Abstract
We previously reported that a basic region/leucine zipper (bZIP) protein, a hybrid of the GCN4 basic region and C/EBP leucine zipper, not only recognizes cognate target sites AP-1 (5′-TGACTCA-3′) and cAMP-response element (CRE) (5′-TGACGTCA-3′) but also binds selectively to noncognate DNA sites: C/EBP (CCAAT/enhancer binding protein, 5′-TTGCGCAA), XRE1 (xenobiotic response element, 5′-TTGCGTGA), HRE (HIF response element, 5′-GCACGTAG), and E-box (5′-CACGTG). In this work, we used electrophoretic mobility shift assay (EMSA) and circular dichroism (CD) for more extensive characterization of the binding of wt bZIP dimer to noncognate sites as well as full- and half-site derivatives, and we examined changes in flanking sequences. Quantitative EMSA titrations were used to measure dissociation constants of this hybrid, wt bZIP, to DNA duplexes: Full-site binding affinities gradually decrease from cognate sites AP-1 and CRE with Kd values of 13 and 12 nM, respectively, to noncognate sites with Kd values of 120 nM to low μM. DNA-binding selectivity at half sites is maintained; however, half-site binding affinities sharply decrease from the cognate half site (Kd = 84 nM) to noncognate half sites (all Kd values > 2 μM). CD shows that comparable levels of α-helical structure are induced in wt bZIP upon binding to cognate AP-1 or noncognate sites. Thus, noncognate sites may contribute to preorganization of stable protein structure before binding target DNA sites. This work demonstrates that the bZIP scaffold may be a powerful tool in the design of small, α-helical proteins with desired DNA recognition properties.
In order to examine the relationship between protein structure and DNA-binding function, we exploit the protein α-helix, a structure used ubiquitously for sequence-selective DNA recognition and one that chemists have successfully manipulated in design and synthesis studies for many years (examples include refs 1-8). GCN4 is a dimeric transcriptional regulator that governs histidine biosynthesis in yeast under conditions of amino acid starvation (9). Crystal structures of the basic region/leucine zipper (bZIP)1 domain of GCN4 bound to the AP-1 site (5′-TGACTCA) (10) and cAMP-response element (CRE) site (5′-TGACGTCA) (11, 12) and the Jun-Fos bZIP heterodimer bound to AP-1 (13) show that a continuous α-helix provides the basic region interface for binding to specific DNA sites, as well as the leucine zipper coiled coil dimerization structure. The full-length GCN4 monomer is 281 amino acids, and the bZIP comprises a dimer of ∼60 residue monomers. We previously generated a series of minimalist bZIP proteins comprising alanine-rich variants of the GCN4 basic region fused to the C/EBP leucine zipper: The wt bZIP comprises the native GCN4 basic region. McKnight and co-workers showed that exchanging basic regions and leucine zippers between GCN4 and C/EBP resulted in functional proteins that targeted cognate DNA sites (14), and likewise, we found our wt bZIP hybrid to be functionally equivalent to the GCN4 bZIP (15, 16).
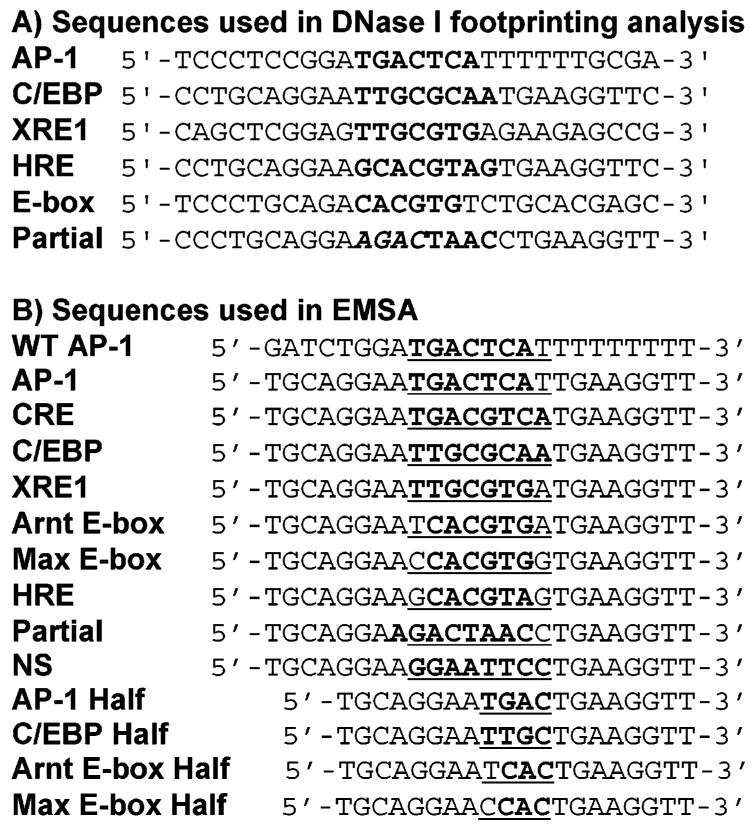
We previously showed that wt bZIP dimer binds strongly and selectively to the full sites of noncognate gene-regulatory sequences (17). In the current work, quantitative electrophoretic mobility shift assay (EMSA) and circular dichroism (CD) were used for more extensive characterization of the binding of wt bZIP dimer to noncognate half sites as well as full sites (Figure 1). These noncognate gene-regulatory sequences include the C/EBP consensus sequence (CCAAT/enhancer binding protein, 5′-TTGCGCAA), XRE1 (xenobiotic response element, 5′-TTGCGTGA), HRE [hypoxia inducible factor (HIF) response element, 5′-GCACGTAG], and canonical enhancer box (E-box) (5′-CACGTG).
FIGURE 1.
(A) Sequences of all of the DNA sites used in DNase I footprinting experiments. All sequences are duplexes. Core target sequences are in bold. The Partial site actually contains a weak half site, 5′-AGAC-3′, which is italicized, and the full Partial site is in bold. The flanking sequences surrounding the AP-1 and XRE1 target sites are from the his3 and CYP1A1 promoter regions, respectively. Flanking sequences surrounding C/EBP, HRE, and the Partial site are the same as those for EMSA. For E-box, the flanking sequences are the same as those used by Agre et al. (14). (B) Sequences of DNA sites used in EMSA experiments. All sequences are 24-mer duplexes for full sites and 20-mer duplexes for half sites. The core target sequences are in bold; the inserts between flanking sequences are underlined. The flanking sequences are identical for all duplexes, except that the flanking sequences of WT AP-1 are from the his3 gene from yeast. Note that for the Partial site, the actual target is shifted by one bp. The NS sequence is a nonspecific DNA control. The two thymines at the 3′-end of each duplex were P32-labeled.
