at either concentration of lovastatin was insignificant (Fig. 5, B and C). Thus, these data demonstrate that 10 μm lovastatin is as effective as 20 μm lovastatin at reducing the TGFβ1-induced expression of CCN2/CTGF in human gingival fibroblasts. It is interesting to note that challenge of cultured gingival fibroblasts retain normal morphological, fibroblastic appearance beyond 24 h of treatment with either concentration of lovastatin.
FIGURE 5.
Lovastatin is equally potent at inhibiting the TGFβ1-stimulated expression of CCN2/CTGF at concentrations of 10 and 20 μm. Cultures of primary human gingival fibroblasts were grown as described above. Cells were then treated for 12 h with increasing concentrations of lovastatin for 12 h or for 12 or 24 h with either 10 or 20 μm lovastatin. Culture media were then aspirated and replaced with equivalent concentrations of lovastatin and 5 ng/ml TGFβ1. Cells were incubated for an additional 6 h prior to harvest of total cell layer protein. The effects of the duration of the inhibition with lovastatin and the concentration on the TGFβ1-induced (5 ng/ml) expression of CCN2/CTGF protein were assessed using Western blot analysis. A, Western blot showing the effects of different concentrations of lovastatin on the TGFβ1-induced expression of CCN2/CTGF protein. B, representative Western blot of the effects of different concentrations of lovastatin and incubation times on CCN2/CTGF protein. C, densitometric analysis of CCN2/CTGF protein levels normalized to total β-actin. Statistical significance was determined comparing lovastatin-treated lanes to those treated with TGFβ1 alone (*, p < 0.0001). Experiments were repeated at least twice with the same findings.
C3 Exotoxin of C. botulinum Significantly Reduces CCN2/CTGF Expression in Response to TGFβ1—The Rho family GTPases are implicated in the TGFβ1-induced expression of CCN2/CTGF in that the inhibition of geranylgeranylation leads to the complete loss of CCN2/CTGF protein expressed and that LPA can itself stimulate CCN2/CTGF expression. To more thoroughly explore this possibility and confirm the hypothesis that Rho-GTPases were critical in the TGFβ1-stimulated expression of CCN2/CTGF in gingival cells, we next used recombinant C. botulinum C3. C3 has been described as a specific inhibitor of RhoA activity by ADP-ribosylation (52). However, there is also evidence that C3 can block the activity of other Rho family GTPases, in particular Rac (the Ras-related C3 botulinum toxin substrate 1) (53).
To determine whether Rho-GTPases were involved in the TGFβ1-induced expression of CCN2/CTGF in primary gingival fibroblasts, the purified form of the C3 exotransferase protein was introduced into gingival cells using the Chariot transfection reagent. We have previously used the Chariot reagent and have determined that it is effective at introducing proteins directly into primary gingival fibroblasts (40). Gingival fibroblasts were then incubated for 12 h with 5 μg of C3 protein/Chariot reagent. Media were then aspirated and replaced with serum-free medium containing TGFβ1. Cultures were incubated for an additional 6 h prior to harvest of total protein for Western blot analysis of CCN2/CTGF protein. Data indicate that C3 strongly reduced the TGFβ1-stimulated expression of CCN2/CTGF protein in primary gingival fibroblasts (Fig. 6A). These data provide further evidence of the importance of Rho-GTPases in mediating the TGFβ1-stimulated expression of CCN2/CTGF in gingival fibroblasts.
FIGURE 6.
The TGFβ1-stimulated expression of CCN2/CTGF is inhibited in gingival cells transfected with the C3 exotransferase of C. botulinum. Gingival fibroblast cultures were grown to near confluence and then placed in serum-free media containing the Chariot small protein transfection reagent and 5 μg of C3 protein. Cultures were incubated for 2 h, and media were replaced with fresh, serum-free media and incubated for 10 h. 5 ng/ml TGFβ1 was then added to cultures with or without C3 and incubated for an additional 6 h prior to harvest of total cell protein. A, representative Western blot demonstrating the inhibition of C3 on the TGFβ1-induced expression of CCN2/CTGF. The ROCK-specific inhibitor Y27632 blocks elevations in CCN2/CTGF protein levels stimulated by TGFβ1 without reducing levels of phosphorylated JNK1. B, Western blot of CCN2/CTGF protein showing the inhibitors Y27632 and H89 block the TGFβ1-induced expression of CCN2/CTGF in gingival cells. Serum-starved cultures of gingival fibroblasts were pretreated with 10 μm of either Y27632 or H89 for 1 h prior to the addition of 5 ng/ml TGFβ1. After a subsequent 6-h incubation, total protein was collected for Western blot analyses of CCN2/CTGF protein. Western blot of phosphorylated JNK1 showing that the inhibition of CCN2/CTGF by Y27632 (C) or H89 (D) was not because of the reduction of JNK1 activation. Cultures were pretreated with either 10 μm Y27632 or H89 for 1 h. Medium was then replaced with fresh medium containing an equivalent concentration of inhibitor and 5 ng/ml TGFβ1 and incubated for times indicated prior to harvest of total protein for analysis of phosphorylated JNK. Each experiment was conducted at least twice with the same results. E, LPA, not TGFβ1, stimulates the activation of RhoA in primary human gingival fibroblasts. Gingival fibroblasts were serum-starved and then treated with either 5 ng/ml TGFβ1, 10 μm LPA, 1 μm PGE2, or 10 μm forskolin for 3 min. One set of cultures was treated with the combination of TGFβ1 and LPA. Cells were collected for pulldown assay for activated RhoA. GDP and GTPγS were used as negative and positive controls, respectively.
