Abstract
There is no systematic examination of affinity tag utility in Gram-positive bacteria, which limits the investigation of protein function in this important group of bacteria as specific antibodies for many of native proteins are generally not available. In this study, we utilized an E. coli-streptococcal shuttle vector pVT1666 and constructed two sets of expression plasmids pVPT-CTag and pVPT-NTag, with each set containing five affinity tags (GST, GFP, HSV, T7 and Nano) that can be fused to either the C- or N-terminus of a target protein. A putative glycosyltransferase (Gtf2) essential for Fap1 glycosylation was used to demonstrate the utility of the cassettes in detection of Gtf2 fusion proteins, and the biological relevance of the proteins in our working strain Streptococcus parasanguinis. GFP and T7 tags were readily expressed in S. parasanguinis as either an N- or C-terminal fusion to Gtf2. Only the C- terminal fusion of GST and HSV were able to be identified in S. parasanguinis. The Nano tag was not detected in either E. coli or S. parasanguinis. Genetic complementation experiments indicated that all the tagged Gtf2 fusion proteins could restore the Gtf2 function in the null mutant except for the Nano-tagged Gtf2 at its N-terminal fusion. Using a T7-tagged Gtf2 fusion construct, we demonstrated that the fusion cassette is also useful in detection of the fusion tag expression in other streptococci including S. mutans, S. pneumoniae and S. sanguinis. Therefore, the expression cassettes we constructed will be a useful tool not only to investigate protein-protein interactions in Fap1 biogenesis in S. parasanguinis, but also to study protein functions in other gram-positive bacteria in which pVT1666 replicates.
Keywords: affinity tags, E. coli and streptococcal shuttle vector, Fap1 glycosylation
1. Introduction
Streptococcus parasanguinis is a primary colonizer of dental plaque and also plays an important role in bacterial endocarditis (Carlsson et al., 1970; Marsh, 1995; Jenkinson and Lamont, 1997). Fap1, a serine-rich glycoprotein of Streptococcus parasanguinis, is required for fimbrial biogenesis and biofilm formation (Wu et al., 1998; Froeliger et al., 2001). A seven-gene cluster downstream of the fap1 locus is involved in Fap1 glycosylation (Wu et al., 2007). We hypothesize these gene products interact with each other to form a protein complex to achieve Fap1 glycosylation. Two putative glycosyltransferases genes, gtf1 and gtf2 are located in the cluster and are required for initiation of Fap1 glycosylation (unpublished). These two gene products interact with each other and the interaction is critical for Fap1 glycosylation. However whether they also interact with other gene products and how they link to the other steps of Fap1 glycosylation are not yet known.
The most common way to detect protein-protein interactions is by use of specific immunological reagents. Unfortunately, many antibodies prepared using recombinant proteins in heterologous hosts fail to react with the endogenous proteins from a native host (http://www.agrisera.se/index.php?p=10&PHPSESSID). A simple and rapid solution is to tag the targeted proteins using different affinity tags and then use tag-special antibody to detect the expression of the fusion proteins. Affinity tags have become a powerful tool for the detection and purification of recombinant proteins, and are also widely used for protein-protein interaction studies (Terpe, 2003; Lichty et al., 2005; Kumada et al., 2007). Currently, the available affinity tags are divided into two groups. One group is composed of small affinity tags such as 6xHis (hexahistidine), c-myc (amino acids 410–419 of human myc protein: EQKLISEEDL), HSV (amino acids 290 300 of an envelop component of herpes simplex virus: QPELAPEDPEA), T7 (the 11 amino acids of the T7 phage gene 10 protein: MASMTGGQQMG) and Nano tag (the 15 amino acids of streptavidin-binding peptide: DVEAWLDERVPLVET). In many instances, the tags don’t interfere with the function of fused proteins because of their small size (Loomis and Moseley, 1998; Varughese et al., 1998; Lamla and Erdmann, 2004; Cazalla et al., 2005). The second group consists of affinity tags of large size such as GFP (green fluorescent protein: 28kDa) and GST (glutathione-S-transferase: 26kDa). These tags are widely used for protein localization or to enhance the solubility of the target protein (Huh et al., 2003; Smyth et al., 2003). Affinity tags have been fused to either the N- or C-terminus of target proteins (Hearn and Acosta, 2001; Arnau et al., 2006) to insure the likelihood of success in protein detection and purification.
Streptococci are Gram-positive bacteria. At present, only a few tags such as GFP, Strep (a nine amino acid peptide with streptavidin-binding activity: AWRHPQFGG) and His tags are used for the detection and purification of recombinant proteins in some gram-positive bacteria (Myscofski et al., 2000; Fujimoto and Ike, 2001; Biedendieck et al., 2007). The GFP tag is mainly used as a reporter to evaluate protein expression (Geoffroy et al., 2000; Marra et al., 2002; Deng et al., 2007). The others have been used randomly. There is no systematic examination of affinity tag utility for Gram-positive bacteria. This deficit limits the investigation of protein function in this important group of bacteria. During the study of Fap1 glycosylation, we successfully tagged the SecA2 protein with GFP in S. parasanguinis (Chen et al., 2006). Since no other tag had been tested for the detection and purification of proteins in S. parasanguinis, we used S. parasanguinis as a model organism. The plasmid pVA838, containing a streptococcal origin of replication (Macrina et al., 1982), was used as the plasmid backbone to systematically examine the utility of affinity tags in expression of the putative glycosyltransferase Gtf2, one important gene product in Fap1 glycosylation. We constructed a series of expression vectors with five different affinity tags (GFP, GST, HSV, Nano and T7) using pVA838 derivative, pVT1666 (Chen et al., 2006). These tags were fused to either the N- or C-terminus of the glycosyltransferase Gtf2. Our results showed that four fusion constructs were expressed in both E. coli and S. parasanguinis. In each case, we detected the expression of Gtf2. The expressed Gtf2 fusion proteins were functional as they could complement the gtf2 mutant in terms of its ability to restore the wild type level of Fap1 glycosylation. Importantly, the T7 affinity tag we constructed in the E. coli-streptococcus shuttle vector was readily expressed in other streptococcal species including S. pneumoniae, S. mutans and S. sanguinis, demonstrating the utility of the affinity tag system in other streptococci.