Like GCN4, C/EBP is a bZIP transcription factor. However, the C/EBP family is broadly distributed and involved in regulation of a variety of functions, whereas GCN4 is native to yeast and regulates histidine biosynthesis. The other regulatory sites are targeted by the basic/helix-loop-helix (bHLH) transcription factor family. Although the bZIP and bHLH both use a dimer of basic α-helices to target specific DNA sites, they are distinctly different families of transcription factors. Within the bHLH are subfamily variants including the basic/helix-loop-helix/zipper (bHLHZ), wherein a leucine zipper contiguous to the bHLH is part of the dimerization domain; bHLHZ proteins includes Max, Mad, and Myc, which target the E-box sequence 5′-CACGTG (18, 19). This transcription factor network comprises widely expressed proteins critical for control of normal cell proliferation and differentiation (20, 21); myc genes are suspected of being among the most frequently affected in human tumors and disease (22).
Another bHLH subfamily is the bHLH/PAS, wherein the PAS domain assists in efficient protein dimerization, such as in aryl hydrocarbon receptor (AhR), aryl hydrocarbon receptor nuclear translocator (Arnt), and HIF-1α (23). AhR (also called the dioxin receptor) (24, 25) and Arnt (26, 27) heterodimerize in the presence of ligands, including dioxins and polychlorinated biphenyls (PCBs); this activated complex then binds XREs and activates gene transcription (28-31). AhR and Arnt bind distinct, unrelated half sites in the XRE1 site, 5′-TTGC·GTG (dot separates half sites) (32). The HIF-1α/Arnt heterodimer targets HRE, which resides in enhancer regions of target genes involved in glycolysis, erythropoiesis, and angiogenesis (upregulation of vascular endothelial growth factor transcription) under the hypoxic conditions surrounding nascent tumor growth (33).
Quantitative EMSA titrations were used to examine the binding behavior of wt bZIP dimer with the cognate AP-1 half site, 5′-TGAC, and noncognate half sites (Figure 1):C/EBP half site (5′-TTGC), Arnt E-box half site (5′-TCAC), and Max E-box half site (5′-CCAC). We also examined different flanking sequences; for example, the core E-box is 5′-CACGTG; yet, Max and Myc prefer 5′-CCACGTGG, and Arnt prefers 5′-TCACGTGA (34). Quantitative EMSA shows that wt bZIP dimer still targets full sites AP-1 and CRE, and their respective cognate half site 5′-TGAC, with strongest affinities. Full-site binding affinities decrease gradually from AP-1 and CRE to the various noncognate sites. In contrast, half-site binding affinities decrease sharply: Binding drops by 3 orders of magnitude between the cognate 5′-TGAC half site and all noncognate half sites. CD shows that these noncognate sites can preorganize the partially folded bZIP into a more stable α-helical structure, inducing the same levels of helicity as does binding to cognate AP-1. Our results demonstrate that the α-helical bZIP molecular recognition scaffold can provide a useful platform for quantitative dissection of protein-DNA recognition and design of new proteins that target desired DNA sequences with tunable levels of binding affinity and sequence selectivity.
EXPERIMENTAL PROCEDURES
Materials
DNA oligonucleotides were purchased from Operon Biotechnologies. Enzymes were supplied by New England Biolabs. Reagents were supplied by Aldrich, Acros/Fisher, or Bioshop. Radioactive nucleotides were purchased from Amersham, and radioactivity was monitored by a Beckman LS 6500 scintillation counter. DNA sequencing was performed on an ABI 3730XL 96 capillary sequencer using a combination of the Big Dye and dGTP Big Dye Sequencing Kits (Applied Biosystems) at the DNA Sequencing Facility in the Centre for Applied Genomics, Hospital for Sick Children (Toronto, ON). Electrospray ionization mass spectrometry (ESI-MS) was performed on a Micromass ZQ model MM1 Mass Spectrometer (Waters) at the University of Toronto at Mississauga.
Preparation of wt bZIP Proteins
The protocol for producing bacterially expressed 96-mer wt bZIP (expressed wt bZIP) has been described in detail previously (35). The following is a brief summary. The gene for expression of wt bZIP was assembled by mutually primed synthesis, amplified by PCR with terminal primers, and purified by nondenaturing polyacrylamide gel electrophoresis (PAGE). Duplex DNA was cloned into vector pTrcHis B (Invitrogen) for expression of proteins with an amino-terminal six-histidine tag for purification purposes. Recombinant plasmids were sequenced and transformed into Escherichia coli strain BL21(DE3) (Stratagene) by electroporation (Bio-Rad E. coli Gene Pulser). Bacterial expression of wt bZIP was performed in LB medium with 100 μg/mL ampicillin. Cells were harvested by centrifugation and lysed by sonication, followed by protein isolation using TALON cobalt metal-ion affinity resin (Clontech).
A chemically synthesized 57-residue peptide (synthetic wt bZIP) was obtained from Biomer Technology (Concord, CA) using standard Fmoc chemistry followed by cleavage and deprotection. Purification of both bacterially expressed and chemically synthesized wt bZIP has been described in detail (17). Protein purity was confirmed by high-performance liquid chromatography (HPLC, Beckman System Gold) on reversed-phase C18 columns (Vydac). ESI-MS analysis: expressed wt bZIP: calculated, 10941.0 g/mol; found, 10939.0 g/mol with the initiating Met at the N terminus of wt bZIP cleaved during post-translational modification (36); synthetic wt bZIP: calculated, 6822.9 g/mol; found, 6821.0 g/mol (see Supporting Information for sequences of expressed and synthetic wt bZIP).
DNase I Footprinting Analysis
Procedures for DNase I footprinting and EMSA were described in more detail previously (17). The 3′- and 5′-flanking sequences surrounding the AP-1 and XRE1 target sites are from the his3 and CYP1A1 promoter regions, respectively. Flanking sequences surrounding C/EBP, HRE, and the Partial site are the same as those for EMSA (discussed below). For E-box, the flanking sequences are the same as those used by Agre et al. in their work on C/EBP bZIP (14). For 5′-endlabeled footprinting, recombinant pUC19 plasmids with correct inserts were first linearized with Hind III and radiolabeled with [γ-32P]ATP, followed by Ssp I digestion resulting in ∼650 bp DNA duplexes (16). The expressed wt bZIP protein was added to reaction mixture containing TKMC buffer (20 mM Tris, pH 7.5, 4 mM KCl, 2 mM MgCl2,and 1 mM CaCl2), 100 μg/mL bovine serum albumin (BSA), 5 mM dithiothreitol (DTT), 1 μg/mL poly(dI-dC), and 5% glycerol.
Footprinting was performed with bacterially expressed wt bZIP, which is a 96-mer comprising the GCN4 basic region, residues 226-252; the C/EBP leucine zipper, residues 310-338; an extra 31 amino acids from the expression vector at the N terminus that include a six-His tag, plus a nine-residue GGCGGYYYY linker at the C terminus (35). Footprinting was also performed with chemically synthetic wt bZIP, which is a 57-mer comprising the GCN4 basic region, C/EBP leucine zipper, and a C-terminal tyrosine for spectroscopic evaluation.