Inhibition Studies with Y27632 or H89 Eliminates CCN2/CTGF Expression without Affecting JNK Activation—The inhibitor Y27632 is a potent inhibitor of the Rho-associated coiled coil forming kinase (ROCK) (54, 55). Y27632 is described as a “ROCK-specific” inhibitor in some studies and by some manufacturers. Y27632 is, however, an even more potent inhibitor of PRK2 activity (54, 55). Another effector protein that shares structural and sequence homology with ROCK is the myotonic dystrophy kinase-related Cdc42-binding kinase (MRCK) (56, 57), and it seems likely that this inhibitor could inhibit the function of MRCK, although the effect of Y27632 on MRCK activity has not been documented. Each of these molecules, ROCK, PRK2, and MRCK, are downstream effectors of RhoA, RhoA/Rac, and Rac/Cdc42, respectively. We hypothesized that if Rho family GTPases were essential in mediating the TGFβ1-stimulated expression of CCN2/CTGF in gingival cells, then Y27632 would diminish TGFβ1-induced CCN2/CTGF expression.
Primary gingival fibroblast cultures were grown and serum-starved as described for previous experiments. Cultures were then pretreated with 10 μm Y27632 for 1 h. Media were aspirated and replaced with fresh medium containing Y27632 and TGFβ1. Cells were incubated for an additional 6 h, and total cell protein was collected for Western blot analysis of CCN2/CTGF protein levels. Data demonstrate that inhibition with Y27632 completely blocks CCN2/CTGF protein expression induced by TGFβ1 in gingival fibroblasts (Fig. 6B), and the data suggest that downstream effectors of Rho-GTPases may be viable targets for slowing the onset and progression in vivo in pro-fibrotic conditions such as phenytoin-induced gingival overgrowth and hereditary gingival fibromatosis where CCN2/CTGF levels are elevated. Because we had previously found that JNK1 activation is critical in the TGFβ1-induced expression of CCN2/CTGF in gingival cells, we investigated the possibility that Y27632 may affect this process. We found that Y27632 did not reduce levels of phosphorylated JNK1 stimulated by TGFβ1 (Fig. 6C).
H89 is commonly used as a PKA inhibitor. However, this compound is more effective than Y27632 at blocking ROCK activity and is actually more effective in inhibiting ROCK activity than it is at reducing the activity of PKA (54, 55). With this knowledge we wished to determine whether H89 was as effective as Y27632 at reducing CCN2/CTGF expression. We found that, like Y27632, H89 completely blocked CCN2/CTGF protein levels (Fig. 6B). Moreover, H89 had no substantial effect on the TGFβ1-stimulated activation of JNK1 (Fig. 6D). It is important to note that other PKA inhibitors, the PKA inhibitory peptide (data not shown) or KT5720 (40), did not reduce the TGFβ1-stimulated expression of CCN2/CTGF in gingival cells.
RhoA Activation Is Induced by LPA but Not TGFβ1 in Human Gingival Fibroblasts—LPA is a strong inducer of RhoA activation in fibroblastic cells. Previous reports indicate that in renal fibroblasts RhoA activity mediates CCN2/CTGF expression in response to stimulation with both LPA and TGFβ1 (8, 9, 42). We hypothesized, therefore, that both LPA and TGFβ1 stimulation could result in RhoA activation in gingival fibroblasts. Gingival fibroblasts were grown to near confluence and serum-starved for 12 h prior to stimulation with 5 ng/ml TGFβ1, 10 μm LPA, 1 μm PGE2 or 10 μm forskolin for 3 min. Cells were lysed, and the assay was conducted using reagents supplied in an EZ-Detect Rho activation assay kit from Pierce capable of detecting activated RhoA, RhoB, and RhoC. Our data show that LPA, and not TGFβ1, was capable of inducing RhoA activation in primary gingival fibroblasts (Fig. 6E) and suggest that the TGFβ1-stimulated expression of CCN2/CTGF was mediated by members of the Rho-GTPase family other than RhoA, RhoB, or RhoC.
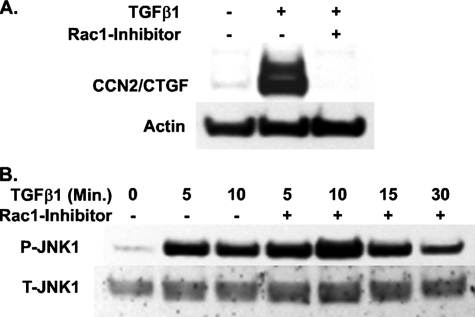
Inhibition of Rac1 Eliminates CCN2/CTGF Expression Induced by TGFβ1 without Effecting JNK Activation—As noted, inhibition of the post-translational modification of cytoplasmic Rho-GTPases with GGTI inhibited CCN2/CTGF protein expression induced by TGFβ1 (Fig. 4C). With the knowledge that TGFβ1 was not stimulating RhoA activation (Fig. 6E), we wished to determine whether another Rho family GTPase, Rac, was involved in mediating this pathway. We therefore used an inhibitor of Rac1 guanine exchange factors Trio and Tiam1 to block Rac activity. This inhibitor, NSC23766, has no effect on inhibiting the activities of RhoA or Cdc42 (58). Primary gingival fibroblasts were grown and serum-starved for 12 h prior to treatment with 100 μm Rac1 inhibitor for 1 h. Culture medium was then removed and replaced with fresh media containing 100 μm Rac1 inhibitor and 5 ng/ml TGFβ1. Cultures were then grown for an additional 6 h prior to harvest of total protein for Western blot analysis of CCN2/CTGF protein. Similar to the effect of inhibitors Y27632 or GGTI, the Rac1 inhibitor prevented CCN2/CTGF protein expression in response to TGFβ1 treatment (Fig. 7A). These data further implicate Rho-GTPases other than RhoA in increasing CCN2/CTGF expression in gingival fibroblasts treated with TGFβ1.