2. Materials and methods
2.1. Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. S. parasanguinis, S. mutans UA159, S. pneumoniae V047 and S. sanguinis SK36 strains were incubated statically in 5% CO2 in Todd–Hewitt (TH) medium or on Todd-Hewitt agar plates at 37°C. Erythromycin (2 or 10 μg/ml) and kanamycin (125μg/ml) were added when required. E. coli was cultured in Luria–Bertani medium or on Luria-Bertani agar plates at 37°C. Erythromycin (300 μg/mL) was added when necessary.
Table 1.
Strains and plasmids used in this study
| Strains | Relevant properties | Source |
|---|---|---|
| E. coli | ||
| Top10 | for propagation of the recombinant plasmids | Invitrogen |
| S. parasanguinis | ||
| FW213 | wild type | Cole et al., 1976 |
| fap1− | wild type; fap1::aphA3 | Chen et al., 2002 |
| gtf2− | Wild type; gtf2::aphA3 | Wu et al. unpublished |
| S. mutans UA159 | wild type | Ajdiæ et al., 2002 |
| S. pneumoniae V047 | wild type | Hollingshead |
| S. sanguinis SK36 | wild type | Xu et al., 2007 |
| Plasmids | ||
| pVT1666 | GFP tag for C-terminal fusion | Chen et al., 2006 |
| PGEX-6P-1 | GST fusion vector | Invitrogen |
| pET27b(+) | HSV tag expression vector | Novagen |
| pVPT-NGFP | GFP tag for N-terminal fusion | This study |
| pVPT-CGST | GST tag for C-terminal fusion | This study |
| pVPT-NGST | GST tag for N-terminal fusion | This study |
| pVPT-CHSV | HSV tag for C-terminal fusion | This study |
| pVPT-NHSV | HSV tag for N-terminal fusion | This study |
| pVPT-Cnano | Nano tag for C-terminal fusion | This study |
| pVPT-Nnano | Nano tag for N-terminal fusion | This study |
| pVPT-CT7 | T7 tag for C-terminal fusion | This study |
| pVPT-NT7 | T7 tag for N-terminal fusion | This study |
| pVPT-Gtf2-CGFP | gtf2 cloned in pVT1666 | This study |
| pVPT-NGFP-Gtf2 | gtf2 cloned in pVPT-NGFP | This study |
| pVPT-Gtf2-CGST | gtf2 cloned in pVPT-CGST | This study |
| pVPT-NGST-Gtf2 | gtf2 cloned in pVPT-NGST | This study |
| pVPT-Gtf2-CHSV | gtf2 cloned in pVPT-CHSV | This study |
| pVPT-NHSV-Gtf2 | gtf2 cloned in pVPT-NHSV | This study |
| pVPT-Gtf2-Cnano | gtf2 cloned in pVPT-Cnano | This study |
| pVPT-NNano-Gtf2 | gtf2 cloned in pVPT-Nnano | This study |
| pVPT-Gtf2-CT7 | gtf2 cloned in pVPT-CT7 | This study |
| pVPT-NT7-Gtf2 | gtf2 cloned in pVPT-NT7 | This study |
2.2. Primers and DNA manipulation
Plasmid DNA was isolated from E. coli using the QIAgen Spin Miniprep kit (Qiagen Inc., Santa Clarita, CA). The primers used in the amplification of tag DNA sequence and gtf2 are listed in Table 2. Restriction endonucleases, alkaline phosphatase, and T4 DNA ligase were purchased from Promega (Madison, WI) and used according to the manufacturer’s instructions. Polymerase chain reaction (PCR) amplifications were performed by using Taq DNA polymerase (Promega) or KOD hot start DNA polymerase (Novagen) according to the manufacturer’s instructions. PCR products were purified using the QIAquick gel extraction Kit (Qiagen Inc.). The appropriate single-stranded oligonucleotides (Table 2) representing small tag (HSV, Nano and T7) genes were synthesized and annealed, respectively, to generate double-stranded oligonucleotides as previously described (Spickofsky and Margolskee, 1991). Transformation of recombinant plasmids into E. coli was performed by using standard methods (Sambrook et al., 1989). Transformation of plasmids into S. parasanguinis was carried out according to previously described method (Fenno et al., 1993). DNA sequencing was performed by using the dideoxynucleotide termination cycle sequencing method. ClustalW and NCBI tools were used for DNA sequence analysis.
Table 2.
Primers used in this study
| Primers | Sequence |
|---|---|
| NGFP-KpnI | GATCAGGTACCATGAGTAAAGGAGAAGAACTTTTC |
| NGFP-BamHI | CAGCTGGATCCTTTGTATAGTTCATCCATGCCATG |
| GST-N/C-KpnI | GATCAGGTACCTCCCCTATACTAGGTTATTGG |
| CGST-BamHI | AGATCGGATCCCTAGGGCCCCTGGAACAGAACTTCC |
| NGST-BamHI | GATCAGGATCCGTCACGATGCGGCCGCTCGAG |
| CHSV-KpnI | GATCAGGTACCAGCCAGCCAGAACTCGCC |
| CHSV-BamHI | GTCATGGATCCCCAACTCAGCTTCCTTTCG |
| NHSV-KpnI | GATCAGGTACCAGCCAGCCAGAACTCGCCCCGGAAGACCCCGAGGATGTCGAGCACCACCACCACCACCACGGATCCCACTG |
| NHSV-BamHI | CAGTGGGATCCGTGGTGGTGGTGGTGGTGCTCGACATCCTCGGGGTCTTCCGGGGCGAGTTCTGGCTGGCTGGTACCTGATC |
| CNano-KpnI | GAGTGAGGTACCGACGTCGAGGCTTGGCTAGATGAACGTGTACCTTTGGTTGAGACTTAGGGATCCGATCGC |
| CNano-BamHI | GCGATCGGATCCCTAAGTCTCAACCAAAGGTACACGTTCATCTAGCCAAGCCTCGACGTCGGTACCTCACTC |
| NNano-KpnI | GAGTGAGGTACCGACGTCGAGGCTTGGCTAGATGAACGTGTACCTTTGGTTGAGACTGGATCCGATCGC |
| NNano-BamHI | GCGATCGGATCCAGTCTCAACCAAAGGTACACGTTCATCTAGCCAAGCCTCGACGTCGGTACCTCACTC |
| CT7-KpnI | GATCAGGGTACCATGGCTAGTATGACAGGAGGTCAACAAATGGGATAGGGATCCGACTGC |
| CT7-BamHI | GCAGTCGGATCCCTATCCCATTTGTTGACCTCCTGTCATACTAGCCATGGTACCCTGATC |
| NT7-KpnI | GATCAGGGTACCATGGCTAGTATGACAGGAGGTCAACAAATGGGAGGATCCGACTGC |