32P-endlabeled DNA fragment was then added to the mixture, followed by incubation at 4 °C for 2 h, 37 °C for 1 h, and room temperature for 30 min. Footprinting reactions were digested by DNase I for 3 min at 22 °C. Increasing wt bZIP concentration may promote soluble aggregates with DNase I and/or nonspecific binding to DNA, which may shield DNA from DNase I cleavage. Therefore, to obtain clear footprints and hypersensitivity bands, final DNase I concentrations varied depending on the concentration of wt bZIP: 0.5 mg/mL DNase I for reactions containing no wt bZIP; 10 mg/mL DNase I for reactions containing 0.5 μM and 1 μM wt bZIP (monomeric concentration); and 50 mg/mL DNase I for reactions containing 3 μM wt bZIP (monomeric concentration). After termination of DNase I cleavage, reaction products were analyzed by PAGE. Gels were dried and autoradiographed using storage phosphor technology with a Molecular Dynamics Storm 840 PhosphorImager System and ImageQuant software (version 5.2).
EMSA
The annealed DNA duplexes were doubly labeled at the 3′ terminus with [α-32P]dTTP by DNA polymerase I, Klenow fragment. Solid wt bZIP was freshly dissolved to a monomeric concentration of 2 μM and sequentially diluted to required concentrations (0.05 nM to 1 μM monomeric concentrations) with EMSA buffer [20 mM Tris and 1 mM phosphate, pH 7.5, 5 mM NaCl, 4 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 1 mM ethylene diamine tetraacetic acid (EDTA), 100 μg/mL non-acetylated BSA, 2 μg/mL poly-(dI-dC), 200 mM guanidine-HCl, and 10% glycerol]. Protein solutions were boiled at 90 °C for 10 min and cooled to room temperature over 4 h, followed by the addition of 3′-endlabeled DNA duplexes (DNA final concentrations <15 pM) on ice and incubation at 4 °C overnight, 37 °C for 1 h, and room temperature for 30 min. Reactions were analyzed by 10% PAGE at 120 V for 90 min. Gels were dried and autoradiographed as above.
Determination of Kd Values
The volumes of the bands corresponding to free and bound DNA were quantified using Molecular Dynamics ImageQuant software (version 5.2). (The “volume of the band” is defined by Molecular Dynamics as the “overall intensity within the area of the band.”) All gels were run under equilibrium binding conditions. The bound DNA fraction (θapp) was calculated as the volume of the band corresponding to the bound DNA, divided by the sum of the volumes of the bands corresponding to free and bound DNA. The bound DNA fractions vs protein monomer concentrations were fit to eq 1, a modified two-state binding equation for determination of apparent dissociation constants, with KaleidaGraph software (version 3.6.4, Synergy Software). This binding model for bZIP-DNA complexation has been used previously by other groups for calculation of dissociation constants (37, 38).
| (1) |
where Kd corresponds to the apparent monomeric dissociation constant, [M] is the concentration of monomeric wt bZIP, θmin is the bound DNA fraction when no wt bZIP is present, and θmax is the bound DNA fraction when DNA binding is saturated. Only data sets fit to eq 1 with R values >0.970 are reported. Each dissociation constant was determined from the average of two independent data sets.
The net bound DNA fractions (Δθapp) are given as references of binding affinities, specifically in those cases where saturation protein binding was not achieved in EMSA titration experiments. The net bound DNA fractions (Δθapp) are the net increases of bound DNA fractions measured in the presence of protein at monomeric concentrations 1 μM or 200 nM from bound DNA fractions measured in the absence of protein. Each net bound DNA fraction was determined from the average of two independent data sets.
CD Spectroscopy
DNA oligonucleotides were the same as those used in EMSA (Figure 1) and annealed without additional purification. The annealed DNA duplexes were purified by Amicon Ultra-4 Centrifugal Filter (Millipore) followed by ethanol precipitation. Purity of DNA duplexes was monitored by ultraviolet/visible (UV/vis) absorbance (Beckman DU 640 spectrometer). Each duplex DNA stock solution (5 μM) was prepared in 12.5 mM potassium phosphate buffer (pH 7.0), 50 mM KCl; synthetic wt bZIP stock solution (10 μM) was prepared likewise. CD samples were freshly prepared from DNA and synthetic wt bZIP stock solutions to obtain final concentrations of 5 μM synthetic wt bZIP monomer and 2.5 μM DNA duplex. We tried different buffer conditions and used expressed wt bZIP; however, the conditions above gave the best results. We also tried a larger excess of duplex (10 μM) and did not observe any difference in helicities. After mixing, CD samples were incubated at 15 °C for 5 min. CD measurements were performed on an Aviv 215 spectrometer with a suprasil, 10 mm path length cell (Hellma). CD spectra were acquired between 175 and 300 nm at 0.2 nm increments and a sampling time of 0.2 s. The DNA CD spectrum was subtracted from each protein spectrum presented in Figure 4. Each spectrum was averaged five times, and curves were not subjected to smoothing. The protein helix content was calculated according to Chau and co-workers (39).
FIGURE 4.
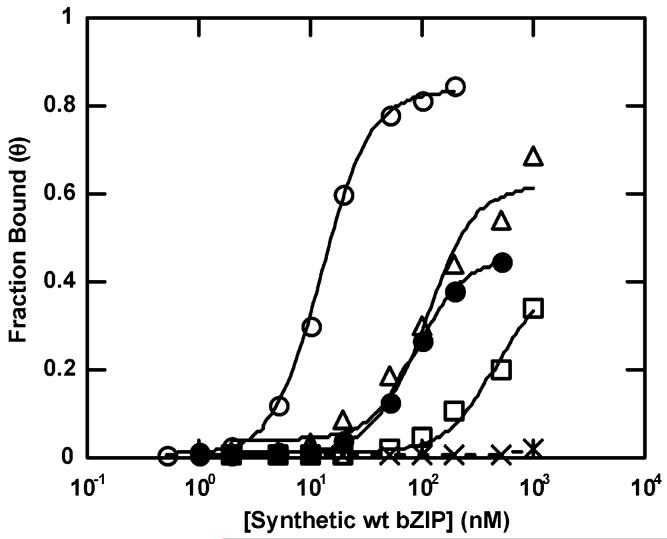
Representative equilibrium binding isotherms for synthetic wt bZIP binding to AP-1 (○), C/EBP (△), Arnt E-box (□), AP-1 half site (●), and C/EBP half site (×). Each isotherm was obtained from an individual EMSA experiment. Kd values shown in Table 1 are averaged from individual isotherms.
RESULTS
We present wt bZIP as being representative of bZIP α-helical structure and DNA-binding function. Although it is an engineered hybrid of the GCN4 basic region and C/EBP leucine zipper, its behavior is comparable to that of native GCN4 bZIP (15, 16); therefore, we extend our results with wt bZIP over the general class of bZIP proteins.