FIGURE 7.
Inhibition of Rac1 activation leads to the complete loss of TGFβ1-induced CCN2/CTGF expression without interfering with JNK1 phosphorylation. Gingival fibroblasts were serum-starved and then pretreated with 100 μm Rac1 inhibitor for 1 h. Cultures were then treated with 5 ng/ml TGFβ1 for 6 h prior to harvest of total cellular protein Western blot of CCN2/CTGF protein expression (A) or for times indicated for assessment of phosphorylation of JNK1 by Western blot (B). The experiment was performed twice with the same results.
We had previously demonstrated that the activation of JNK1 by TGFβ1 was essential for maximal expression of CCN2/CTGF mRNA and protein (40). Because there is substantial evidence that suggests a potential for Rac involvement in the stimulation of JNK1 activation in some cell types, we next examined the effect of the Rac1 inhibitor on the activation of JNK1 induced by TGFβ1 in our gingival cells. We found that the inhibition of Rac1 with NSC23766 had no effect on the TGFβ1-stimulated phosphorylation of JNK1 (Fig. 7B). These data, together with our finding that the inhibition of geranylgeranyltransferase had no effect on JNK1 phosphorylation (Fig. 4D), strongly suggest that JNK1 is stimulated by TGFβ1 independent of Rho-GTPase involvement.
Overexpression of Dominant-negative Cdc42 or Rac1, but Not RhoA, Dramatically Inhibits CCN2/CTGF Expression Induced by TGFβ1—Thus far our data collected with regard to the regulation of CCN2/CTGF expression by Rho-GTPases has been based on pharmacological inhibitors. To examine more fully the role of different small GTPases in stimulating CCN2/CTGF expression, we chose to utilize recombinant adenovirus expressing dominant-negative forms of RhoA, Rac1, Cdc42, and Ras. The optimal level of virus to achieve nearly 100% infection of gingival fibroblasts in our cultures was determined using recombinant adenovirus expressing the β-galactosidase gene (Fig. 8A). Identical concentrations of virus containing all other genes were used in subsequent assays. Gingival fibroblast cultures were infected with dominant-negative adenovirus for 48 h. Culture medium was then replaced with fresh medium containing 5 ng/ml TGFβ1. Following another 6-h incubation, cells were harvested for Western blot analysis for CCN2/CTGF protein.
FIGURE 8.
Overexpression of dominant-negative Rac1 or Cdc42, and not RhoA, leads to the near complete loss of CCN2/CTGF expression induced by TGFβ1 in gingival cells. Gingival cell cultures were infected with recombinant adenovirus (Ad) expressing dominant-negative forms of either Rac1, RhoA, Ras, or Cdc42 and incubated for 48 h. Cells were then fed with media containing 5 ng/ml TGFβ1. Cultures were incubated for an additional 6 h, and total cell layer protein was harvested for Western blot analysis of CCN2/CTGF protein levels. A, levels of recombinant adenovirus expressing β-galactosidase were used to optimize viral load in primary gingival fibroblasts. Near 100% infection of cells in culture was achieved with no obvious alteration in cell morphology. The same titer of each dominant-negative (DN) virus was used to infect cultures. B, representative Western blot of CCN2/CTGF protein levels in response to the overexpression of dominant-negative GTPases. C, densitometric analysis of Western blot data obtained from three separate experiments. Bars represent the mean CCN2/CTGF protein levels normalized to total β-actin, ±S.D., and statistical evaluation per each experimental condition determined versus control, Ad-β-galactosidase-infected (Ad-β-Gal) cultures (*, p < 0.001; **, p < 0.05; ***, p < 0.0005).
These data indicate that the expression of dominant-negative Cdc42 or Rac1 blocks CCN2/CTGF expression stimulated by TGFβ1 in primary human gingival fibroblasts. Moreover, the expression of the dominant-negative form of RhoA had no significant effect on CCN2/CTGF protein levels (Fig. 8B). It was interesting to note that infection with the recombinant adenovirus expressing the dominant-negative form of Ras led to a statistically significant reduction in CCN2/CTGF expression, although not as dramatic as with Cdc42 or Rac1 (Fig. 8B). Densitometric analysis of CCN2/CTGF normalized to total β-actin is shown in Fig. 8C. Interestingly, we have found that the overexpression of recombinant, dominant-negative Cdc42 or Rac1 does not significantly alter levels of phosphorylated JNK1 in response to TGFβ1 (data not shown).
DISCUSSION
Platelets are critical components involved in the initial clotting mechanism at a wound site. Platelets are rich in LPA and CCN2/CTGF protein (35, 39). Moreover, both LPA and TGFβ are components of the normal wound healing processes, and both are potent inducers of CCN2/CTGF expression by fibroblastic cells (8). Early exposure to platelet-derived LPA and CCN2/CTGF, in combination with the latent production of TGFβ at the wound site, may work in concert to drive the production of extracellular matrix macromolecules and stimulate tissue remodeling.