| NT7-BamHI | GCAGTCGGATCCTCCCATTTGTTGACCTCCTGTCATACTAGCCATGGTACCCTGATC |
| Gtf2-SalI | GCAGCGTCGACATGATTAGGTTGTTTGAATG |
| Gtf2-BamHI-F | GATCAGGATCCATGATTAGGTTGTTTGAATGG |
| Gtf2-KpnI | CGGCCCGGTACCATCTACATTACTAACCAATAC |
| Gtf2-BamHI-R | GATCAGGATCCCTAATCTACATTACTAACCAATAC |
| VPT1 | ACCTTAGGATTAGATAGT |
| VPT2 | TCCCATTTAGCCGTCATT |
2.3. Constructions of fusion vectors
A DNA fragment of GFP tag used for the N-terminal fusion was amplified from pVT1666 using primer pairs, NGFP-KpnI and NGFP-BamHI. The DNA sequence of GST tag used for N- and C-terminal fusion was amplified from pGEX-6P-1 using primer pairs GST-N/C-KpnI and NGST-BamHI, and GST-N/C-KpnI and CGST-BamHI, respectively. The DNA sequence of HSV tag used for the C-terminal fusion was amplified from pET27b(+) using primers pairs CHSV-KpnI and CHSV-BamHI, whereas the DNA sequence for N-terminal fusion was obtained by annealing primer pairs NHSV-KpnI and NHSV-BamHI. The DNA sequence of Nano tag used for the N- and C-terminal fusion was obtained by annealing primer pairs NNano-KpnI and NNano-BamHI, and CNano-KpnI and CNano-BamHI, respectively. The DNA sequence of T7 tag used for the N- and C-terminal fusion was obtained by annealing primer pairs NT7-KpnI and NT7-BamHI, and CT7-KpnI and CT7-BamHI, respectively. The resulting DNA fragments coding for those tags were cloned into KpnI and BamHI restriction enzyme sites of pVT1666 vector to produce pVPT-CTag and pVPT-NTag vectors in which the engineered tags replaced the GFP tag. The full-length gtf2 gene, amplified from genomic DNA of S. parasanguinis, was then fused with tags at its C-terminus using SalI and KpnI restriction enzyme sites, or at its N-terminus using BamHI sites, respectively, to produce plasmids pVPT-Gtf2-CTag and pVPT-NTag-Gtf2. E. coli strain Top10 was used as a host for all constructions.
2.4. Detection of expression of fusion tags in E. coli and S. parasanguinis
To detect the expressions of fusion tags in E. coli, 1ml of exponentially grown Top10 cells carrying different tag constructs (OD600≈0.6–0.7) were harvested and lysed in 100μl 1X SDS loading buffer (50mM Tris-HCl, pH 6.8; 2% SDS; 0.1% bromophenol blue; 10% glycerol; 100mM β-mercaptoethanol). To detect the expressions of fusion tags in S. parasanguinis, the plasmids pVPT-CTag, pVPT-Gtf2-CTag and pVPT-NTag-Gtf2 were electroporated into a gtf2 mutant respectively. 1 ml of the gtf2 mutant cells transformed with different tag constructs were grown to exponential phase (OD470≈0.6–0.7), harvested and lysed in 40μl lysis buffer [1:10 dilution of LambdaSa2 lysin (GenBank accession number: AE014275) in PBS buffer (0.1 M NaCl, 0.01 M NaH2PO4, pH 7.4)] for 10 min at room temperature before adding 5X SDS loading buffer. The cell lysates of E. coli and S. parasanguinis were subsequently subjected to electrophoresis on 4–12% commercially available precast gradient gels (Cambrex) and electrotransferred onto nitrocellulose membranes for Western blot analysis (Wu et al., 2007). Tag special antibodies were used for immunodetection. Anti-GFP (Covance), Anti-T7, anti-HSV and anti-GST (Novagen) monoclonal antibodies were used at 1:4000, 1:10000, 1:10000 and 1:2000 dilution, respectively. Horse Radish Peroxidase-conjugated streptavidin (1:1000 dilution) was used for Nano tag detection (Pierce).
2.5. Genetic complementation of the gtf2 mutant
The gtf2 mutants transformed with different tag fusion constructs were used to detect the expression of mature Fap1 by Western blot analyses. In brief, 0.5ml of culture supernatants from those transformants grown to exponential phase (OD470≈0.6–0.7) were harvested, and subject to ice cold ethanol precipitation as described (Wu et al., 2007). The precipitated protein samples were dissolved in 30μl 1 × SDS-loading buffers. 15μl of the culture supernatant samples were subjected to Western blot analysis using mature Fap1 specific antibody MAb F51 (Elder and Fives-Taylor, 1986).
2.6. Immunodetection of T7-tagged Gtf2 fusion protein in other streptococcal species
To examine the expression of T7 fusion tag in other streptococcal species, plasmids pVPT-CT7 and pVPT-Gtf2-CT7 were transformed into S. mutans UA159, S. pneumoniae V047 and S. sanguinis SK36, respectively. The transformation methods of the three strains were performed as previously described (Perry and Kuramitsu, 1981; Correia et al., 1996; Bricker and Camilli, 1999). The three host strains and their corresponding transformants were grown to exponential phase (OD470≈0.6–0.7) and 1ml cells of all tested strains were harvested, and then lysed as described in previous section. The resulting cell lysates were subjected to Western blot analysis using Anti-T7 monoclonal antibody. The S. parasanguinis gtf2− and its transformants with pVPT-CT7 or pVPT-Gtf2-CT7 were used as the controls for the expression of T7 fusion tag in S. parasanguinis.