DNase I Footprinting Analysis
Figure 2 shows DNase I footprinting analysis of wt bZIP targeting AP-1, C/EBP, XRE1, E-box, HRE, and Partial site, performed with bacterially expressed wt bZIP. Footprinting was also performed with chemically synthetic wt bZIP. However, the footprints with bacterially expressed wt bZIP were more distinct than those with chemically synthetic wt bZIP. We suspect that the six-His tag that aids in protein purification is the cause of stronger footprints exhibited by expressed wt bZIP, as well as stronger DNA-binding affinities discussed below. The His tag is attached to the amino terminus of the basic region of expressed wt bZIP and has been found to promote aggregation (ref 40 provides an excellent review) and is therefore the likely source of experimental difficulties encountered maintaining protein solubility. Therefore, we suspect that while expressed wt bZIP binds to DNA target, the two His tags from the each arm of wt bZIP dimer can aggregate; such aggregation may be enhanced by metal ion chelation between His tags on each arm, for instance. Hence, the basic regions can form a “locked” dimer on the DNA binding site. Partial aggregation of His tags should not affect the selectivity of the basic region; however, it appears to enhance the ability of expressed wt bZIP to bind DNA targets with higher affinity and display more distinct footprints.
FIGURE 2.
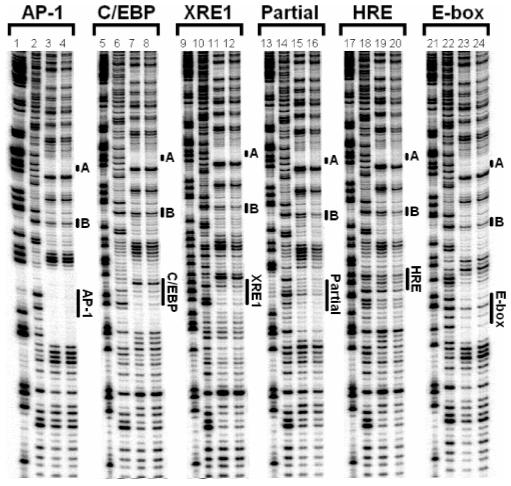
DNase I footprinting analysis on expressed wt bZIP bound to AP-1, C/EBP, XRE1, E-box, HRE, and Partial full sites, as well as cognate and E-box half sites. Data presented for 5′ 32P-endlabeled DNA (∼20000 cpm/lane). Lanes 1, 5, 9, 13, 17, and 21: chemical sequencing G reactions. Lanes 2, 6, 10, 14, 18, and 22: DNase I cleavage control reactions. Lanes 3, 7, 11, and 15: DNase I cleavage reactions with 0.5 μM wt bZIP. Lanes 19 and 23: DNase I cleavage reactions with 1 μM wt bZIP. Lanes 4, 8, 12, 16, 20, and 24: DNase I cleavage reactions with 3 μM wt bZIP. The positions of the corresponding DNA targets are indicated. A indicates footprints at cognate half site TGAC, and B indicates footprints at E-box half site CAC. The results were reproduced at least three times.
The footprint at the Partial site was unexpected, although its 5′-AGAC half site is similar to the native TGAC half site (17). Qualitatively, DNA binding at the canonical AP-1 site is strongest; expressed wt bZIP shows a clean, strong footprint at AP-1 at 0.5 μM protein concentration (Figure 2, lane 3). Among the noncognate sites examined by footprinting, expressed wt bZIP targets the C/EBP site the strongest; 0.5 μM wt bZIP shows a clear footprint at C/EBP target site (Figure 2, lane 7). Footprints of wt bZIP binding to XRE1 and the Partial site are somewhat weaker than to C/EBP; 0.5 μM wt bZIP still shows distinguishable footprints at these sites (Figure 2, lanes 11 and 15). Footprints at E-box and HRE are the weakest; 1-3 μM wt bZIP shows distinguishable, light footprints at these sites (Figure 2, lanes 19, 20, 23, and 24).
Figure 2 also shows DNase I footprints of wt bZIP dimer targeting the cognate (TGAC) and E-box (CAC) half sites (labeled A and B, respectively). An additional footprint coincides with an A tract that lies between these two half sites and appears to be an artifact. Hypersensitivity bands flank these footprints. Further up the footprinting lanes, more potential footprints and hypersensitivity can be detected, and these likely originate from the same reasons. Footprints and hypersensitivity at these sites are indicative of selective low-affinity protein-DNA complexes that can form under our low-stringency conditions (note the low salt concentrations in Experimental Procedures). Detailed discussion of these sites was previously published (17).
EMSA
Figure 3 shows EMSA of synthetic wt bZIP with all of the full and half sites listed in Figure 1 (see Supporting Information, Figure S2, for the same EMSA with expressed wt bZIP): The concentration of wt bZIP is 1.5 μM in all lanes containing protein. In the EMSA shown in Figures 3 and S2, binding to AP-1 and CRE is clearly the strongest, and we have previously characterized the interaction of wt bZIP to both AP-1 and CRE by footprinting and fluorescence anisotropy titration, which we used to show that the dissociation constant of expressed wt bZIP to AP-1 is 10 nM (15, 16). Relatively weaker binding is seen at C/EBP, followed by XRE1, Partial site, both E-boxes, and weakest binding at HRE. Both the synthetic and expressed versions of wt bZIP exhibit this same trend of decreasing binding affinities, and these qualitative EMSA results parallel the footprinting results. Binding is still selective at these non-cognate sites, for no binding is observed at the nonspecific DNA control site; note also that all duplexes used in EMSA contain the same flanking sequences with the target site embedded centrally, and no binding is observed in the flanking regions as well.
FIGURE 3.
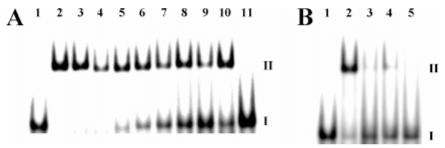
EMSA on synthetic wt bZIP bound to all full and half sites. Each lane contains ∼3000 cpm 32P-endlabeled DNA 24-mer duplex (full sites) or 20-mer duplex (half sites). I indicates free DNA; II indicates a band shift from dimeric wt bZIP complexation. Parts A and B represent different EMSA gels. (A) EMSA on synthetic wt bZIP bound to full sites. Lane 1, free AP-1 DNA; lanes 2-11, DNA in the presence of 1500 nM synthetic wt bZIP. DNA target sites used in lanes 2-11 are WT AP-1, AP-1, CRE, C/EBP, XRE1, Arnt E-box, Max E-box, HRE, Partial site, and nonspecific control, respectively. (B) EMSA on synthetic wt bZIP bound to half sites. Lane 1, free AP-1 half site DNA; lanes 2-5, DNA in the presence of 1500 nM synthetic wt bZIP. DNA target sites used in lanes 2-5 are the half sites of AP-1, C/EBP, Arnt E-box, and Max E-box, respectively.
Quantitative Measurement of Dissociation Constants at Full and Half Sites
Data presented are from EMSA with the synthetic wt bZIP, which is less prone to aggregation than the expressed version; however, some EMSA were performed on both the expressed and the synthetic versions of wt bZIP for comparison (Tables 1 and 2). Most of the quantitative EMSA titrations were performed with a synthetic version of wt bZIP. Additionally, when EMSA is run with the expressed wt bZIP, higher band shifts are seen, and these likely correspond to higher-order multimers or soluble aggregates of wt bZIP (Figure S2); no higher band shifts are observed with synthetic wt bZIP. These additional band shifts observed with expressed wt bZIP pose difficulty in accurate measurement and calculation of apparent monomeric dissociation constants, but we still present data obtained with expressed wt bZIP in Tables 1 and 2 for additional comparison of binding affinities. Thus, more straightforward and accurate quantitative EMSA with synthetic wt bZIP was pursued.
