Abstract
Summary
Independent of the platform and the analysis methods used, the result of a microarray experiment is, in most cases, a list of differentially expressed genes. An automatic ontological analysis approach has been recently proposed to help with the biological interpretation of such results. Currently, this approach is the de facto standard for the secondary analysis of high throughput experiments and a large number of tools have been developed for this purpose. We present a detailed comparison of 14 such tools using the following criteria: scope of the analysis, visualization capabilities, statistical model(s) used, correction for multiple comparisons, reference microar-rays available, installation issues and sources of annotation data. This detailed analysis of the capabilities of these tools will help researchers choose the most appropriate tool for a given type of analysis. More importantly, in spite of the fact that this type of analysis has been generally adopted, this approach has several important intrinsic drawbacks. These drawbacks are associated with all tools discussed and represent conceptual limitations of the current state-of-the-art in ontological analysis. We propose these as challenges for the next generation of secondary data analysis tools.
Contact
sod@cs.wayne.edu
1 INTRODUCTION
Microarrays are at the center of a revolution in biotechnology, allowing researchers to simultaneously monitor the expression of tens of thousands of genes. Independent of the platform and the analysis methods used, the result of a microarray experiment is, in most cases, a list of genes found to be differentially expressed. The common challenge faced by the researchers is to translate such lists of differentially regulated genes into a better understanding of the underlying biological phenomena. A first step in this direction can be the translation of the list of differentially expressed genes into a functional profile able to offer insight into the cellular mechanisms relevant in the given condition. As recently as 2002, an automatic ontological analysis approach using Gene Ontology (GO) has been proposed to help with this task (Khatri et al., 2002). From 2003 to 2005, 13 other tools have been proposed for this type of analysis and more tools continue to appear every day (Fig. 1). Currently, this approach is the de facto standard for the secondary analysis of high throughput experiments and a large number of tools have been developed for this purpose. Although these tools use the same general approach, they differ greatly in many respects that influence in an essential way the results of the analysis. In most cases, researchers using such tools are either unaware of, or are confused about certain crucial features. There is no unified analysis of this field available in the literature.
Fig. 1.
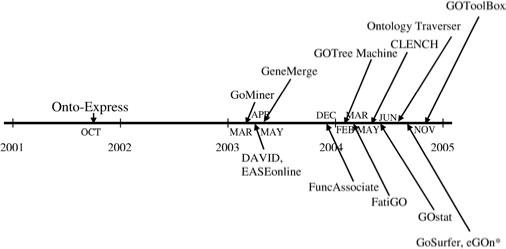
Evolution history of GO-based functional analysis software. The tool marked with a star has not been published in a peer-reviewed journal.
This paper presents a comparison of 14 current tools in this area. A detailed analysis of the capabilities of these tools, of the statistical models deployed as well as of their back-end annotation databases (if applicable), is included here in order to help researchers choose the most appropriate tool for a given type of analysis.
More importantly, we include a discussion of some of the issues associated with the current ontological analysis approach. Since all existing tools implement the same approach, these drawbacks are also associated with all tools discussed and represent conceptual limitations of the current state-of-the-art in ontological analysis. We propose these as the challenges for the next generation of secondary data analysis tools.
2 A COMPARISON OF EXISTING FUNCTIONAL PROFILING TOOLS
The comparison between the tools currently available for the ontological analysis of high throughput gene expression experiments is summarized in Tables 1 and 2. The criteria used in these tables are described in detail in the following.
Table 1.
A comparison of the tools reviewed. Scope of the analysis refers to the number of GO categories that can be analyzed simultaneously. Level of abstraction refers to the depth in GO at which genes are associated with annotations. Note that some tools (e.g. GoMiner) allow the user to expand and collapse nodes in the results, but the analysis is only performed once, without reassigning the genes as nodes are collapsed or expanded by the user. This is described as a static global analysis. The other columns are self-explanatory
| Tool | Scope of the analysis | Level of abstraction | User interface | Application type | Platform | Supported input IDs |
|---|---|---|---|---|---|---|
| Onto-Express | All GO categories | Fully flexible; different levels of abstractions in different GO subtrees | Java GUI | Web-based | Any | GenBank, UniGene, Entrez Gene, Affymetrix, Gene symbol |
| GoMiner | All GO categories | Static global analysis | Java GUI | Stand-alone | Windows only | Organism specific IDs in GO |
| DAVID | All GO categories | Only lowest level of GO | HTML GUI | Web-based | Any | GenBank, UniGene, Entrez Gene, Affymetrix, RefSeq, UniProt, PIR |
| EASEonline | All GO categories | User-selected, fixed level | HTML GUI | Both | Any | Affymetrix, GenBank, UniGene, Entrez Gene |
| GeneMerge | One category | Only lowest level of GO | HTML GUI | Both | Any | Only supports organism specific IDs used in GO |
| FuncAssociate | All GO categories | Only lowest level of GO | HTML GUI | Web-based | Any | MODB gene products |
| GOTM | All GO categories | Only lowest level of GO | HTML GUI | Web-based | Any | Affymetrix, UniGene, ENSEMBL, Swiss-Prot, Entrez Gene |
| FatiGO | One category | User-selected, fixed level and static global analysis | HTML GUI | Web-based | Any | Affymetrix, GenBank |
| CLENCH | All GO categories | Static global analysis | Command-line input, HTML output | Stand-alone | Windows only | A.thaliana MIPS IDs |
| GOstat | All GO categories | User-selected, fixed level | HTML GUI | Web-based | Any | GenBank, UniGene, Gene symbol, Organism specific IDs in GO |
| GOToolBox | All GO categories | User-selected, fixed level | HTML GUI | Web-based | Any | Only organism specific IDs in GO |
| GoSurfer | All GO categories | Only lowest level of GO | C/C++ GUI | Stand-alone | Windows only | Affymetrix, UniGene, Entrez Gene |
| Ontology Traverser | One category | Only lowest level of GO | HTML GUI | Web-based | Any | Affymetrix |
| eGOn | One category | Only lowest level of GO | HTML GUI | Web-based | Any | GenBank, UniGene, Clone |
Table 2.