3. Results and discussion
3.1. Strategies for construction of fusion vectors
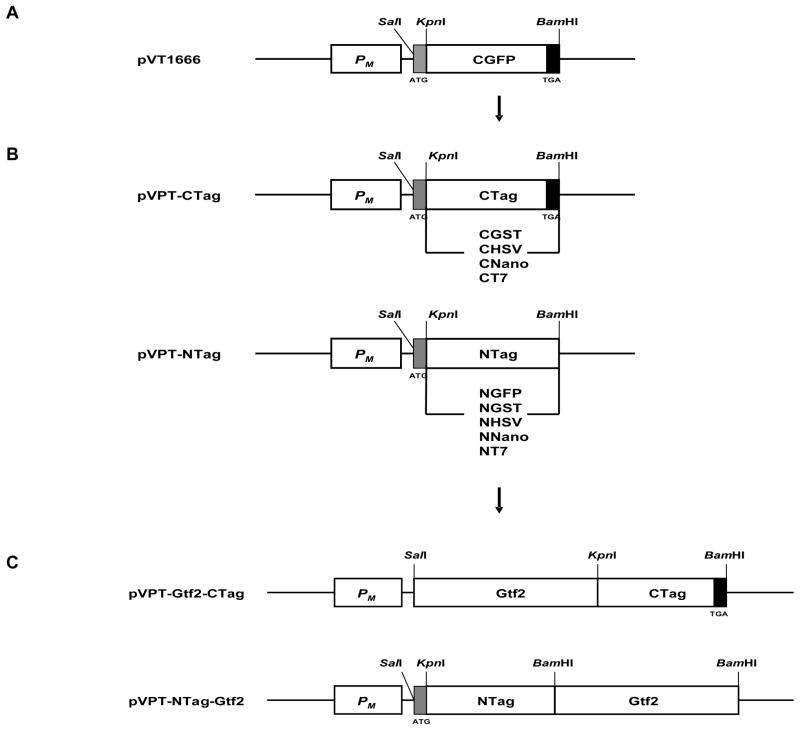
We utilized an expression vector, pVT1666, as a template to investigate the expression of different affinity tags in E. coli and S. parasanguinis. The plasmid pVT1666 is a derivative of a broad-host-range shuttle vector pVA838 that replicates in both E. coli and Streptococcal species (Macrina et al., 1982; Chen et al., 2006). It possesses a maltose promoter PM (Fig. 1A), derived from a tightly regulated plasmid pLS1RGFP of S. pneumoniae, which can drive the expression of GFP protein in S. pneumoniae (Nieto et al., 2000) and S. parasanguinis (Chen et al., 2006). The plasmid pVT1666 carries a GFP tag with a stop codon TGA (Fig. 1A), which was amplified from plasmid pLS1RGFP. The SecA2-GFP fusion protein cloned into pVT1666 is readily expressed in S. parasanguinis (Chen et al., 2006).
Fig. 1.
Schematic diagrams of fusion constructs. pVT1666 (A) was used as a backbone to generate fusion vectors pVPT-CTag and pVPT-NTag (B) with five tags fused to Gtf2 at either N-terminus or C-terminus (C).PM, maltose promoter; CTag (CGST, CHSV, CNano or CT7) stands for GST, HSV, Nano or T7 tag fused to the C-terminus of Gtf2 protein by using SalI and KpnI sites. NTag (NGFP, NGST, NHSV, NNano or NT7) stands for GFP, GST, HSV, Nano or T7 tag fused to the N-terminus of Gtf2 protein by using BamHI site. Start codon (ATG) and stop codon (TGA) are indicated with grey rectangle and black rectangle, respectively.
We constructed a series of affinity tags (GFP, GST, HSV, Nano and T7) in the plasmid pVT1666 to explore the utility of pVT1666. These constructs were utilized to generate fusion proteins with the glycosyltransferase Gtf2. GST and HSV tags were obtained from the widely used expression vectors pGEX-6P-1 and pET27b(+). Two small affinity tags, 6xHis and c-myc, were previously used for tagging SecA2 in S. parasanguinis, but failed to generate detectable fusion proteins using His and c-myc special antibodies (data not shown). Therefore, we decided to try other small affinity tags to examine their ability to form detectable fusion proteins in S. parasanguinis. Based on common genetic codes appearing in S. parasanguinis, we designed and synthesized DNA sequences for two small tags, Nano and T7. The GFP tag in pVT1666 was replaced by GST, HSV, Nano and T7 tags using KpnI and BamHI sites in pVT1666 (Fig. 1B). We constructed two sets of plasmids pVPT-CTag and pVPT-NTag to increase the flexibility of the expression vectors. These constructs allowed us to fuse the tags to the N- and C-terminus of the target protein respectively. pVPT-CTag was constructed by insertion of corresponding tags into KpnI and BamHI sites. A stop codon was engineered at the end of each tag (Fig. 1B). pVPT-NTag was constructed in the same manner except that no stop code was added at the end of each tag (Fig. 1B). To tag the C-terminal Gtf2, Gtf2 was cloned into SalI and KpnI sites of pVPT-CTag. Its stop codon TGA was removed to allow fusion with GST, HSV, Nano and T7 tags (Fig. 1C). To tag the N-terminus of Gtf2, Gtf2 with its own stop codon was cloned into the BamHI site of pVPT-NTag in the correct orientation (Fig. 1C). The resulting expression vectors pVPT-CTag and pVPT-NTag, and Gtf2 expression plasmids pVPT-Gtf2-CTag and pVPT-NTag-Gtf2 were confirmed by restriction enzyme digestion and DNA sequencing analysis (using primers VPT1 and VPT2).