The kinetics of LPA compared with TGFβ-induced expression of CCN2/CTGF mRNA in primary cultures of gingival fibroblasts differs from that reported for immortalized human renal fibroblasts. In renal fibroblasts, LPA appeared to be more potent than TGFβ in stimulating CCN2/CTGF mRNA expression (8). In contrast, primary gingival fibroblasts treated with identical concentrations of LPA stimulated a rapid yet transient induction of CCN2/CTGF expression in gingival cells, whereas TGFβ1 promotes a stronger, longer lasting elevation in CCN2/CTGF mRNA and protein. Moreover, LPA and TGFβ1 together appear to produce an additive effect in promoting CCN2/CTGF expression in cultures of primary gingival fibroblasts. Thus, upon the introduction of TGFβ to the wound site, CCN2/CTGF expression induced by both LPA and TGFβ1 would be expected to enhance levels of various extracellular matrix macromolecules. Gingival cells may therefore begin to produce CCN2/CTGF and proteinase-activated receptors components very early after wounding following surgical excision of gingival tissues and may represent a consequence of the aggregation and activation of platelets and increased local levels of LPA. Therefore, these data may provide a novel insight into the temporal effects of LPA and/or TGFβ1 in the healing processes in gingival tissues. Blood- and platelet-derived LPA and TGFβ1 may stimulate gingival CCN2/CTGF production and the rapid regrowth of gingival tissues that is observed following conventional gingi-vectomy surgery. Based on our studies in vitro, it is interesting to speculate that procedures such as electrocauterization or laser cauterization may be preferential to the traditional surgical excision of fibrotic gingival overgrowth tissues. We suggest that these techniques may serve to slow the re-initiation of fibrous overgrowth induced by the recruitment of platelets and the stimulation of the expression of extracellular matrix macromolecules indirectly by limiting CCN2/CTGF induction by gingival cells via LPA and directly by the recruitment of extra-gingival CCN2/CTGF carried on platelets.
The LPA- and TGFβ1-induced expression of CCN2/CTGF involves the activation of similar critical signaling molecules, including JNK1 (40) and Smad3. This study shows that LPA and TGFβ1 each stimulate the phosphorylation of JNK1 to a similar degree, whereas LPA is a less effective inducer of Smad3 nuclear localization compared with TGFβ1. The observed weak Smad3 nuclear localization caused by LPA in gingival fibroblasts may account for this tissue-specific response to LPA regulation of CCN/CTGF levels compared with published studies in kidney cells (8). Please see Fig. 9 for a summary of our current understanding of signaling in gingival fibroblasts.
FIGURE 9.
Several signaling pathways work together in the TGFβ1-induced expression of CCN2/CTGF in primary human gingival fibroblasts. Cdc42, Rac1, and Ras are small GTPases involved in the TGFβ1-stimulated expression of CCN2/CTGF whose activity relies upon the availability of products of the cholesterol biosynthesis pathway. Proposed stimulation of protease secretion in response to stimulation with TGFβ1 may result in the activation of protease-activated receptors and ultimately lead to the activation of Ras to prolong CCN2/CTGF expression in these cells (hypothetical pathway indicated by broken arrows). TGFβ1 and LPA both stimulate CCN2/CTGF expression in gingival cells with TGFβ1 being the more potent inducer. Both TGFβ1 and LPA stimulate the activating phosphorylation of JNK1 and nuclear localization of phosphorylated Smad3. JNK1 does not, however, enhance Smad3 nuclear localization in gingival fibroblasts but is required for TGFβ1 stimulation of CCN2/CTGF expression (40). LPA induces only a weak nuclear localization of Smad3. Data support that although LPA stimulates signaling pathways also stimulated by TGFβ1, LPA and TGFβ1 each uniquely stimulate a subset of complementary parallel signaling events that result in additive up-regulation of CCN2/CTGF levels in gingival fibroblasts.
Arresting the platelet-driven increase in CCN2/CTGF levels by nonsurgical methodologies may not be sufficient to completely block the gingival overgrowth associated with the overexpression of CCN2/CTGF. Although platelets containing LPA and CCN2/CTGF protein may help stimulate an early rapid pro-fibrotic gingival overgrowth in susceptible post-surgical patients, the prolonged stimulation of CCN2/CTGF expression by TGFβ1 in gingival fibroblasts could be involved in perpetuating this response. This is true in scleroderma lesions of the skin in which CCN2/CTGF levels are also high. In studies of CCN2/CTGF and TGFβ mRNA expression in scleroderma lesions, TGFβ mRNA was found to be relegated to the leading edges of the lesion and not in the body of the lesion. Conversely, CCN2/CTGF mRNA was highly expressed predominantly within the lesion. Together, these data suggest that TGFβ provides the impetus that drives the formation and progression of scleroderma lesions through the subsequent overexpression of CCN2/CTGF (12, 59-61). We therefore designed our studies to determine the specific signaling requirements of TGFβ1 required for stimulating CCN2/CTGF expression in primary gingival fibroblasts, which may lead to the persistence and expansion of gingival overgrowth lesions.
LPA is also a strong activator of Rho family GTPases. The LPA- or TGFβ1-stimulated expression of CCN2/CTGF in cultures of immortalized human renal fibroblasts was previously reported to require the activation of the Rho family GTPase, RhoA. Although our lab had previously demonstrated that gingival fibroblasts are unique in their ability to resist the inhibitory effects of PGE2 on the TGFβ1-stimulated expression of CCN2/CTGF (40), we now report that human gingival fibroblasts are also unique in that Cdc42 and Rac1, but not RhoA, represent the essential Rho family GTPases mediating the TGFβ1-induced expression of CCN2/CTGF in primary human gingival fibroblasts.