In the GO visualization column, ‘flat’ indicates that the tool does not represent the hierarchical structure of the GO when displaying the results, ‘tree’ indicates that the tool displays the GO hierarchy as a tree, whereas ‘DAG’ indicates that the tool displays the GO as a directed acyclic graph. The other columns are self-explanatory
| Tool | Statistical model | Correction for multiple experiments | GO Visualization | Microarrays supported | Time to process 200 genes (s) |
|---|---|---|---|---|---|
| Onto-Express | χ2, binomial, hypergeometric, Fisher's exact test | Šidák, Holm, Bonferroni, FDR | Flat, Tree | 172 commercial arrays (Affymetrix, SuperArray, Sigma-Genonsys, ClonTech, PerkinElmer, Operon, Takara, NIA); can also upload a user-defined list | 7, 8, 16, 28 |
| GoMiner | Fisher's exact test | Relative enrichment | Tree, DAG | uploads from user | 77, 123, 223, 340 |
| DAVID | None | None | Not available | Not applicable | 15, 17, 27, 54 |
| EASEonline | Fisher's exact test | Bonferroni | Not available | 27 arrays (Affymetrix only); can also upload a user-defined list | 15, 19, 34, 74 |
| GeneMerge | Hypergeometric | Bonferroni | Flat, no hierarchical structure | Uploads from user | 6, 6, 6, 8 |
| FuncAssociate | Fisher's exact test | Not available | Uploads from user | 22, 27, 29, 50 | |
| GOTM | Hypergeometric | None | Tree | 37 arrays (Affymetrix only); uploads from user | 59, 60, 157, |
| FatiGO | Percentage | Step-down minP, FDR (Benjamini and Hochberg, 1995), FDR (Benjamini and Yekutieli, 2001) | Flat, Tree | Uploads from user | 15, 49, 69, 105 |
| CLENCH | Hypergeometric, χ2, binomial | None | DAG | Uploads from user | NA |
| GOstat | χ2, Fisher's exact test | FDR, Holm | Not available | Uploads from user | 12, 20, 46, 80 |
| GOToolBox | Hypergeometric, binomial, Fisher's exact test | Bonferroni, Holm, Hochberg, Hommel, FDR | Not available | Uploads from user | 22, 81, 145, 270 |
| GoSurfer | χ2 | q-value | DAG | 22 arrays (Affymetrix only); uploads from user | 2, 2, 2, 3 |
| Ontology Traverser | Hypergeometric | FDR | Not available | 5 arrays (Affymetrix); uploads from user | NA |
| eGOn | Binomial | None | Tree | Uploads from user | 20, 45, 80, 95 |
2.1 The statistical model
The ontological analysis can be performed with a number of statistical models including hypergeometric (Cho et al., 2001), binomial, χ2 (chi-square) (Fisher and van Belle, 1993), and Fisher's exact test (Man et al., 2000). The probability that a certain category occurs x times just by chance in the list of differentially regulated genes is appropriately modeled by a hypergeometric distribution. However, the hypergeometric distribution can be more difficult to calculate when large arrays (e.g. Affymetrix HGU133A) are involved. However, the hypergeometric distribution tends to the binomial distribution when the number of genes is large. Therefore, the binomial model is perfectly usable when larger arrays are used. Alternative approaches include a χ2 test for equality of proportions and Fisher's Exact test. In most cases, the differences between the models will not be dramatic. These tests are discussed in detail in the literature (Drăghici, 2003; Drăghici et al., 2003a,b). FatiGO does not use a statistical model as such but does calculate percentages with respect to the genes annotated with GO terms or all known genes in an organism. GoMiner, EASEonline, GeneMerge, FuncAssociate, GOTree Machine (GOTM), GOSurfer, Ontology Traverser, and eGOn only support one statistical test. GOstat allows the user to choose between two tests (χ2, and Fisher's exact test), CLENCH and GOToolBox allow a choice between three tests (χ2, hypergeometric, and binomial for CLENCH and hypergeometric, binomial, and Fisher's exact test for GOToolBox), while Onto-Express implements all four tests (χ2, hypergeometric, binomial, and Fisher's exact test).
2.2 The set of reference genes
An important consideration when identifying statistically significant GO terms is the choice of the reference list of genes against which the P-values for each GO term in the results are calculated. Several tools such as GOToolBox, GOstat, GoMiner, FatiGO, and GOTM1 use the total set of genes in a genome as the reference (Beissbarth and Speed, 2004; Martin et al., 2004; Zeeberg et al., 2003; Zhang et al., 2004) or the set of genes with GO annotations (Al-Shahrour et al., 2004). Either of these may be an inappropriate choice when the input list of genes to these tools is a list of differentially expressed genes obtained from a microarray experiment, since the genes that are not present on a microarray do not ever have a chance of being selected as differentially regulated. The fundamental idea is to assign significance to various functional categories by comparing the observed number of genes in a specific category with the number of genes that might appear in the same category if a selection performed from the same pool were completely random. If the whole genome is considered as the reference, the pool considered when calculating the random choice includes all genes in the genome. At the same time, the pool available when actually selecting differentially regulated genes includes only the genes represented on the array used, since a gene that is not on the array can never be found to be differentially regulated. This represents a flagrant contradiction of the assumptions of the statistical models used.