Table 1.
Dissociation Constants for Synthetic and Expressed wt bZIP Bound to DNA Sites
|
Kd (10-9 M)a |
||
|---|---|---|
| binding site | synthetic wt bZIP | expressed wt bZIPb |
| WT AP-1 | 13 ± 0.38 | |
| AP-1 | 13 ± 0.59e | 4.8 ± 0.12b |
| CRE | 12 ± 0.28 | |
| C/EBP | 120 ± 16e | 29 ± 3.6b |
| XRE1 | 240 ± 18e | |
| Arnt E-box | 570 ± 0.69c,e | |
| Max E-box | 840 ± 26c | |
| HRE | 1400 ± 160c,e | |
| Partial | 280 ± 49e | |
| AP-1 half | 84 ± 0.01 | 23 ± 8.7b |
| C/EBP half | >5000c | >1500c |
| Arnt E-box half | >4000c | |
| Max E-box Half | no activityd | |
| NS | no activityd | no activityd |
Average values of dissociation constants were obtained from two independent experiments; R values >0.97.
Higher-order bandshifts were not included in calculation of dissociation constants (see text and Supporting Information).
Saturation protein binding was not achieved in these titrations.
The concentration of wt bZIP ranged from 0 to 1000 nM.
From ref 17.
Table 2.
Net Bound DNA Fractions for Synthetic and Expressed wt bZIP Bound to DNA Sites
| synthetic wt bZIP |
expressed wt bZIPc |
|||
|---|---|---|---|---|
| binding site | Δθapp,200nMa | Δθapp,1μMb | Δθapp,200nMa | Δθapp,1μMb |
| WT AP-1 | saturationd | saturationd | ||
| AP-1 | saturationd | saturationd | saturationc,d | saturationc,d |
| CRE | saturationd | saturationd | ||
| C/EBP | 0.37 ± 0.081 | saturationd | saturationc,d | saturationc,d |
| XRE1 | 0.20 ± 0.015 | saturationd | ||
| Arnt E-box | 0.10 ± 0.00047 | 0.36 ± 0.031 | ||
| Max E-box | 0.036 ± 0.0023 | 0.28 ± 0.008 | ||
| HRE | 0.028 ± 0.0052 | 0.15 ± 0.029 | ||
| Partial | 0.16 ± 0.026 | saturationd | ||
| AP-1 half | 0.39 ± 0.016 | saturationd | saturationc,d | saturationc,d |
| C/EBP half | no activity | 0.015 ± 0.005 | no activity | 0.079 ± 0.026c,b |
| Arnt E-box half | no activity | 0.021 ± 0.0002 | ||
| Max E-box half | no activity | no activity | ||
| NS | no activity | no activity | no activity | no activity |
Average values of net bound DNA fractions were obtained from two independent experiments (from same experiments presented in Table 1); wt bZIP at 200 nM protein monomer concentration.
Average values of net bound DNA fractions were obtained from two independent experiments (from same experiments presented in Table 1); wt bZIP at 1 μM protein monomer concentration.
Higher-order band shifts were not included in calculation of net bound DNA fractions (see text and Supporting Information).
Saturation protein binding was achieved at 1 μM protein concentration.
Kd values reveal protein-DNA binding at full and half sites in the nanomolar to micromolar range, although some sites exhibited binding too weak for accurate measurement, as indicated in Table 1. In these cases, net bound DNA fractions at 1 μM and 200 nM monomeric protein concentrations are provided in Table 2. Figure 4 shows representative binding isotherms and graphically demonstrates that binding affinities for weak binders cannot be accurately measured. No binding was observed at the nonspecific control duplex or flanking sequences.
As measured by EMSA, wt bZIP dimer binds to AP-1 and CRE with very similar affinities as determined previously by fluorescence anisotropy titrations (16). We note that those Kd values obtained with expressed wt bZIP generally show tighter binding affinities than those with synthetic wt bZIP (Table 1), although the values are very close and reproducible. This observation is in agreement with the footprinting results. In the case of AP-1, we measured binding with two different flanking sequences: The “wild-type” flanking sequence (referred to as duplex WT AP-1 in Figure 1) is that flanking the AP-1 site in the his3 promoter of the yeast genome (9), whereas we also measured AP-1 binding with the same arbitrarily chosen flanking sequence used in all other duplexes. We observe no difference in binding affinities due to changes in sequences flanking AP-1.
AP-1 and CRE are the tightest binding sites to synthetic wt bZIP at Kd = 12 nM; their half site, TGAC, is bound at Kd = 84 nM, which is a decrease in binding of 7-fold. This is in agreement with measurements by Hollenbeck and Oakley, who measured a 10-fold decrease in half-site binding (38). In related studies, other labs have found that monomeric and dimeric GCN4 bZIP bind similarly and strongly to their target half and full sites (41-43). The C/EBP full site is bound by wt bZIP dimer at Kd = 120 nM, which is notably strong binding at a noncognate full site and only 10-fold decreased from native AP-1 and CRE full sites. However, half-site binding of wt bZIP dimer at noncognate C/EBP drops sharply, by around 2-3 orders of magnitude, from that at the C/EBP full site: This result contrasts markedly with the modest 5-10-fold drop in binding between the canonical full and half sites. We were surprised to find that the C/EBP half site shows very weak binding with Kd > 5μM, which is >50 times weaker than binding at the full site, given that the full site is bound well.
We observe a similar pattern of very weak or no half-site binding at the noncognate sequences. The full sites XRE1, Max, and Arnt E-boxes, and Partial site exhibit binding of wt bZIP dimer at 240 nM, >500 nM (both E-boxes), and 280 nM, respectively. These noncognate full sites are bound reasonably well; yet, their half sites are very weakly bound in the micromolar range; binding affinities could not be accurately obtained for any of the noncognate half sites (Table 1); therefore, binding fractions are presented in Table 2. This extremely weak binding of noncognate half sites contrasts with the strong binding at the cognate AP-1 and CRE half site, TGAC.
CD Shows Induction of Helicity at Noncognate Sites
Nonspecific DNA can serve to preorganize α-helical protein structure in the bZIP: That is, the disordered or partially ordered DNA-binding basic region can be more fully ordered into the appropriate α-helical structure capable of binding the target DNA site by preorganizing first on nonspecific DNA. We therefore used CD to measure the induction of α-helicity in synthetic wt bZIP upon addition of short DNA duplexes containing target full sites: AP-1, C/EBP, Arnt E-box, and nonspecific DNA control (Figure 5). The duplexes are the same as those used in EMSA, and all have the same flanking sequences (Figure 1).
FIGURE 5.