Individual Rho family GTPases have different effects on the cytoskeleton. RhoA activation stimulates the formation of stress fibers and focal adhesions (62), and Rac activity leads to the formation of lamellipodia and membrane ruffles (63), and Cdc42 promotes the formation of filopodia (64). ROCK-II has been shown to mediate the effects of activated RhoA (65). However, the downstream effectors of Cdc42 and Rac1 required for transducing cytoskeletal alterations largely remain unidentified. Our finding of the requirement of Cdc42 and Rac1 in the TGFβ1-stimulated expression underscores the importance of identifying the downstream effectors of Cdc42 and Rac1 that may mediate CCN2/CTGF expression in response to TGFβ1 in gingival cells.
The role of Ras in the TGFβ1-induced expression of CCN2/CTGF is intriguing. Although the inhibition of Ras with either the inhibitor of farnesylation (FTI) or the overexpression of dominant-negative Ras (Fig. 8, B and C) does not inhibit CCN2/CTGF protein levels to the extent as do inhibitors of Cdc42 or Rac1 (GGTI or dominant-negative forms) (Fig. 4C and Fig. 8C), Ras is clearly involved in some aspect of CCN2/CTGF regulation. A possible mechanism could be that Ras may be activated secondarily to the initial cellular response to TGFβ1. This would imply that other factors may be expressed and secreted by gingival fibroblasts with the capacity for stimulating Ras. It is conceivable that the secretion of CCN2/CTGF protein by gingival cells could act in a paracrine manner to stimulate Ras-mediated pathways and perpetuate the expression of CCN2/CTGF in these cells. One possibility includes the activation of proteinase-activated receptors. Thrombin, for instance, produced as a consequence of TGFβ1 stimulation could stimulate CCN2/CTGF expression through the action of proteinase-activated receptors and potentially Ras-mediated pathways. The ability of thrombin to stimulate the synthesis of pro-collagen in fibroblasts and smooth muscle cells (66, 67) and Cyr61/CCN1 and CCN2/CTGF (68) in human fetal lung cells has been reported. If thrombin expression and secretion are induced in gingival cells by TGFβ1, it could serve to recruit platelets (69) to the gingival tissues. As we mentioned before, the platelets carrying LPA and CCN2/CTGF could then further enhance and/or perpetuate the fibrotic response.
Several potential mediators exist that may link Cdc42 and/or Rac1 to their cytoskeletal effects. These include MRCK, WASP (Wiskott-Aldrich syndrome protein), and PRK2/PKN2 (protein kinase C-related kinase 2). Unfortunately, there are many obstacles experimentally in clearly defining the roles of these molecules in GTPase signaling networks. For instance, ROCK is described as a RhoA/B/C-specific effector that is susceptible to inhibition by a commonly used “ROCK-II-specific” inhibitor Y27632. Yet it has been reported that PRK2 is inhibited more potently by Y27632 than ROCK-II (54, 55) and that both PRK2 and MRCK are potential downstream effectors of Cdc42/Rac1 (56, 70). Moreover, MRCK contains a high level of homology with ROCK (56), and PRK2 contains coiled coil domains as do MRCK and ROCK (71). Thus, like PRK2 and ROCK-II, MRCK may also be susceptible to inhibition by Y27632. However, this has not yet been determined. It remains to be seen as to which effector(s) is (are) responsible for regulating CCN2/CTGF expression stimulated by TGFβ1 and Cdc42/Rac1. Investigations are currently in progress to explore these possibilities summarized in Fig. 9.
Inhibition of the cholesterol pathway with HMG-CoA reductase inhibitors (statins) has been shown to reduce the expression of CCN2/CTGF mRNA in IMR90 (human lung) fibroblasts, fibroblasts derived from patients with idiopathic pulmonary fibrosis, and immortalized human renal fibroblasts in response to LPA and/or TGFβ (8, 72). In primary gingival fibroblasts, we found that challenge with 10 or 20 μm lovastatin was equally potent at down-regulating CCN2/CTGF protein levels. The inhibitory effect of lovastatin on the TGFβ1-induced expression of CCN2/CTGF was rapid, and no significant difference was observed if cells were treated for 12 or 24 h. Inhibition of specific transferase enzymes essential in the activation of cytosolic Rho-GTPases yielded an even stronger inhibition of CCN2/CTGF. Isoprene transferase inhibitors such as GGTI are highly toxic and do not represent useful clinical applications (73). The inhibition of TGFβ signaling with antibodies against TGFβ or its receptors for use in clinical applications could also have numerous adverse consequences. In contrast, HMG-CoA reductase inhibitors, statins, represent a valuable alternative to reducing the expression of genes reliant on GTPase signaling. These drugs are primarily used to reduce high cholesterol levels in patients with hypercholesterolemia. The use of these drugs in treating fibrotic conditions associated with the overexpression of CCN2/CTGF may be useful and may provide a relatively harmless, yet effective, treatment modality for patients with phenytoin-induced gingival overgrowth and/or hereditary gingival fibromatosis.