2.3 Correction for multiple experiments
Another crucial factor in the assessment of a functional category is the correction for multiple experiments [see for instance Chapter 9 in Drăghici (2003)]. This type of correction must be performed in all situations in which the functional category is not selected a priori and many such categories are considered at the same time. The importance of this step cannot be overstated and has been well recognized in the literature (Al-Shahrour et al., 2004; Beissbarth and Speed, 2004; Berriz et al., 2003; Castillo-Davis and Hartl, 2002; Drăghici, 2003; Shah and Fedoroff, 2004; Zeeberg et al., 2003). In spite of this, several of the tools reviewed here do not perform such a correction: GoMiner, DAVID, GOTM, CLENCH, and eGOn. GoMiner provides a ‘relative enrichment’ statistic calculated as Re = (nf/n)/(Nf/N), where n and N are the numbers of genes in the selected and reference sets, respectively, and nf and Nf are the number of genes in the functional category of interest in the selected and reference sets, respectively (Zeeberg et al., 2003). However, this relative enrichment cannot be used in any way as a correction for multiple experiments2 but rather as another indication of the significance of the given category, somewhat redundant to, but less informative than the P-value. This statistic can be misleading because the user will be tempted to assign biological meanings to all those categories that are enriched. In reality, any particular relative enrichment value can actually appear with a non-zero probability just by chance. It is the magnitude of the probability that should be used to decide whether a category is significant or not, rather than the relative enrichment.
All remaining tools deal with the problem of multiple comparisons in some way. EASEonline, and GeneMerge support the Bonferroni correction. Bonferroni and Šidák are perfectly suitable in many situations, in particular, when not very many functional categories are involved (e.g. fewer than 50). However, these corrections are known to be overly conservative if more categories are involved (Drăghici, 2003). A family of methods that allow less conservative adjustments of the P-values is the Holm step-down group of methods (Hochberg and Tamhane, 1987; Holland and Copenhaver, 1987; Holm, 1979; Shaffer, 1986).
Bonferroni, Šidák, and Holm's step-down adjustment are statistical procedures that assume the variables are independent, which is known to be false for this type of analysis.3 When it is known that dependencies exist, methods such as false discovery rate (FDR) are more appropriate (Benjamini and Hochberg, 1995; Benjamini and Yekutieli, 2001; Drăghici, 2003). Another suitable approach is that of bootstrapping which actually calculates the null distribution by performing many resamplings from the same data, thus taking into consideration all existing dependencies. Great care should be taken in those situations in which only few categories are involved because the number of distinct resamplings may be insufficient for a reliable conclusion [see Chap. 9 in Drăghici (2003)]. In those instances, even Bonferroni or Šidák may be a better choice instead of bootstrapping.
The tools offering more than one correction method effectively allow the researcher to adapt the analysis to the number of categories and degree of known dependencies between them. Bonferroni and Šidák are suitable if few, not directly related categories are involved. If more unrelated categories are involved, Holm's may be a good compromise. If there are several functional categories that are clearly related, FDR is probably the best choice. If the dependencies are very strong (e.g. several sub-processes of the same larger process), a bootstrap or Monte-Carlo simulation approach may be better able to capture these dependencies, but only if enough categories are present to make the simulation meaningful.
The one tool standing out regarding this criterion is FuncAssociate which uses a more original Monte-Carlo simulation. FatiGO and GOstat implement Holm's and FDR corrections. Onto-Express offers Bonferroni, Šidák, Holm's and FDR, whereas GOToolBox offers FDR, Bonferroni, Holm, Hochberg, and Hommel corrections.
2.4 The scope of the analysis
An important factor in assessing the usefulness of a tool is its ability to provide a complete picture of the phenomenon studied. In terms of functional profiling using GO, a complete analysis should include all three primary GO categories: molecular function, biological process, and cellular component as well as other information if available. Among the tools reviewed, eGOn, FatiGO, GeneMerge, and Ontology Traverser only analyze one category at a time. The other tools allow the user to analyze all three categories simultaneously. Extra features are present in GeneMerge and Onto-Express. GeneMerge shows KEGG metabolic and signaling pathways for yeast and fruit fly, and deletion viability data for yeast. Onto-Express also shows KEGG signaling pathway data, as well as a chromosome location of differentially regulated genes (linked to NCBI's Mapviewer for further analysis).
2.5 Performance issues
We compared the speed of the tools by submitting four sets of 100, 200, 500, and 1000 human genes, respectively, to each of the tools (Fig. 2). We started with a list of genes containing gene symbols because this type of ID is accepted by most tools (four tools). Since several tools work only with specific types of IDs, we had to translate these lists of genes into the appropriate type. This was done with Onto-Translate (Drăghici et al., 2003a; Khatri et al., 2004). We translated the lists into Entrez gene IDs for three tools, TrEMBL IDs for three tools and GeneBank accessions for two other tools. The times shown here do not include such translations. We do not report response times for CLENCH and OntologyTraverser because CLENCH only supports Arabidopsis thaliana, and Onto-logyTraverser was unavailable in spite of our numerous attempts over several weeks.4 The three fastest tools, GoSurfer, GeneMerge and Onto-Express, perform the analysis of 200 genes in 2, 6 and 8 s, respectively. Interestingly, two of these top three tools (GeneMerge and Onto-Express) are web-based which is somewhat counter-intuitive since one would have expected the stand-alone tools to be faster.