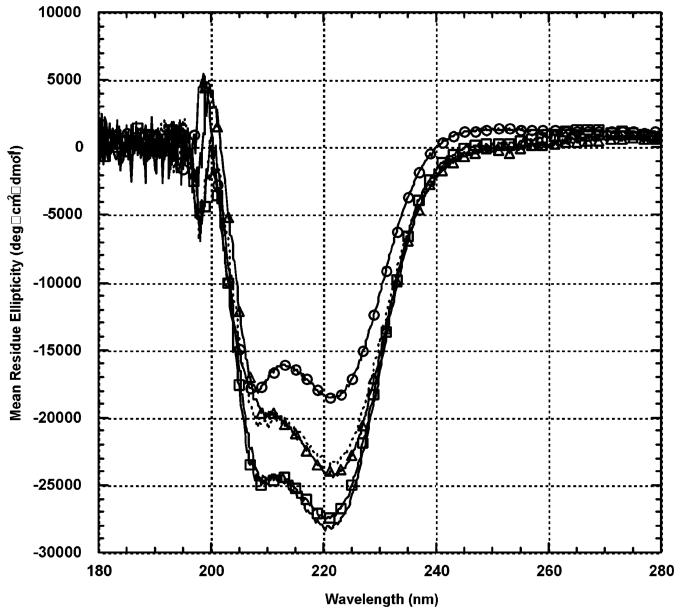
CD on synthetic wt bZIP in the presence of DNA duplexes: synthetic wt bZIP control (○), C/EBP (△), Arnt E-box (□), nonspecific control (···), and AP-1 (-). Protein dimer was placed in 200 μL of 12.5 mM potassium phosphate buffer (pH 7.0), 50 mM KCl, such that final concentrations of protein dimer and DNA duplex were 2.5 μM each. Samples were equilibrated for 5 min at 15 °C prior to measurement. Each spectrum was averaged five times, and curves were not subjected to smoothing. The DNA control spectrum (including buffer) was subtracted from each protein-DNA spectrum.
Figure 5 shows the CD difference spectra that were obtained by subtracting the corresponding DNA control spectrum (including buffer) from the synthetic wt bZIP-DNA complex spectrum; CD spectra for the different DNA duplexes confirmed their structures to be B-form DNA and virtually identical. For an ideal α-helix, the mean residue ellipticity was calculated to be -37700 deg cm2 dmol-1 (39). According to this ideal value, synthetic wt bZIP in the absence of DNA is 49% helical as measured at 222 nm. Upon addition of duplex DNA (1:1 ratio, protein dimer:DNA duplex), helicity of synthetic wt bZIP increases to approximately 60-75%: the strongly bound AP-1 (74%), reasonably bound C/EBP (65%), weakly bound Arnt E-box (73%), and unbound nonspecific DNA control (62%) all display significant induction of helicity. There were no major changes in helicities at 222 nm when the protein dimer:DNA duplex ratio was changed to 1:4 (data not shown).
For the selected sites, the increased helicities differed only modestly; yet, the thermodynamic binding affinities varied dramatically (Table 1): Synthetic wt bZIP binds to AP-1 approximately 10 times stronger than to C/EBP and binds to C/EBP almost five times stronger than to Arnt E-box. CD with the nonspecific DNA control, which shows no detectable binding by wt bZIP, shows significant induction of helicity, similar to that at C/EBP. Thus, under our conditions, we were intrigued to observe comparable induction of helical structure in wt bZIP regardless of DNA duplex sequence; such a result may indicate that preorganization of bZIP transcription factors on genomic DNA can be part of the mechanism for bZIP binding its target in vivo. Induction of helicity by nonspecific duplexes appears to be variable and dependent on the different conditions and sequences of protein and DNA used. For example, little or no induction of helicity in bZIP derivatives upon addition of nonspecific DNA has been observed in some cases (44, 45). However, Hollenbeck and Oakley found that their nonspecific DNA control induced comparable helicity in their GCN4 bZIP derivative as did the native AP-1 and CRE sites (38). Regardless of strength of thermodynamic binding affinity, all of the sites that we examined by CD can preorganize α-helical structure, and there appears to be no clear relationship between binding affinities and structural stabilization.
DISCUSSION
The bZIP domain of GCN4 has been found to be capable of targeting noncognate DNA sequences, including XRE1, C/EBP, E-box, and HRE. These results indicate that likely the GCN4 bZIP, as represented by our related wt bZIP, may also be capable of binding other DNA sequences. None of the noncognate sites are bound as tightly as the canonical AP-1 and palindromic CRE sites; yet, binding at noncognate sites still shows sequence selectivity and significant binding affinities.
We suggest that these noncognate sites can be considered weak, yet selective, sites, and likely such sites abound in the cell and may assist in protein prefolding before binding the targeted site buried within a genome. Although the various DNA duplexes examined by CD show comparable levels of induction of helicity, the contribution of noncognate sites to preordering and stabilization of protein structure is expected to be greater than that of truly nonspecific DNA: Protein may spend more time residing, and therefore prefolding, at noncognate sites, if protein-DNA complexation occurs under thermodynamic equilibrium conditions in the cell. This point may be more relevant from a protein design perspective or in vitro experiments under equilibrating conditions. The von Hippel group demonstrated that non-specific duplex DNA can serve as a template for ordering short Ala- and Lys-rich peptides into α-helices (46). Similarly, Padmanabhan et al. found that Ala-rich 17-mers containing four lysines assumed compact, helical structures upon binding DNA; the folding penalty was worth 1-2 orders of magnitude in binding affinities (47). In nuclear magnetic resonance (NMR) studies on the GCN4 bZIP/AP-1 complex, Palmer and co-workers found that although the GCN4 basic region is substantially helical, it is highly dynamic; restriction of the conformational space accessible to the basic region may significantly reduce the entropic cost associated with helix formation consequent to DNA binding (48).
Preorganization of α-helical conformations by nonspecific DNA can be thermodynamically advantageous for highly selective and rapid targeting of a particular DNA site within a genome. If the helix folds only upon recognition of target DNA, the binding energy diverted to folding will reduce selective binding affinity. Additionally, a flexible DNA-binding domain may be more capable of making unfavorable interactions at nonspecific sites; unfavorable interactions can be avoided with a rigid DNA-reading head (46). Thus, bZIP proteins may use binding at noncognate sites to assist in preorganizing stable, helical structure during the quest toward target DNA sites.
Binding at noncognate sequences of DNA in an organism’s genome may not be of significance if these affinities are too weak to be of consequence or if kinetics, rather than thermodynamics, contributes more significantly to cellular binding activity. We obtain binding affinities that are reduced by 10-fold, for example, in the case of the C/EBP full site; this is still a significant degree of sequence-selective binding affinity. These results may lead us to consider that GCN4, as well as other transcription factors, while finding its canonical target site, can spend a significant part of its time in nonproductive complexes along the DNA in vivo.