Forskolin is a compound derived from the plant Coleus forskohlii that belongs to the mint and lavender family. Forskolin is a direct stimulator of adenylate cyclase and promotes the increase in cAMP and activation of the cAMP-responsive element-binding protein. This herbal derivative is a common dietary supplement with few documented side effects. We previously showed that 10 μm forskolin reduced JNK1 phosphorylation stimulated by TGFβ1, through a process mediated by PKA, and reduced CCN2/CTGF mRNA and protein levels 30-50% in gingival fibroblasts (40). Blocking the activation step for cytosolic Rho-GTPases with GGTI, which completely inhibits CCN2/CTGF expression by interfering with Cdc42 and Rac1, has no effect on JNK1 phosphorylation induced by TGFβ1 (Fig. 4D). Our finding that the combination of lovastatin with forskolin resulted in an enhanced inhibition on CCN2/CTGF mRNA and protein levels suggests that this combination produces additive inhibitory effects through their directed inhibition of different pathways, each independently essential for CCN2/CTGF expression in response to stimulation with TGFβ1. Statins in combination with forskolin may have enhanced therapeutic potential than statins alone by the targeted inhibition multiple pathways essential in stimulating CCN2/CTGF expression in gingival cells. Taken together, results from analyses of parallel signal transduction pathways that together mediate CCN2/CTGF regulation in gingival fibroblasts point to possible new therapeutic strategies to address fibrosis in gingiva and perhaps in other tissues as well. Preliminary data in dermal fibroblasts suggest that some pathways identified in gingival fibroblasts may also mediate CCN2/CTGF expression in skin.
This work was supported by National Institutes of Health NIDCR Grants R01 DE11004, K08 DE016609, and M01 RR00533. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TGFβ1, transforming growth factor-β1; LPA, lysophosphatidic acid; GGTI, geranylgeranyltransferase inhibitor; FTI, farnesyltransferase inhibitor; qPCR, quantitative PCR; GTPγS, guanosine 5′-3-O-(thio)triphosphate; HMG, hydroxymethylglutaryl; JNK, c-Jun N-terminal kinase; PGE2, prostaglandin E2; MRCK, myotonic dystrophy kinase-related Cdc42-binding kinase; PKA, cAMP-dependent protein kinase; ROCK, Rho-associated coiled coil forming kinase; MRCK, myotonic dystrophy kinase-related Cdc42-binding kinase.
References
- 1.Bork, P. (1993) FEBS Lett. 327 125-130 [DOI] [PubMed] [Google Scholar]
- 2.Oemar, B. S., and Luscher, T. F. (1997) Arterioscler. Thromb. Vasc. Biol. 17 1483-1489 [DOI] [PubMed] [Google Scholar]
- 3.Bradham, D. M., Igarashi, A., Potter, R. L., and Grotendorst, G. R. (1991) J. Cell Biol. 114 1285-1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brigstock, D. R. (1999) Endocr. Rev. 20 189-206 [DOI] [PubMed] [Google Scholar]
- 5.Brigstock, D. R. (2003) J. Endocrinol. 178 169-175 [DOI] [PubMed] [Google Scholar]
- 6.Grotendorst, G. R. (1997) Cytokine Growth Factor Rev. 8 171-179 [DOI] [PubMed] [Google Scholar]
- 7.Grotendorst, G. R., Okochi, H., and Hayashi, N. (1996) Cell Growth & Differ. 7 469-480 [PubMed] [Google Scholar]
- 8.Heusinger-Ribeiro, J., Eberlein, M., Wahab, N. A., and Goppelt-Struebe, M. (2001) J. Am. Soc. Nephrol. 12 1853-1861 [DOI] [PubMed] [Google Scholar]
- 9.Eberlein, M., Heusinger-Ribeiro, J., and Goppelt-Struebe, M. (2001) Br. J. Pharmacol. 133 1172-1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blom, I. E., Goldschmeding, R., and Leask, A. (2002) Matrix Biol. 21 473-482 [DOI] [PubMed] [Google Scholar]
- 11.Abraham, D. J., Shiwen, X., Black, C. M., Sa, S., Xu, Y., and Leask, A. (2000) J. Biol. Chem. 275 15220-15225 [DOI] [PubMed] [Google Scholar]
- 12.Leask, A., Sa, S., Holmes, A., Shiwen, X., Black, C. M., and Abraham, D. J. (2001) Mol. Pathol. 54 180-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, Y., Aten, J., Bende, R. J., Oemar, B. S., Rabelink, T. J., Weening, J. J., and Goldschmeding, R. (1998) Kidney Int. 53 853-861 [DOI] [PubMed] [Google Scholar]
- 14.Riser, B. L., Denichilo, M., Cortes, P., Baker, C., Grondin, J. M., Yee, J., and Narins, R. G. (2000) J. Am. Soc. Nephrol. 11 25-38 [DOI] [PubMed] [Google Scholar]
- 15.Oemar, B. S., Werner, A., Garnier, J. M., Do, D. D., Godoy, N., Nauck, M., Marz, W., Rupp, J., Pech, M., and Luscher, T. F. (1997) Circulation 95 831-839 [DOI] [PubMed] [Google Scholar]