Fig. 2.
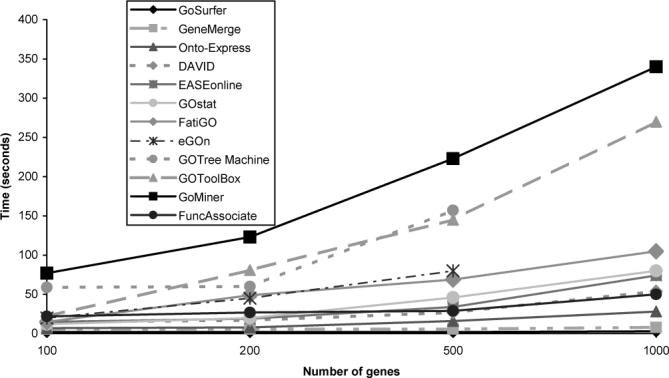
A speed comparison of the tools reviewed here. Four sets of 100, 200, 500 and 1000 human genes were submitted to each tool. The three fastest tools, GoSurfer, GeneMerge and Onto-Express, are all able to perform the analysis of up to 200 genes in under 8 seconds. Note that GOTM allows upload of only up to 500 genes.
2.6 Visualization capabilities
The GO is organized as a directed acyclic graph (DAG), which is a hierarchical structure similar to a tree. Unlike a tree, a DAG allows a node to have several parents. However, the DAG structure may not be the best choice for navigational purposes in GO since it tends to clutter the display (Zeeberg et al., 2003). An alternative way to visualize the DAG structure of the GO is to represent and visualize it as a tree structure in which a node with several parents is represented in the tree multiple times, once under each parent. Any tool using GO for functional profiling of a list of genes should be able to represent the hierarchical relationships between various functional categories. A graphical representation of the analysis results in the hierarchical context of GO allows the user to better understand the phenomenon studied. Furthermore, the functional analysis can be continued and refined by exploring certain interesting sub-graphs of the GO hierarchy.
Among the tools reviewed, DAVID, EASEonline, FuncAssociate, GOstat, GOToolBox and OntologyTraverser do not show the results in the context of the hierarchical structure of GO. Onto-Express, eGOn, FatiGO, CLENCH, GoMiner and GOTM represent the results in their GO context. Onto-Express, GOMiner. GOTM and eGOn allow the user to manually collapse/expand nodes. Onto-Express also allows sorting and searching operations in the hierarchy, automatically expanding and/or collapsing nodes if necessary.
2.7 Custom level of abstraction
In the hierarchical structure of the GO, the genes are annotated at various levels of abstraction (Fig. 3). For instance, ‘induction of apoptosis by hormones’ is a type of ‘induction of apoptosis’ which in turn is a part of ‘apoptosis’. Apoptosis represents a higher level of abstraction, more general, whereas induction of apoptosis by hormones represents a lower level of abstraction, more specific. When annotating the genes with the GO terms, efforts are made to annotate the genes with the highest level of details possible which corresponds to the lowest level of abstraction. For example, if a gene is known to induce apoptosis in response to hormones, it will be annotated with the term ‘induction of apoptosis by hormones’ and not merely with one of the higher level terms such as ‘induction of apoptosis’ or ‘apoptosis’. A very valuable capability of a functional profiling tool is to let the user select a custom level of abstraction. From this point of view, the tools reviewed fall into one of the following three categories. The first category includes the tools able to perform the analysis only with the specific terms associated with each gene. This corresponds to an analysis undertaken at the lowest possible level of abstraction or the highest level of specificity (see the dash-and-dot line in Fig. 3). This type of analysis is essentially a one-shot look-up into the annotation database used. Each dataset can only be analyzed once, since any further analysis can only provide exactly the same results. The analysis cannot be directed to answer specific biological questions and cannot be refined in any way.
Fig. 3.
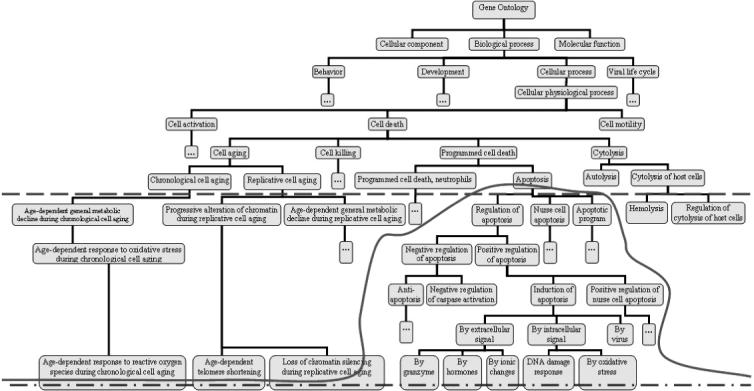
Levels of abstraction. The analysis can be performed at a lowest level of abstraction (dash-and-dot line), at a fixed level of abstraction chosen by the user (dashed line) or at a custom level of abstraction that can go to different depths in various sub-trees of the GO (continuous line).