3.2. Expression of tagged Gtf2 in both E. coli and S. parasanguinis
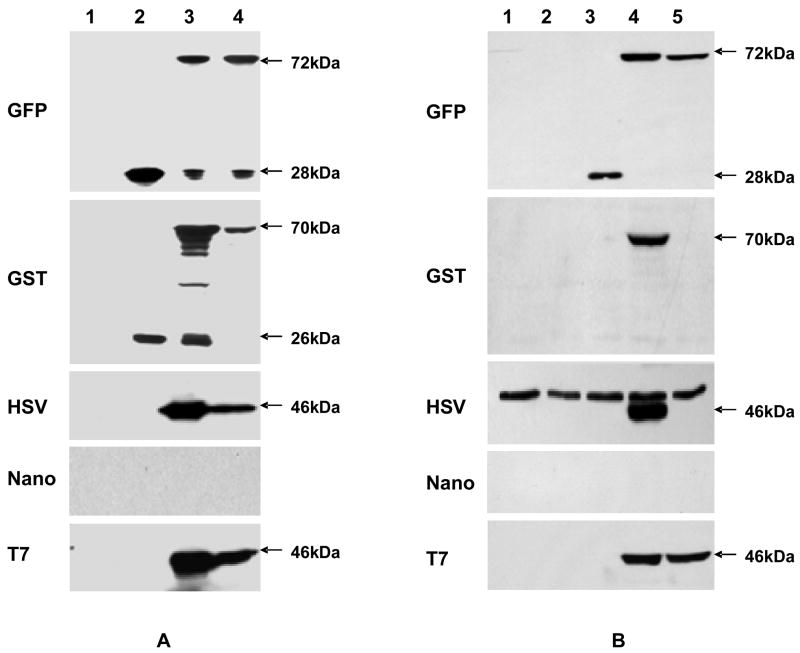
Western blotting analyses using special antibodies to the tags were performed to determine if the tagged fusion constructs expressed the fusion proteins. To detect the expression of tagged Gtf2 in E. coli cells harboring pVPT-Gtf2-CTag and pVPT-NTag-Gtf2, we used E. coli Top10 cells and E. coli Top10 cells harboring pVPT-CTag as controls. No signal was detected in E. coli Top10 cells (Fig. 2A, lane 1) using tag-specific antibodies. GFP (28 kDa) and GST (26 kDa) tag alone could be detected in E. coli Top10 cells harboring pVPT-CGFP or pVPT-CGST (Fig. 2A, panels 1 and 2, lane 2), but HSV, Nano, and T7 tag alone couldn’t be detected because of their small size (Fig. 2A, panels 3, 4 and 5, lane 2). Both the N- and C-terminus constructs of GFP, GST, HSV and T7 fusion tags were expressed in E. coli, displayed 72kDa GFP-tagged Gtf2 (Fig. 2A, panel 1, lanes 3 and 4), 70kDa GST-tagged Gtf2 (Fig. 2A, panel 2, lanes 3 and 4), 46kDa HSV-tagged and T7-tagged Gtf2s (Fig. 2A, panels 3 and 5, lanes 3 and 4), respectively. No signal was detected for the Nano tag fusion in E. coli (Fig. 2A, panel 4, lanes 3 and 4), even though the sequence of the Nano-tagged Gtf2 fusion was correct. This result suggested the Nano tag is not properly presented at either termini of Gtf2. To detect the expression of tagged Gtf2 in S. parasanguinis, pVPT-CTag, pVPT-Gtf2-CTag and pVPT-NTag-Gtf2 were transformed into the gtf2 mutant, respectively. The resulting transformants were analyzed by Western blotting. The S. parasanguinis FW213 (wild type) bacteria, gtf2 mutant and gtf2 mutant harboring pVPT-CTag were used as controls. No specific signal was detected in wild type and gtf2 mutant cells using tag-specific antibodies (Fig. 2B, lanes 1 and 2). GFP tag (28 kDa) alone could be detected in S. parasanguinis (Fig. 2B, panel 1, lane 3). HSV, Nano and T7 tag alone couldn’t be detected in S. parasanguinis due to their small size (Fig. 2B, panels 3–5, lane 3). GFP and T7 tag fusions (72kDa and 46kDa) were readily expressed in S. parasanguinis, independent of their fusion position (Fig. 2B, panels 1 and 5, lanes 4 and 5). The expression of the GST and HSV tag fusions (70kDa and 46kDa) in S. parasanguinis was dependent on fusion position as only the C-terminal fusion could be detected (Fig. 2B, panels 2 and 3, lane 4). Interestingly, we can detect the expression of the Gtf2-GST fusion protein in S. parasanguinis (Fig. 2B, panel 2, lane 4), but failed to detect the expression of the GST tag in the absence of Gtf2 (Fig. 2B, panel 2, lane 3). It is possible that the presence of Gtf2 aids in presenting the appropriate conformation of GST suitable for immunodetection in S. parasanguinis. A further investigation of the application of the GST tag in S. parasanguinis is warranted. The Nano tag fusion protein, as noted for E. coli, was not detected in S. parasanguinis (Fig. 2A and B, panel 4, lanes 4 and 5), indicating the tag is not properly presented in either species. We conclude the Nano tag is not a good choice for protein tagging in S. parasanguinis.
Fig. 2.
Immunodetection of fusion constructs. (A) Western blot analyses of expression of fusion tags in E. coli. Lane 1, Top10; lane 2, Top10/pVPT-CTag; lane 3, Top10/pVPT-Gtf2-CTag; lane 4, Top10/pVPT-NTag-Gtf2. (B) Western blot analyses of expression of fusion tags in S. parasanguinis. Lane 1, wild type; lane 2, gtf2−; lane 3, gtf2−/pVPT-CTag; lane 4, gtf2−/pVPT-Gtf2-CTag; lane 5, gtf2−/pVPT-NTag-Gtf2. Arrows point to the position corresponding to tag or tagged Gtf2 fusion proteins.
The successful construction of these Gtf2-tag fusion proteins will guide us in selecting appropriate tags to construct other fusion proteins of interest to us. We will be able to determine protein subcellular localization and to isolate functional protein complexes involved in Fap1 glycosylation as well as other biological processes. Proteins implicated in Fap1 glycosylation, such as GalT1, GalT2, Gap1, Gap2, Gap3 and Gtf1, were chosen to fuse with GFP, T7 or the HSV tag. These constructs will aid in the verification of the utility of these tags for other fusion protein expression in S. parasanguinis. The fusion proteins, GalT1-T7, GalT2-HSV, GFP-Gap1, Gap1-GFP, Gap2-GFP, Gap2-T7, Gap3-GFP, Gap3-HSV and Gtf1-GFP were successfully expressed in both E. coli and S. parasanguinis (data not shown). Successful construction of pVPT-NTag and pVPT-CTag expression vectors for immunodetection of a variety of fusion proteins indicates that these constructs are useful and can be applied for general use of protein fusion in streptococci which will facilitate the investigation of protein-protein interactions in streptococci.