As depicted in Figure 6, GCN4 is capable of binding to specific DNA targets utilizing three different binding modes: dimeric binding observed when GCN4 binds its canonical full sites AP-1 and CRE, dimeric binding to its canonical DNA half site TGAC in which one monomer binds specifically to the canonical half site and the other monomer binds to DNA nonspecifically (38), and monomeric binding to a single canonical half site (41, 49). The wt bZIP is capable of selective dimeric binding to noncognate DNA full sites (Figure 6A) (17). In this work, we found that wt bZIP is also capable of selective dimeric binding to noncognate DNA half sites utilizing the binding mode depicted in Figure 6B, as shown by EMSA band shifts consistent with dimeric, not monomeric, protein binding to noncognate half sites (Figure 3B). For wt bZIP, we find strong and specific binding to the canonical half site (Kd = 84 nM), but binding to noncognate half sites is extremely weak. This discrimination occurring at the level of monomer binding to half site may be how GCN4 avoids being trapped in nonproductive protein-DNA complexes as it searches for its target site.
FIGURE 6.
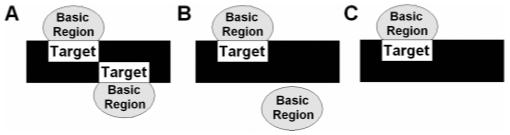
GCN4 bZIP-DNA binding schemes. (A) Dimeric binding of basic regions on DNA full site. Both basic regions of the dimer bind to target DNA half sites selectively. (B) Dimeric binding of basic regions on DNA half site. Only one basic region of the dimer binds selectively to the target DNA half site; the other basic region interacts nonspecifically with DNA. (C) Monomeric binding of basic regions on DNA half site; no protein dimerization occurs.
A mechanism for avoiding such nonproductive binding is for the bZIP monomer to bind using the monomer (sequential binding) pathway, in which monomer binds the targeted DNA half site first, followed by cooperative binding of the second monomer to generate dimer. In stopped-flow experiments, Bosshard and co-workers showed that dimeric bZIP had not even formed by the time DNA binding was observed (43, 49). Schepartz and co-workers showed that dimerization of the Jun-Fos bZIP is much slower than monomer binding to its DNA target, despite Jun-Fos heterodimerization occurring rapidly in the absence of DNA and being notably strong with a dimerization constant of 54 nM (42, 50). In these cases, kinetic data support the monomer over dimer pathway, in which the bZIP dimer forms first before binding to the full target site.
The monomeric binding pathway has been observed in other protein families. Sauer and co-workers found that the Arc repressor monomer’s folding is stimulated by templating nonspecifically with nucleic acid and polyanion polymers and then completing folding in this bound state. Similarly, nonspecific interactions with abundant cellular DNA and RNA can aid Arc folding and DNA association (51). Locker and co-workers examined dimeric homeodomain HNF1 (52). Strongly bound half sites averaged 6.3/7 bp matches, and some half sites matched only 1/7 bp. Two complications are that substitutions are dependent on other substitutions and sequence context: Even a weakly recognized DNA site is functional if adjacent factors, including those in the transcriptional activation complex, stabilize the HNF1-DNA complex (52).
A number of proposals describe how proteins can locate their specific DNA targets with rates at, or even, exceeding the rate of diffusion: These include facilitated diffusion, one-dimensional sliding, intersegmental transfer, short-range diffusive hopping, and processivity (see refs 53 and 54 for reviews). The monomeric binding pathway for protein recognition of DNA can be accommodated in all of these models for rapid, specific recognition of target sites. Our thermodynamic data also provide evidence for monomeric binding being critical for discrimination among target sites. Although noncognate full sites, including regulatory sequences C/EBP, XRE1, E-box, and HRE, can be targeted with sequence selectivity and significant binding affinities by the GCN4 bZIP, half-site binding varies dramatically: Only the cognate half site TGAC is bound strongly at low nanomolar affinity (Kd = 84 nM), whereas all other half sites show binding at least 3 orders of magnitude weaker.
This work demonstrates that the bZIP is a versatile and flexible protein motif for recognition of DNA, as it can also target noncognate sequences with high to low affinities. The bZIP naturally exists as a dimer, but monomeric binders can also be designed. Although we show that monomeric binding affinities are extremely weak aside from the native half site, the bZIP can serve as a platform for design of short α-helical monomers with fine-tuned binding specificities and affinities. Such monomers can also be homo- or heterodimerized, thereby further exploiting the utility of the bZIP platform for design. Because in vitro work is often performed under thermodynamic conditions and the experimental environment can be controlled, the bZIP may be an ideal platform for dissection of protein-DNA recognition, further manipulation, and design. The data presented here may be useful in design of new α-helical proteins capable of targeting specific DNA sites: Binding affinities and sequence selectivities can be tuned as desired, whether for use as a scientific tool or possible therapeutic.
ACKNOWLEDGMENT
We thank Alevtina Pavlenco for technical assistance.
Footnotes
We express gratitude for funding from the U.S. National Insitutes of Health (RO1GM069041), Canadian Foundation for Innovation/Ontario Innovation Trust (CFI/OIT), Premier’s Research Excellence Award (PREA), and the University of Toronto.
- bZIP
- basic region/leucine zipper
- bHLHZ
- basic/helix-loop-helix/zipper
- CRE
- cAMP-response element
- C/EBP
- CCAAT/enhancer binding protein
- XRE1
- xenobiotic response element
- HIF
- hypoxia inducible factor
- HRE
- HIF response element
- E-box
- enhancer box
- EMSA
- electrophoretic mobility shift assay
- CD
- circular dichroism
- bp
- base pair
- Kd
- apparent monomeric equilibrium dissociation constant
- AhR
- aryl hydrocarbon receptor
- Arnt
- aryl hydrocarbon receptor nuclear translocator
- PCB
- polychlorinated biphenyl
- ESI-MS
- electrospray ionization-mass spectrometry
- DTT
- dithiothreitol
- BSA
- bovine serum albumin
- EDTA
- ethylene diamine tetraacetic acid
- HPLC
- high-performance liquid chromatography
- UV/vis
- ultraviolet/visible
- PAGE
- polyacrylamide gel electrophoresis
- NMR
- nuclear magnetic resonance.