The second category of the tools include those tools which allow the user to select a predetermined depth, or level of abstraction in GO. Once this level is selected, these tools will consider any genes below the chosen level associated with the corresponding category at the chosen level. This is illustrated by the dashed line in Figure 3. Care should be taken here in order to make sure that each category is propagated up through all its parents, by following the DAG structure and not the tree structure that may be used for visualization. The capability of choosing a pre-determined depth allows the user to refine this analysis by performing it repeatedly, at various levels of abstraction, thus forcing various very specific terms to be grouped into more general, and perhaps more informative categories. When this is done, several genes that are associated with very specific categories (e.g. induction of apoptosis by X, Y and Z) are now grouped together under a more general category such as ‘positive regulation of apoptosis’. It is often the case that each specific category does not appear to be significant because there are only few genes associated with it, while the more general category becomes highly significant once all genes associated with specific sub-categories are analyzed together as representing the more general category. Tools having this capability allow a more complex and detailed analysis that can be directed to ask specific biological questions.
Finally, the third category includes tools that allow a completely custom cut through the GO, at different levels of abstraction in different directions. If the analysis is performed at a fixed depth of 9 for instance, the analysis can distinguish between the various sub-types of apoptosis induction: by hormones, by extracellular signals, by intra-cellular signals, etc. (Fig. 3). However, for a fixed depth, the same analysis will also be performed on several other thousands of functional categories situated at the same level. If the results are presented in a bar graph, the interesting categories will be cluttered by all the extra categories that just happened to be at the same depth in GO, even though they may not be interesting to the researcher. At the same time, other phenomena may be missed because the chosen level may be too specific for those GO categories. A tool that allows full customization is most powerful since it will allow the user to perform the analysis at different depths in various parts of the GO hierarchy, as required by the specific biological hypothesis investigated. This is illustrated by the continuous red line in Figure 3.
Most of the existing tools only perform the analysis at the lowest level in GO, with the specific categories that genes have been annotated with, and do not allow any further refinement. Among the tools reviewed, FatiGO, EASEonline, GOToolBox, and GOstat allow the user to select a specific level of abstraction before submitting the input list of genes. FatiGo, CLENCH and GOMiner also calculate a P-value for all nodes throughout the GO. This corresponds to a global static analysis in which all genes under a certain node are considered to be associated with that node. Onto-Express is, at this time, the only tool that allows a fully customized analysis by allowing any node to be collapsed or expanded in the GO. Collapsing a node is equivalent to re-assigning to this node all genes associated with any of its descendants. The P-value calculated for a collapsed node in Onto-Express corresponds to the P-value calculated in the global static analysis performed by GoMiner and FatiGo. Expanding a node will distinguish between genes associated with the node itself and the genes associated to any of it descendants. The P-value of an expanded node will be based only on the genes directly associated with it. This P-value is not provided by any other tool from those reviewed here. A current drawback in Onto-Express is that if a user wishes to perform the analysis at a fixed depth throughout GO, the user is required to manually expand the nodes up to this level.
2.8 Prerequisites and installation issues
Another important factor is the amount of effort necessary in order to install and use a tool. The web-based services provide the experimental biologists a convenient solution by avoiding the problems usually associated with a local installation of a program (Zhang et al., 2004). On the other hand, tools available over the web may be initially obstructed by security issues. For instance, if the tool uses a specific TCP/IP port and the researcher is behind a firewall, the required port must be open on the firewall before the tool can be used.
Stand-alone tools such as CLENCH, GoMiner and GoSurfer force the user to understand the complexities of a software installation. For example, prerequisites for CLENCH include the prior installation of perl modules for: (1) HTTP request handling, (2) file and console access, (3) common gateway interface, (4) database access, (5) statistical computation and (6) graphical display (http://www.personal.psu.edu/faculty/n/h/nhs109/Clench/Clench_2.0/Prerequisites.txt). As another example, GoMiner requires the user to install the Adobe scalable vector graphics plug-in in order to view the results as a DAG, and the NCBI Cn3D browser plug-in in order to view molecular structures from the Entrez structure database (http://discover.nci.nih.gov/gominer/requirements.jsp). However, the core GoMiner application works without the plug-ins. In principle, web-based tools such as Onto-Express, EASEonline, DAVID, Gene-Merge, Ontology Traverser, GOTM, FuncAssociate, FatiGO only require that the user has a web-browser with an Internet connection. In practice though, even the web-based tools suffer from some platform compatibility issues. For instance, the Microsoft Virtual Machine included in the Internet Explorer browser does not fully implement the Java standard (Lindholm and Yellin, 1999). In consequence, some Java-based tools such as Onto-Express, will require the installation of the Sun Java Runtime Environment.
Another issue is related to the availability of the tool and the requirement for an Internet connection. Web-based tools can be used from any computer, but they cannot be used without an Internet connection. Stand-alone tools require local installations on all computers from which they are to be used, but in principle, they can be used without network access. In practice though, among the stand-alone tools in Table 1 GoSurfer is the only tool that allows the user to actually analyze data without network access, after the initial download of the application and the required data files. The other stand-alone tools in Table 1, either use a local database server (GoMiner, EASE, DAVID) or actually retrieve annotation data at runtime (CLENCH). Unless both the client and the database are on the same computer, these stand-alone tools are essentially the same as web-based tools inasmuch as the client tool requires some network access to connect to the database server. The specific requirements of an application as well as the user preferences will probably be the determining factors from this perspective.
The most important problem in this category is the version control. From this point of view, the web-based tools are far superior because the researcher can always be assured that they are using the very latest version of the software. The software or database updates are always done on the server, by the team who initially wrote the tool. For stand-alone tools, the burden of version control usually rests upon the user who is required to check periodically for new releases and updates. Once such an update becomes available, the burden of software or data update rests again with the user who has to go over the installation process again. In many cases, the updated version works worse than the older version due to issues related to the local environment. In principle, stand-alone tools can try to address this issue by providing automatic software updates. However, this approach means that the updating software must correctly identify, and appropriately deal with, various local software environment issues that tend to be slightly different over the potential hundreds or thousands of different installations.