3.3. Genetic complementation of the gtf2 mutant using tagged Gtf2 constructs
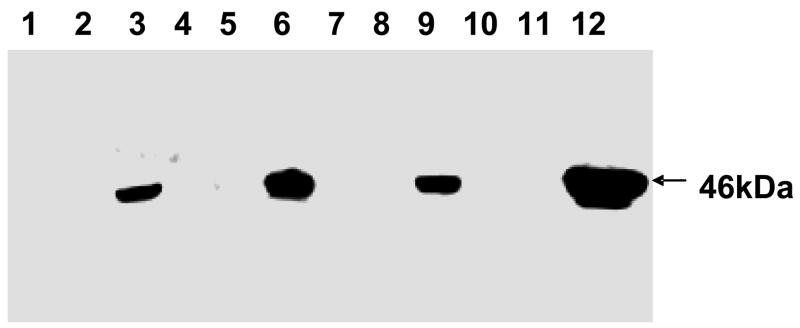
We previously determined Gtf2 is involved in Fap1 glycosylation as disruption of gtf2 renders the mutant unable to produce mature Fap1 (200kDa) that is detected by the Fap1 glycan-specific antibody MAb F51 (Elder and Fives-Taylor, 1986). The functionality of the tagged proteins and hence their biological relevance, was determined by genetic complementation experiments and examination of the ability of the Gtf2 fusion proteins to restore mature Fap1 production. The gtf2 mutant was transformed with the tagged Gtf2 constructs, and analyzed by Western blotting (Fig. 3). The S. parasanguinis wild type strain, fap1 mutant, gtf2 mutant and gtf2 mutant harboring pVPT-CTag were used as controls. The wild type bacteria express mature 200kDa Fap1 (lane 1), the fap1 mutant, gtf2 mutant and gtf2 mutant harboring pVPT-CTag failed to express mature Fap1 (lanes 2–4), all the tagged Gtf2 fusion proteins were able to restore the expression of mature Fap1 (lanes 5 and 6) except for the Nano tag placed at the N-terminus of Gtf2 (panel 4, lane 6). These results demonstrated that the tagged constructs are biologically relevant to study protein complexes involved in Fap1 glycosylation.
Fig. 3.
Genetic complementation of the gtf2 mutant using Gtf2 with different fusion tags. Lane 1, wild type; lane 2, fap1−; lane 3, gtf2−; lane 4, gtf2−/pVPT-CTag; lane 5, gtf2−/pVPT-Gtf2-CTag; lane 6, gtf2−/pVPT-NTag-Gtf2. Arrows point to the position corresponding to mature Fap1.
3.4. Immunodetection of T7-tagged Gtf2 in other streptococcal species
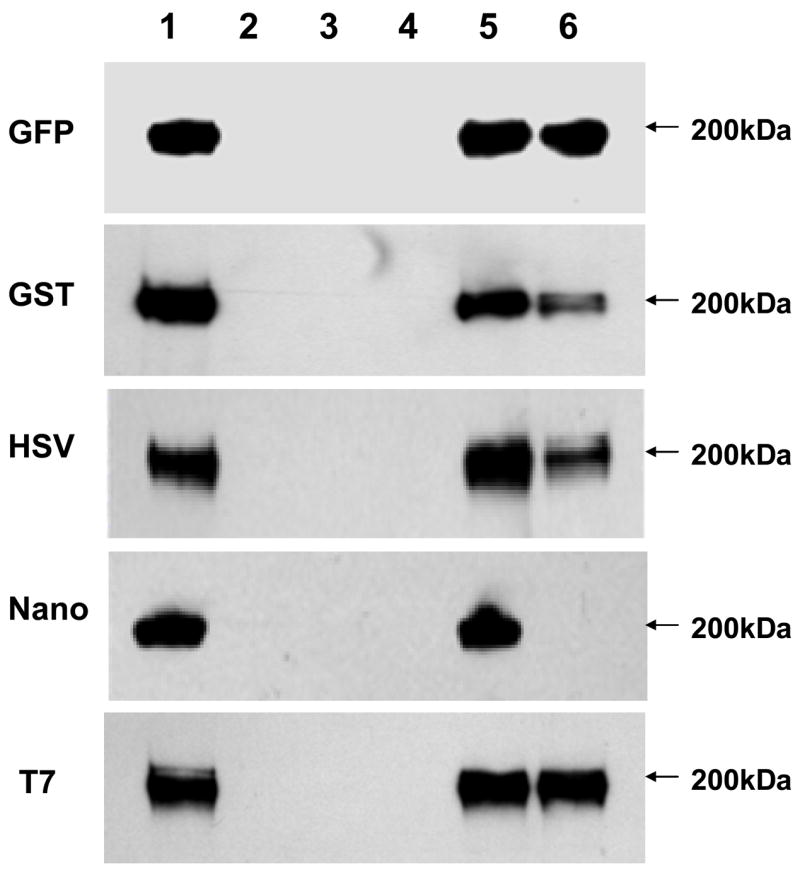
Since the expression of different fusion tags is readily detected in S. parasanguinis and the fusion constructs we generated contain a universal streptococcal replicon (Macrina et al., 1982; Chen et al., 2006), we hypothesize that the expression of these tags would be readily detectable in other streptococcal species. We transformed the plasmid pVPT-Gtf2-CT7 into some representative streptococcal strains as an example of the ability to detect fusion tag expression in other streptococcal species (Fig. 4). We used the S. parasanguinis gtf2 mutant, S. mutans UA159, S. pneumoniae V047 and S. sanguinis SK36, and their transformants harboring pVPT-CT7 as negative controls. No signal was found in the negative controls (lanes 1, 2, 4, 5, 7, 8, 10 and 11). The expression of T7-tagged Gtf2 (46kDa) in S. parasanguinis strain was used as a postitive control (lane 3). The expression of Gtf2-CT7 (46kDa) was also detected in S. mutans UA159, S. pneumoniae V047 and S. sanguinis SK36 transformed with pVPT-Gtf2-CT7 (lanes 6, 9 and 12), suggesting T7 was an excellent affinity tag for protein immunodetection in these streptococci. Interestingly, we found that the expression level of T7-tagged Gtf2 in S. sanguinis SK36 (lane 12) was significantly higher than that in S. parasanguinis (lane 3), S. mutans UA159 (lane 6) and S. pneumoniae V047 (lane 9), suggesting the shuttle vector pVA838, replicating in S. sanguinis, may have a higher copy number than when replicating in other streptococcal species. Successful detection of T7-tagged Gtf2 in other distinct streptococci demonstrated that the tag cassettes we constructed are useful for protein fusion expression in general and will facilitate the investigation of protein-protein interactions in gram-positive bacteria as specific antibodies for many of native proteins are generally not available.