- 16.Ohnishi, H., Oka, T., Kusachi, S., Nakanishi, T., Takeda, K., Nakahama, M., Doi, M., Murakami, T., Ninomiya, Y., Takigawa, M., and Tsuji, T. (1998) J. Mol. Cell. Cardiol. 30 2411-2422 [DOI] [PubMed] [Google Scholar]
- 17.Schober, J. M., Chen, N., Grzeszkiewicz, T. M., Jovanovic, I., Emeson, E. E., Ugarova, T. P., Ye, R. D., Lau, L. F., and Lam, S. C. (2002) Blood 99 4457-4465 [DOI] [PubMed] [Google Scholar]
- 18.di Mola, F. F., Friess, H., Martignoni, M. E., Di Sebastiano, P., Zimmermann, A., Innocenti, P., Graber, H., Gold, L. I., Korc, M., and Buchler, M. W. (1999) Ann. Surg. 230 63-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dammeier, J., Brauchle, M., Falk, W., Grotendorst, G. R., and Werner, S. (1998) Int. J. Biochem. Cell Biol. 30 909-922 [DOI] [PubMed] [Google Scholar]
- 20.Paradis, V., Dargere, D., Vidaud, M., De Gouville, A. C., Huet, S., Martinez, V., Gauthier, J. M., Ba, N., Sobesky, R., Ratziu, V., and Bedossa, P. (1999) Hepatology 30 968-976 [DOI] [PubMed] [Google Scholar]
- 21.Rachfal, A. W., and Brigstock, D. R. (2003) Hepatol. Res. 26 1-9 [DOI] [PubMed] [Google Scholar]
- 22.Sedlaczek, N., Jia, J. D., Bauer, M., Herbst, H., Ruehl, M., Hahn, E. G., and Schuppan, D. (2001) Am. J. Pathol. 158 1239-1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamatani, T., Kobayashi, H., Tezuka, K., Sakamoto, S., Suzuki, K., Nakanishi, T., Takigawa, M., and Miyano, T. (1998) Biochem. Biophys. Res. Commun. 251 748-752 [DOI] [PubMed] [Google Scholar]
- 24.Williams, E. J., Gaca, M. D., Brigstock, D. R., Arthur, M. J., and Benyon, R. C. (2000) J. Hepatol. 32 754-761 [DOI] [PubMed] [Google Scholar]
- 25.Ueberham, U., Ueberham, E., Gruschka, H., and Arendt, T. (2003) Neuroscience 116 1-6 [DOI] [PubMed] [Google Scholar]
- 26.Lee, E. H., and Joo, C. K. (1999) Investig. Ophthalmol. Vis. Sci. 40 2025-2032 [PubMed] [Google Scholar]
- 27.Allen, J. T., Knight, R. A., Bloor, C. A., and Spiteri, M. A. (1999) Am. J. Respir. Cell Mol. Biol. 21 693-700 [DOI] [PubMed] [Google Scholar]
- 28.Lasky, J. A., Ortiz, L. A., Tonthat, B., Hoyle, G. W., Corti, M., Athas, G., Lungarella, G., Brody, A., and Friedman, M. (1998) Am. J. Physiol. 275 L365-L371 [DOI] [PubMed] [Google Scholar]
- 29.Hong, H. H., Uzel, M. I., Duan, C., Sheff, M. C., and Trackman, P. C. (1999) Lab. Investig. 79 1655-1667 [PubMed] [Google Scholar]
- 30.Kantarci, A., Black, S., Xydas, C., Murawel, P., Uchida, Y., Yucekal-Tuncer, B., Atilla, G., Emingil, G., Uzel, M., Lee, A., Firatli, E., Sheff, M., Hasturk, H., Van Dyke, T., and Trackman, P. (2006) J. Pathol. 210 59-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uzel, M. I., Kantarci, A., Hong, H. H., Uygur, C., Sheff, M. C., Firatli, E., and Trackman, P. C. (2001) J. Periodontol. 72 921-931 [DOI] [PubMed] [Google Scholar]
- 32.Hall, E. E. (1997) Curr. Opin. Periodontol. 4 59-63 [PubMed] [Google Scholar]
- 33.Hassell, T. M. (1981) Monogr. Oral Sci. 9 1-205 [PubMed] [Google Scholar]
- 34.Hassell, T. M., and Hefti, A. F. (1991) Crit. Rev. Oral Biol. Med. 2 103-137 [DOI] [PubMed] [Google Scholar]
- 35.Gerrard, J. M., and Robinson, P. (1989) Biochim. Biophys. Acta 1001 282-285 [DOI] [PubMed] [Google Scholar]
- 36.Fischer, D. J., Liliom, K., Guo, Z., Nusser, N., Virag, T., Murakami-Murofushi, K., Kobayashi, S., Erickson, J. R., Sun, G., Miller, D. D., and Tigyi, G. (1998) Mol. Pharmacol. 54 979-988 [DOI] [PubMed] [Google Scholar]
- 37.Moolenaar, W. H., Kranenburg, O., Postma, F. R., and Zondag, G. C. (1997) Curr. Opin. Cell Biol. 9 168-173 [DOI] [PubMed] [Google Scholar]
- 38.Jedsadayanmata, A., Chen, C. C., Kireeva, M. L., Lau, L. F., and Lam, S. C. (1999) J. Biol. Chem. 274 24321-24327 [DOI] [PubMed] [Google Scholar]
- 39.Kubota, S., Kawata, K., Yanagita, T., Doi, H., Kitoh, T., and Takigawa, M. (2004) J. Biochem. (Tokyo) 136 279-282 [DOI] [PubMed] [Google Scholar]
- 40.Black, S. A., Jr., Palamakumbura, A. H., Stan, M., and Trackman, P. C. (2007) J. Biol. Chem. 282 15416-15429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Essig, M., Vrtovsnik, F., Nguyen, G., Sraer, J. D., and Friedlander, G. (1998) J. Am. Soc. Nephrol. 9 1377-1388 [DOI] [PubMed] [Google Scholar]
- 42.Goppelt-Struebe, M., Hahn, A., Iwanciw, D., Rehm, M., and Banas, B. (2001) Mol. Pathol. 54 176-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lakos, G., Takagawa, S., Chen, S. J., Ferreira, A. M., Han, G., Masuda, K., Wang, X. J., DiPietro, L. A., and Varga, J. (2004) Am. J. Pathol. 165 203-217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonniaud, P., Margetts, P. J., Ask, K., Flanders, K., Gauldie, J., and Kolb, M. (2005) J. Immunol. 175 5390-5395 [DOI] [PubMed] [Google Scholar]