2.9 Data sources
Most of the available tools use annotation data from a single public database. This has the advantage that the data is always as up to date as the database used. The disadvantage is that no single database offers a complete picture. For primary GO annotation data, the GO database is a comprehensive and up-to-date source since the contributing databases commit their data directly there. Other sources such as Entrez Gene derive their data from the GO database, so there is little advantage from the point of view of GO annotations to derive this data from several sources. However, the secondary analysis discussed here is more powerful if more types of data are integrated in a coherent way. A dedicated annotation database that integrates various types of data from various sources (e.g. KEGG pathways) is potentially more useful than any single database. The drawback is that such a database is: (1) difficult to design and (2) will need to be updated every time any one of its source databases is. This places a heavy burden on the shoulders of the team maintaining it. Given this, it is understandable that most tools use only one of the available annotation databases and most of them use only GO annotations. EASEonline, GOSurfer, eGOn and GOTM use Entrez Gene, whereas GeneMerge, GoMiner, and GOToolBox use the GO database. Onto-Express uses its own Onto-Tools database which is currently the only attempt to integrate resources from several annotation databases. Currently, Onto-Tools uses data from, and is linked to: GenBank, dbEST, Uni-Gene, Entrez Gene, RefSeq, GO and KEGG. Onto-Tools also uses data from NetAffx and Wormbase without being linked to them.
2.10 Supported input IDs
Each probe on a microarray identifies a specific nucleotide sequence, which in turn identifies a specific gene. The annotation databases typically use genes to provide functional annotations. Hence, in order to create a functional profile for a list of differentially expressed genes, one first needs to convert the list of probe IDs into a list of genes. A similar condition also exists when the functional annotations are provided using proteins, where one needs to further map genes to proteins. An ontological analysis tool that supports more than one type of IDs as input will be more useful since it will relieve the user from translating one type of IDs (e.g. Affymetrix probe IDs) to appropriate IDs (e.g. gene IDs).
Onto-Express, GoMiner and GeneMerge provide separate tools (Onto-Translate, MatchMiner and the Gene Name Converter, respectively) that allow the user to convert from other ID types (e.g. GenBank accession number, RefSeq IDs, etc.) to the type(s) of ID used by the application. Although in principle, these tools support more types of IDs as input, this design adds a separate step to the analysis pipeline since the user has to manually take the results from the conversion tool and submit them to the ontological analysis tool. For the purpose of this comparison, we only considered the capabilities of the ontological analysis tool itself.
GoMiner, GeneMerge, and GOToolBox only allow the user to submit organism specific IDs used in the GO database as input. FuncAssociate, Ontology Traverser, and CLENCH only allow one type of ID as input. These tools support MODB gene products, Affymetrix probe IDs, and A.thaliana MIPS IDs, respectively. FatiGO supports Affymetrix probe IDs and GenBank accession numbers, and GOToolBox supports GenBank accession numbers and gene symbols. Onto-Express, DAVID, EASEonline and GOTM support the most different types of IDs: Affymetrix probe IDs, GenBank accession numbers, UniGene cluster IDs and Entrez Gene IDs. In addition, Onto-Express and GOTM also support gene symbols, whereas DAVID supports GenPept, PIR and UniProt protein IDs, and RefSeq IDs.
A tool supporting more than one type of ID must use an appropriate and correct type of identifiers to create functional profiles. For instance, the analysis performed by eGOn is centered around Uni-Gene clusters which assumes that each UniGene cluster corresponds to distinct genes. However, this assumption may not always be accurate. UniGene has been created by comparing expressed sequence tags (ESTs) in the dbEST database (Schuler, 1997). However, due to the alternative splicing of the mRNA, it is entirely possible that ESTs from the same gene cluster in different groups, which will result in several UniGene clusters being associated with the same gene. As an example, the SET8 gene (SET8: PR/SET domain containing protein 8, Entrez Gene ID: 387893) is associated to UniGene clusters Hs.443735 and Hs.536369. If a study includes any such genes, treating each UniGene cluster as a distinct gene may not always be appropriate. In such circumstances, the results can be skewed towards those GO terms that are associated to genes from which more than one UniGene cluster is derived. This may become particularly important if further research will confirm the current estimates that more than half of the human genes may have alternative splice variants.
3 DRAWBACKS AND LIMITATIONS OF THE CURRENT APPROACH
Each of these tools uses one or more annotation databases and creates a list of function categories in which the genes from the input list are known to be involved in. The functional categories that are overly represented in a statistically significant way in the list of differentially regulated genes are inferred to be meaningfully related to the condition under study. However, this approach of translating a list of differentially expressed genes into a list of functional categories using annotation databases suffers from a few important limitations. Since these limitations are related to the approach itself, all current tools exhibit them.
Firstly, the existing annotations databases are incomplete. For virtually all sequenced organisms only a subset of known genes are functionally annotated (King et al., 2003). Furthermore, most annotation databases are built by curators who manually review the existing literature. Although unlikely, it is possible that certain known facts might get temporarily overlooked. For instance, we found references in literature published in the early 90s, for 65 functional annotations that are yet not included in the current functional annotation databases. As an example, the gene HMOX2 was shown to be involved in the process of pigment biosynthesis in 1992 (McCoubrey et al., 1992) and is still not annotated as such today. More commonly, recent annotations are not in the databases yet because of the time lag necessary for the manual curation process.