Fig. 4.
Immunodetection of T7-tagged Gtf2 in other streptococcal species. Plasmid pVPT-Gtf2-CT7 was transformed into S. mutans UA159, S. pneumoniae V047 and S. sanguinis SK36, respectively. The expressions of T7-tagged Gtf2 with a C-terminus fusion were detected by Western blot analysis using anti-T7 monoclonal antibody. Lane 1, gtf2−; lane 2, gtf2−/pVPT-CT7; lane 3, gtf2−/pVPT-Gtf2-CT7; lane 4, UA159; lane 5, UA159/pVPT-CT7; lane 6, UA159/pVPT-Gtf2-CT7; lane 7, V047; lane 8, V047/pVPT-CT7; lane 9, V047/pVPT-Gtf2-CT7; lane 10, SK36; lane 11, SK36/pVPT-CT7; lane 12, SK36/pVPT-Gtf2-CT7. Arrows point to the position corresponding to T7-tagged Gtf2.
In summary, we have constructed two sets of expression plasmids pVPT-CTag and pVPT-NTag, which have five affinity tags that can be fused to either N- or C-termini of target proteins. We demonstrated the utility of the cassettes in detection of Gtf2 fusion proteins, and the biological relevance of the tag fused proteins in our working strain S. parasanguinis. We found GFP and T7 tag are excellent for Gtf2 immunodetection in both E. coli and S. parasanguinis. We demonstrated that the fusion cassette also was useful in detection of the Gtf2 fusion tag expression in other streptococci including S. mutans, S. pneumoniae and S. sanguinis. Therefore, the expression cassettes we constructed will be a useful tool to help investigate protein-protein interactions in Fap1 biogenesis in S. parasanguinis. Further, as pVA838 and its derivatives have been used as expression vectors in many Gram-positive bacteria including Streptococcus gordonii, Streptococcus bovis and Lactococcus lactis etc (Kubo et al., 1993; Ozkose et al., 2004; Gerber and Solioz, 2007), we envision our tag cassettes will also have application in studying protein function in those and other gram-positive bacteria.
Acknowledgments
We thank Dr. Susan Hollingshdead for providing the clinical isolate, S. pneumoniae V047. This study was supported by R01DE11000, R01DE017954 and K22 DE014726 from the National Institute of Dental and Craniofacial Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ajdiæ D, McShan WM, McLaughlin RE, Saviæ G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A. 2002;99:14434–9. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnau J, Lauritzen C, Petersen GE, Pedersen J. Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein Expr Purif. 2006;48:1–13. doi: 10.1016/j.pep.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Biedendieck R, Yang Y, Deckwer WD, Malten M, Jahn D. Plasmid system for the intracellular production and purification of affinity-tagged proteins in Bacillus megaterium. Biotechnol Bioeng. 2007;96:525–37. doi: 10.1002/bit.21145. [DOI] [PubMed] [Google Scholar]
- Bricker AL, Camilli A. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol Lett. 1999;172:131–5. doi: 10.1111/j.1574-6968.1999.tb13460.x. [DOI] [PubMed] [Google Scholar]
- Carlsson J, Grahnén H, Jonsson G, Wikner S. Establishment of Streptococcus sanguis in the mouths of infants. Arch Oral Biol. 1970;15:1143–8. doi: 10.1016/0003-9969(70)90005-1. [DOI] [PubMed] [Google Scholar]
- Chen Q, Wu H, Kumar R, Peng Z, Fives-Taylor PM. SecA2 is distinct from SecA in immunogenic specificity, subcellular distribution and requirement for membrane anchoring in Streptococcus parasanguinis. FEMS Microbiol Lett. 2006;264:174–81. doi: 10.1111/j.1574-6968.2006.00455.x. [DOI] [PubMed] [Google Scholar]
- Cole RM, Calandra GB, Huff E, Nugent KM. Attributes of potential utility in differentiating among “group H” streptococci or Streptococcus sanguis. J Dent Res. 1976;55:A142–A153. doi: 10.1177/002203457605500106011. [DOI] [PubMed] [Google Scholar]
- Correia FF, McKay TL, Farrow MF, Rosan B, DiRienzo JM. Natural transformation of Streptococcus crista. FEMS Microbiol Lett. 1996;143:13–8. doi: 10.1111/j.1574-6968.1996.tb08454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Sanford JR, Cáceres JF. A rapid and efficient protocol to purify biologically active recombinant proteins from mammalian cells. Protein Expr Purif. 2005;42:54–8. doi: 10.1016/j.pep.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Deng DM, Liu MJ, ten Cate JM, Crielaard W. The VicRK system of Streptococcus mutans responds to oxidative stress. J Dent Res. 2007;8:606–10. doi: 10.1177/154405910708600705. [DOI] [PubMed] [Google Scholar]
- Elder BL, Fives-Taylor P. Characterization of monoclonal antibodies specific for adhesion: isolation of an adhesin of Streptococcus sanguis FW213. Infect Immun. 1986;54(2):421–7. doi: 10.1128/iai.54.2.421-427.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno JC, Shaikh A, Fives-Taylor P. Characterization of allelic replacement in Streptococcus parasanguinis: transformation and homologous recombination in a “nontransformable” streptococcus. Gene. 1993;130:81–90. doi: 10.1016/0378-1119(93)90349-8. [DOI] [PubMed] [Google Scholar]
- Froeliger EH, Fives-Taylor P. Streptococcus parasanguinis fimbriae-associated adhesin Fap1 is required for biofilm formation. Infect Immun. 2001;69:2512–9. doi: 10.1128/IAI.69.4.2512-2519.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S, Ike Y. pAM401-based shuttle vectors that enable overexpression of promoterless genes and one-step purification of tag fusion proteins directly from Enterococcus faecalis. Appl Environ Microbiol. 2001;67:1262–7. doi: 10.1128/AEM.67.3.1262-1267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy MC, Guyard C, Quatannens B, Pavan S, Lange M, Mercenier A. Use of green fluorescent protein to tag lactic acid bacterium strains under development as live vaccine vectors. Appl Environ Microbiol. 2000;J66:383–91. doi: 10.1128/aem.66.1.383-391.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber SD, Solioz M. Efficient transformation of Lactococcus lactis IL1403 and generation of knock-out mutants by homologous recombination. J Basic Microbiol. 2007;47:281–6. doi: 10.1002/jobm.200610297. [DOI] [PubMed] [Google Scholar]