- 45.Blom, I. E., van Dijk, A. J., de Weger, R. A., Tilanus, M. G., and Goldschmeding, R. (2001) Mol. Pathol. 54 192-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gohla, A., Harhammer, R., and Schultz, G. (1998) J. Biol. Chem. 273 4653-4659 [DOI] [PubMed] [Google Scholar]
- 47.Jackson, S. M., Ericsson, J., and Edwards, P. A. (1997) Subcell. Biochem. 28 1-21 [DOI] [PubMed] [Google Scholar]
- 48.Chan, K. K., Oza, A. M., and Siu, L. L. (2003) Clin. Cancer Res. 9 10-19 [PubMed] [Google Scholar]
- 49.Zhang, F. L., and Casey, P. J. (1996) Annu. Rev. Biochem. 65 241-269 [DOI] [PubMed] [Google Scholar]
- 50.Wong, W. W., Dimitroulakos, J., Minden, M. D., and Penn, L. Z. (2002) Leukemia (Baltimore) 16 508-519 [DOI] [PubMed] [Google Scholar]
- 51.Xia, Z., Tan, M. M., Wong, W. W., Dimitroulakos, J., Minden, M. D., and Penn, L. Z. (2001) Leukemia (Baltimore) 15 1398-1407 [DOI] [PubMed] [Google Scholar]
- 52.Aktories, K., Braun, U., Rosener, S., Just, I., and Hall, A. (1989) Biochem. Biophys. Res. Commun. 158 209-213 [DOI] [PubMed] [Google Scholar]
- 53.Didsbury, J., Weber, R. F., Bokoch, G. M., Evans, T., and Snyderman, R. (1989) J. Biol. Chem. 264 16378-16382 [PubMed] [Google Scholar]
- 54.Davies, S. P., Reddy, H., Caivano, M., and Cohen, P. (2000) Biochem. J. 351 95-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bain, J., McLauchlan, H., Elliott, M., and Cohen, P. (2003) Biochem. J. 371 199-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leung, T., Chen, X. Q., Tan, I., Manser, E., and Lim, L. (1998) Mol. Cell. Biol. 18 130-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan, I., Seow, K. T., Lim, L., and Leung, T. (2001) Mol. Cell. Biol. 21 2767-2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao, Y., Dickerson, J. B., Guo, F., Zheng, J., and Zheng, Y. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 7618-7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holmes, A., Abraham, D. J., Sa, S., Shiwen, X., Black, C. M., and Leask, A. (2001) J. Biol. Chem. 276 10594-10601 [DOI] [PubMed] [Google Scholar]
- 60.Holmes, A., Abraham, D. J., Chen, Y., Denton, C., Shi-wen, X., Black, C. M., and Leask, A. (2003) J. Biol. Chem. 278 41728-41733 [DOI] [PubMed] [Google Scholar]
- 61.Querfeld, C., Eckes, B., Huerkamp, C., Krieg, T., and Sollberg, S. (1999) J. Dermatol. Sci. 21 13-22 [DOI] [PubMed] [Google Scholar]
- 62.Ridley, A. J., and Hall, A. (1992) Cell 70 389-399 [DOI] [PubMed] [Google Scholar]
- 63.Ridley, A. J., Paterson, H. F., Johnston, C. L., Diekmann, D., and Hall, A. (1992) Cell 70 401-410 [DOI] [PubMed] [Google Scholar]
- 64.Nobes, C. D., and Hall, A. (1995) Cell 81 53-62 [DOI] [PubMed] [Google Scholar]
- 65.Ishizaki, T., Naito, M., Fujisawa, K., Maekawa, M., Watanabe, N., Saito, Y., and Narumiya, S. (1997) FEBS Lett. 404 118-124 [DOI] [PubMed] [Google Scholar]
- 66.Chambers, R. C., Dabbagh, K., McAnulty, R. J., Gray, A. J., Blanc-Brude, O. P., and Laurent, G. J. (1998) Biochem. J. 333 121-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dabbagh, K., Laurent, G. J., McAnulty, R. J., and Chambers, R. C. (1998) Thromb. Haemostasis 79 405-409 [PubMed] [Google Scholar]
- 68.Pendurthi, U. R., Allen, K. E., Ezban, M., and Rao, L. V. (2000) J. Biol. Chem. 275 14632-14641 [DOI] [PubMed] [Google Scholar]
- 69.Macfarlane, S. R., Seatter, M. J., Kanke, T., Hunter, G. D., and Plevin, R. (2001) Pharmacol. Rev. 53 245-282 [PubMed] [Google Scholar]
- 70.Vincent, S., and Settleman, J. (1997) Mol. Cell. Biol. 17 2247-2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maesaki, R., Ihara, K., Shimizu, T., Kuroda, S., Kaibuchi, K., and Hakoshima, T. (1999) Mol. Cell 4 793-803 [DOI] [PubMed] [Google Scholar]
- 72.Watts, K. L., Sampson, E. M., Schultz, G. S., and Spiteri, M. A. (2005) Am. J. Respir. Cell Mol. Biol. 32 290-300 [DOI] [PubMed] [Google Scholar]
- 73.Lobell, R. B., Omer, C. A., Abrams, M. T., Bhimnathwala, H. G., Brucker, M. J., Buser, C. A., Davide, J. P., deSolms, S. J., Dinsmore, C. J., Ellis-Hutchings, M. S., Kral, A. M., Liu, D., Lumma, W. C., Machotka, S. V., Rands, E., Williams, T. M., Graham, S. L., Hartman, G. D., Oliff, A. I., Heimbrook, D. C., and Kohl, N. E. (2001) Cancer Res. 61 8758-8768 [PubMed] [Google Scholar]