Secondly, certain pieces of information may also be imprecise or incorrect. In the GO, out of 19 490 total biological process annotations available for Homo sapiens, 11 434 associations are inferred exclusively from electronic annotations (i.e. without any expert human involvement) (http://www.geneontology.org/GO.current.annotations.shtml). The vast majority of such electronic annotations are reasonably accurate (Camon et al., 2005). However, many such annotations are often made at very high-level GO terms which limits their usefulness. Furthermore, some of these inferences are incorrect (King et al., 2003; Wang et al., 2004). Even though in some cases the error is very conspicuous to a human expert, currently, there are no automated techniques that could analyze, discover and correct such erroneous assignments. At the present time, none of the tools allows any type of weighting by the type of evidence which is a limitation since experimentally derived annotations are more trustworthy than electronically inferred ones.
The current approach used for ontological analysis is limited to looking up existing annotations and performing a significance analysis for the categories found. This approach cannot discover previously unknown functions for known genes even if there is data justifying such inferences. For example, the gene SLC13A2 [solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 (H.sapiens)] encodes the human Na(+)-coupled citrate transporter and is annotated in GO for the molecular function organic anion transporter activity. However, it is not annotated for the corresponding biological process, organic anion transport. This is not a problem for the curator, and the human expert querying GO for this specific gene. For them, it is obvious that a gene that has organic anion transporter activity will be involved in the organic anion transport. However, a query that tries to find all genes involved in the process of organic anion transport will fail to retrieve this gene. Similarly, any ontological analysis software trying to find out what underlying processes are represented by a given list of genes containing this gene, will either fail to consider the organic anion transport if no other genes are involved in it, or will calculate its statistical significance incorrectly by ignoring this gene.
Another limitation is related to those genes that are involved in several biological processes. For such genes, all current tools weight all the biological processes equally. At the moment, it is not possible to single out the more relevant one by using the context of the other genes differentially expressed in the current experiment. BRCA1 for instance, is a well known tumor suppressor but is also known to be involved in carbohydrate metabolism. If most other genes found to be changed in the current experiment are involved in processes such as DNA damage response, apoptosis, induction of apoptosis, and signal transduction, it is perhaps more likely that in this experiment BRCA1 is playing its usual tumor suppressor role. However, if most other genes are involved in carbohydrate mediated signaling, carbohydrate transport and metabolism, etc., then it is perhaps more likely that BRCA1's role in the carbohydrate metabolism is more relevant.
The existing GO based functional profiling approaches are currently decoupled from the gene expression data obtained from the microarray experiment in the previous step. In any given biological phenomenon, different genes are regulated to different extents. The data providing information about different amount of regulation for one gene versus another gene can be useful in assigning different weights to the corresponding biological processes they are involved in and hence, can help in inferring if one biological process is more relevant than the other(s).
The usefulness of the existing functional profiling approaches is impacted by the annotation bias present in the ontological annotation databases. Some biological processes are studied in more detail than the others (e.g. apoptosis), thus generating more data. If more data about a specific biological process is available, more of the genes associated with it will be known and hence, the process is more likely to appear as significant than the others.
An important issue related to the ontological analysis is the name–space mapping from one resource to another. At the moment, the existing knowledge about known genes is spread out over a number of databases and other resources. Different databases are maintained by various independent groups that many times have very different interest and research foci. Each such resource often uses its own type of identifiers. For instance, GenBank uses accession numbers, UniGene uses cluster identifiers (IDs), Entrez Gene uses gene IDs, SWISSPROT uses protein IDs, TrEMBL accession numbers, etc. Furthermore, genes are also represented by various company-specific gene IDs. A typical example would be Affymetrix which uses its own probe IDs to represent various genes. Various resources try to address the problem by maintaining other types of IDs together with their own and by providing ad hoc tools able to map from one type of ID to another. For instance, besides its own gene names, Entrez-Gene database also contains UniGene cluster IDs, and Affymetrix's NetAffyx provides RefSeq and GenBank accession numbers, besides its own array specific probe IDs. For example, the gene beta actin in mouse is referred to as MGI:87904 in Mouse Genome Informatics (MGI), Actb (Gene ID: 11461) in Entrez Gene, Mm.297 in UniGene, ACTB_MOUSE (primary accession number: P60710) in UniProt, and TC1242885 in the TIGR gene index. In addition, the beta actin gene in mouse is referred to by 29 mRNA sequences and 4552 ESTs in dbEST, 5 secondary accession numbers in UniProt, 4 other accession IDs in MGI, and 5 probe IDs on 4 different Affymetrix mouse arrays. The burden of mapping various types of ID on each other is left entirely on the shoulders of the researchers, who often have to revert to cutting-and-pasting lists of IDs from one database to another.
The name–space issue becomes crucial when trying to translate from lists of differentially regulated genes to functional profiles because the mapping from one type of identifier to another is not one-to-one. In consequence, the type of IDs used to specify the list of differentially regulated genes can potentially affect the results of the analysis (Drăghici, 2003; Khatri et al., 2004). While GO represents a viable, long term solution to the problem of inconsistent vocabulary, the name–space problem is yet to be solved.