- Hearn MT, Acosta D. Applications of novel affinity cassette methods: use of peptide fusion handles for the purification of recombinant proteins. J Mol Recognit. 2001;14:323–69. doi: 10.1002/jmr.555. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF, Lamont RJ. Streptococcal adhesion and colonization. Crit Rev Oral Biol. 1997;8:175–200. doi: 10.1177/10454411970080020601. [DOI] [PubMed] [Google Scholar]
- Kubo S, Kubota H, Ohnishi Y, Morita T, Matsuya T. Matsushiro A. Expression and secretion of an Arthrobacter dextranase in the oral bacterium Streptococcus gordonii. Infect Immun. 1993;61:4375–81. doi: 10.1128/iai.61.10.4375-4381.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumada Y, Zhao C, Ishimura R, Imanaka H, Imamura K, Nakanishi K. Protein-protein interaction analysis using an affinity peptide tag and hydrophilic polystyrene plate. J Biotechnol. 2007;128:354–61. doi: 10.1016/j.jbiotec.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Lamla T, Erdmann VA. The Nano-tag, a streptavidin-binding peptide for the purification and detection of recombinant proteins. Protein Expr Purif. 2004;33:39–47. doi: 10.1016/j.pep.2003.08.014. [DOI] [PubMed] [Google Scholar]
- LeBlanc DJ, Lee LN, Abu-Al-Jaibat A. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid. 1992;28:130–45. doi: 10.1016/0147-619x(92)90044-b. [DOI] [PubMed] [Google Scholar]
- Lichty JJ, Malecki JL, Agnew HD, Michelson-Horowitz DJ, Tan S. Comparison of affinity tags for protein purification. Protein Expr Purif. 2005;41:98–105. doi: 10.1016/j.pep.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Loomis WP, Moseley SL. Translational control of mRNA processing in the F1845 fimbrial operon of Escherichia coli. Mol Microbiol. 1998;30:843–53. doi: 10.1046/j.1365-2958.1998.01117.x. [DOI] [PubMed] [Google Scholar]
- Macrina FL, Tobian JA, Jones KR, Evans RP, Clewell DB. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene. 1982;19:345–53. doi: 10.1016/0378-1119(82)90025-7. [DOI] [PubMed] [Google Scholar]
- Marra A, Asundi J, Bartilson M, Lawson S, Fang F, Christine J, Wiesner C, Brigham D, Schneider WP, Hromockyj AE. Differential fluorescence induction analysis of Streptococcus pneumoniae identifies genes involved in pathogenesis. Infect Immun. 2002;70:1422–33. doi: 10.1128/IAI.70.3.1422-1433.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh PD. The role of microbiology in models of dental caries. Adv Dent Res. 1995;9:244–269. doi: 10.1177/08959374950090030901. [DOI] [PubMed] [Google Scholar]
- Myscofski DM, Dutton EK, Bolken TC, Franke CA, Hruby DE. Expression and purification of histidine-tagged proteins from the gram-positive Streptococcus gordonii SPEX system. Protein Expr Purif. 2000;20:112–23. doi: 10.1006/prep.2000.1275. [DOI] [PubMed] [Google Scholar]
- Nieto C, Fernández de Palencia P, López P, Espinosa M. Construction of a tightly regulated plasmid vector for Streptococcus pneumoniae: controlled expression of the green fluorescent protein. Plasmid. 2000;43:205–13. doi: 10.1006/plas.2000.1465. [DOI] [PubMed] [Google Scholar]
- Ozkose E, Akyol I, Ekinci MS. Molecular study on cloned endoglucanase gene from rumen bacterium. J Mol Microbiol Biotechnol. 2004;8:111–6. doi: 10.1159/000084566. [DOI] [PubMed] [Google Scholar]
- Perry D, Kuramitsu HK. Genetic transformation of Streptococcus mutans. Infect Immun. 1981;32:1295–1297. doi: 10.1128/iai.32.3.1295-1297.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Smyth DR, Mrozkiewicz MK, McGrath WJ, Listwan P, Kobe B. Crystal structures of fusion proteins with large-affinity tags. Protein Sci. 2003;12:1313–22. doi: 10.1110/ps.0243403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spickofsky N, Margolskee RF. A highly efficient directional cDNA cloning method utilizing an asymmetrically tailed linker-primer plasmid. Nucleic Acids Res. 1991;19:7105–11. doi: 10.1093/nar/19.25.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2003;60:523–33. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- Varughese M, Chi A, Teixeira AV, Nicholls PJ, Keith JM, Leppla SH. Internalization of a Bacillus anthracis protective antigen c-Myc fusion protein mediated by cell surface anti-c-Myc antibodies. Molecular Medicine. 1998;4:87–95. [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mintz KP, Ladha M, Fives-Taylor PM. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguinis FW213. Mol Microbiol. 1998;28:487–500. doi: 10.1046/j.1365-2958.1998.00805.x. [DOI] [PubMed] [Google Scholar]
- Wu H, Bu S, Newell P, Chen Q, Fives-Taylor P. Two gene determinants are differentially involved in the biogenesis of Fap1 precursors in Streptococcus parasanguinis. J Bacteriol. 2007;189:1390–8. doi: 10.1128/JB.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Alves JM, Kitten T, Brown A, Chen Z, Ozaki LS, Manque P, Ge X, Serrano MG, Puiu D, Hendricks S, Wang Y, Chaplin MD, Akan D, Paik S, Peterson DL, Macrina FL, Buck GA. Genome of the opportunistic pathogen Streptococcus sanguinis. J Bacteriol. 2007;189:3166–75. doi: 10.1128/JB.01808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]