Novel ideas have started to appear in this area addressing some of the issues above. Onto-Semantics has been proposed as a tool able to analyze the semantic content of annotation databases and find incomplete and incorrect annotations (Khatri et al., 2005b). GoToolBox offers a different tool (GO-Proxy) to identify clusters of related terms. MAPPFinder (Doniger et al., 2003), Pathway-Express (Khatri et al., 2005a), Cytoscape (Shannon et al., 2003), Pathway Tools (Karp et al., 2002) and Pathway Processor (Grosu et al., 2002) are only a few of the tools trying to expand the secondary analysis by including metabolic or regulatory pathway information. Other related tools can be found on the tools page of the GO (http://www.geneontology.org/GO.tools.shtml).
4 CONCLUSIONS
This paper presents a comparison of several ontological analysis tools. This comparison emphasizes characteristics of each tool as well as a number of limitations and drawbacks of the approach as a whole. Currently, there is a large number of tools implementing a very similar approach. At the same time, this approach is severely limited in certain regards. It would be more beneficial if future tools expand the current approach by trying to address some of these limitations rather than providing endless variations of the same idea.
ACKNOWLEDGEMENTS
This work has been supported by the following grants: NSF DBI-0234806, DOD DAMD 17-03-02-0035, NIH(NCRR) 1S10 RR017857-01, MLSC MEDC-538 and MEDC GR-352, NIH 1R21 CA10074001, 1R21 EB00990-01 and 1R01 NS045207-01.
Footnotes
GOTM also allows the users to upload their own list of genes or use one of 37 Affymetrix arrays as the set of reference genes.
Note that this statistic does not take into consideration the number of experiments performed in parallel.
The very hierarchy of the GO on which this type of analysis relies, shows that many biological categories are very closely related, sometimes as children of the same node on the next level up.
Every attempt produced: “Error unmarshaling return header; nested exception is: java.io.EOFException.”
Conflict of Interest: none declared.
REFERENCES
- Al-Shahrour F, et al. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- Berriz GF, et al. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- Camon EB, et al. An evaluation of GO annotation retrieval for BioCreAtIvE and GOA. BMC Bioinformatics. 2005;6:S17. doi: 10.1186/1471-2105-6-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis CI, Hartl DL. GeneMerge-post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2002;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- Cho RJ, et al. Transcriptional regulation and function during the human cell cycle. Nat. Genet. 2001;27:48–54. doi: 10.1038/83751. [DOI] [PubMed] [Google Scholar]
- Doniger SW, et al. Mappfinder: using gene ontology and genmapp to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drăghici S. Data Analysis Tools for DNA Microarrays. Chapman and Hall/CRC Press; 2003. [Google Scholar]
- Drăghici S, et al. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 2003a;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drăghici S, et al. Global functional profiling of gene expression. Genomics. 2003b;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- Fisher LD, van Belle G. Biostatistics: A Methodology for Health Sciences. John Wiley and Sons; New York: 1993. [Google Scholar]
- Grosu P, et al. Pathway processor: a tool for integrating whole-genome expression results into metabolic networks. Genome Res. 2002;12:1121–1126. doi: 10.1101/gr.226602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Tamhane AC. Multiple Comparison Procedures. John Wiley and Sons, Inc.; New York: 1987. [Google Scholar]
- Holland B, Copenhaver MD. An improved sequentially rejective Bonferroni test procedure. Biometrica. 1987;43:417–423. [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979;6:65–70. [Google Scholar]
- Karp PD, et al. The pathway tools software. Bioinformatics. 2002;18:225–232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- Khatri P, et al. Profiling gene expression using Onto-Express. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- Khatri P, et al. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 2004;32:W449–W456. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P, et al. Recent additions and improvements to the onto-tools. Nucleic Acids Res. 2005a;33:W762–W765. doi: 10.1093/nar/gki472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P, et al. A semantic analysis of the annotations of the human genome. Bioinformatics. 2005b doi: 10.1093/bioinformatics/bti538. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, et al. Predicting gene function from patterns of annotation. Genome Res. 2003;13:896–904. doi: 10.1101/gr.440803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm T, Yellin F. The Java™Virtual Machine Specification. 2nd edition Addison-Wesley Professional; 1999. [Google Scholar]
- Man MZ, et al. POWER_SAGE: comparing statistical tests for SAGE experiments. Bioinformatics. 2000;16:953–959. doi: 10.1093/bioinformatics/16.11.953. [DOI] [PubMed] [Google Scholar]
- Martin D, et al. GOToolBox: functional analysis of gene datasets based on gene ontology. Genome Biol. 2004;5:R101. doi: 10.1186/gb-2004-5-12-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoubrey WK, Jr, et al. Human heme oxygenase-2: characterization and expression of a full-length cDNA and evidence suggesting that the two HO-2 transcripts may differ by choice of polyadenylation signal. Arch. Biochem. Biophys. 1992;295:13–20. doi: 10.1016/0003-9861(92)90481-b. [DOI] [PubMed] [Google Scholar]
- Schuler GD. Pieces of puzzle: Expressed sequence tags and the catalog of human genes. J. Mol. Med. 1997;75:694–698. doi: 10.1007/s001090050155. [DOI] [PubMed] [Google Scholar]
- Shaffer JP. Modified sequentially rejective multiple test procedures. J. Am. Stat. Assoc. 1986;81:826–831. [Google Scholar]
- Shah N, Fedoroff NV. CLENCH: a program for calculating Cluster ENriCHment using the Gene Ontology. Bioinformatics. 20:1196–1197. doi: 10.1093/bioinformatics/bth056. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Azuaje F, Bodenreider O, Dopazo J. Gene expression correlation and gene ontology-based similarity: An assessment of quantitative relationships.. Proceedings of the 2004 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology; San Diego, CA. 2004. pp. 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, et al. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]