SUPPORTING INFORMATION AVAILABLE
Sequences of expressed and synthetic wt bZIP and additional representative EMSA (PDF). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Dawson PE, Kent SBH. Convenient total synthesis of a four-helix TASP molecule by chemoselective ligation. J. Am. Chem. Soc. 1993;115:7263–7266. [Google Scholar]
- 2.Grove A, Mutter M, Rivier JE, Montal M. Template-assembled synthetic proteins designed to adopt a globular, four-helix bundle conformation form ionic channels in lipid bilayers. J. Am. Chem. Soc. 1993;115:5919–5924. [Google Scholar]
- 3.Marqusee S, Baldwin RL. Helix stabilization by Glu-Lys salt bridges in short peptides of de novo design. Proc. Natl. Acad. Sci. U.S.A. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sasaki T, Kaiser E. Helichrome: Synthesis and enzymatic activity of a designed hemiprotein. J. Am. Chem. Soc. 1989;111 [Google Scholar]
- 5.Wharton RP, Ptashne M. Changing the binding specificity of a repressor by redesigning an a-helix. Nature. 1985;316:601–605. doi: 10.1038/316601a0. [DOI] [PubMed] [Google Scholar]
- 6.O’Neil KT, Hoess RH, DeGrado WF. Design of DNA-binding peptides based on the leucine zipper motif. Science. 1990;249:774–778. doi: 10.1126/science.2389143. [DOI] [PubMed] [Google Scholar]
- 7.Ghadiri MR, Soares C, Choi C. Design of an artificial four-helix bundle metalloprotein via a novel ruthenium (II)-assisted self-assembly process. J. Am. Chem. Soc. 1992;114:4000–4002. [Google Scholar]
- 8.Bishop P, Ghosh I, Jones C, Chmielewski J. Basic-helix-loop-helix region of Tal: Evaluation of structure and DNA affinity. J. Am. Chem. Soc. 1995;117:8283–8284. [Google Scholar]
- 9.Hill DE, Hope IA, Macke JP, Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: Requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986;234:451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- 10.Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted R helices: Crystal stucture of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 11.König P, Richmond TJ. The X-ray structure of the GCN4-bZIP bound to ATF/CREB site DNA shows the complex depends on DNA flexibility. J. Mol. Biol. 1993;233:139–154. doi: 10.1006/jmbi.1993.1490. [DOI] [PubMed] [Google Scholar]
- 12.Keller W, König P, Richmond TJ. Crystal structure of a bZIP/DNA complex at 2.2 Å: Determinants of DNA specific recognition. J. Mol. Biol. 1995;254:657–667. doi: 10.1006/jmbi.1995.0645. [DOI] [PubMed] [Google Scholar]
- 13.Glover JNM, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 14.Agre P, Johnson PF, McKnight SL. Cognate DNA binding specificity retained after leucine zipper exchange between GCN4 and C/EBP. Science. 1989;246:922–926. doi: 10.1126/science.2530632. [DOI] [PubMed] [Google Scholar]
- 15.Lajmi AR, Lovrencic ME, Wallace TR, Thomlinson RR, Shin JA. Minimalist, alanine-based, helical protein dimers bind to specific DNA sites. J. Am. Chem. Soc. 2000;122:5638–5639. [Google Scholar]
- 16.Bird GH, Lajmi AR, Shin JA. Sequence-specific recognition of DNA by hydrophobic, alanine-scanning mutants of the bZIP motif investigated by fluorescence anisotropy. Biopolymers. 2002;65:10–20. doi: 10.1002/bip.10205. [DOI] [PubMed] [Google Scholar]
- 17.Fedorova AV, Chan I, Shin JA. The GCN4 bZIP can bind to noncognate gene regulatory sequences. Biochim. Biophys. Acta. 2006;1764:1252–1259. doi: 10.1016/j.bbapap.2006.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair SK, Burley SK. X-ray structure of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 19.O’Hagan RC, Schreiber-Agus N, Chen K, David G, Engelman JA, Schwab R, Alland L, Thomson C, Ronning DR, Sacchettini JC, Meltzer P, DePinho RA. Gene-target recognition among members of the Myc superfamily and implications for oncogenesis. Nat. Genet. 2000;24:113–119. doi: 10.1038/72761. [DOI] [PubMed] [Google Scholar]
- 20.Amati B, Land H. Myc-Max-Mad: A transcription factor network controlling cell cycle progression, differentiation and death. Curr. Opin. Gene Dev. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 21.Orian A, van Steensel B, Delrow J, Bussemaker HJ, Li L, Sawado T, Williams E, Loo LWM, Cowley SM, Yost C, Pierce S, Edgar BA, Parkhurst SM, Eisenman RN. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nesbit CD, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 23.Gradin K, McGuire J, Wenger RH, Kvietikova I, Whitelaw ML, Toftgård R, Tora T, Gassmann M, Poellinger L. Functional interference between hypoxia and dioxin signal transduction pathways: Competition for recruitment of the Arnt transcription factor. Mol. Cell. Biol. 1996;16:5221–5231. doi: 10.1128/mcb.16.10.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burbach KM, Poland A, Bradfield CA. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc. Natl. Acad. Sci. U.S.A. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ema M, Sogawa K, Watanabe N, Chujoh Y, Matsushita N, Gotoh O, Funae Y, Fujii-Kuriyama Y. cDNA cloning and structure of mouse putative Ah receptor. Biochem. Biophys. Res. Commun. 1992;184:246–253. doi: 10.1016/0006-291x(92)91185-s. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman EC, Reyes H, Chu F, Sander F, Conley LH, Brooks BA, Hankinson O. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 1991;252:954. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 27.Reyes H, Reisz-Porszasz S, Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992;256:1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- 28.Cuthill S, Wilhelmsson A, Poellinger L. Role of the ligand in intracellular receptor function: Receptor affinity determines activation in vitro of the latent dioxin receptor to a DNA-binding form. Mol. Cell. Biol. 1991;11:401–411. doi: 10.1128/mcb.11.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitelaw M, Pongratz I, Wilhelmsson A, Gustafsson J-A, Poellinger L. Ligand-dependent recruitment of the Arnt coregulator determines DNA recognition of the dioxin receptor. Mol. Cell. Biol. 1993;13:2504–2514. doi: 10.1128/mcb.13.4.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu L, Whitlock JP. Mechanism of dioxin action: Receptor-enhancer interactions in intact cells. Nucleic Acids Res. 1993;21:119–125. doi: 10.1093/nar/21.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujisawa-Sehara A, Sogawa K, Yamane M, Fujii-Kuriyama Y. Characterization of xenobiotic responsive elements upstream from the drug-metabolizing cytochrome P-450c gene: A similarity to glucocorticoid regulatory elements. Nucleic Acids Res. 1987;15:4179–4191. doi: 10.1093/nar/15.10.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bacsi SG, Reisz-Porszasz S, Hankinson O. Orientation of the heterodimeric aryl hydrocarbon (dioxin) receptor complex on its asymmetric DNA recognition sequence. Mol. Pharmacol. 1995;47:432–438. [PubMed] [Google Scholar]
- 33.Lando D, Gorman JJ, Whitelaw ML, Peet DJ. Oxygen-dependent regulation of hypoxia-inducible factors by prolyl and asparaginyl hydroxylation. Eur. J. Biochem. 2003;270:781–790. doi: 10.1046/j.1432-1033.2003.03445.x. [DOI] [PubMed] [Google Scholar]
- 34.Swanson HI, Yang J-H. Specificity of DNA binding of the c-Myc/Max and ARNT/ARNT dimers at the CACGTG recognition site. Nucleic Acids Res. 1999;27:3205–3212. doi: 10.1093/nar/27.15.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lajmi AR, Wallace TR, Shin JA. Short, hydrophobic, alanine-based proteins based on the bZIP motif: Overcoming inclusion body formation and protein aggregation during overexpression, purification, and renaturation. Protein Expression Purif. 2000;18:394–403. doi: 10.1006/prep.2000.1209. [DOI] [PubMed] [Google Scholar]
- 36.Bird GH, Shin JA. MALDI-TOF mass spectrometry characterization of hydrophobic basic region/leucine zipper proteins. Biochim. Biophys. Acta. 2002;1597:252–259. doi: 10.1016/s0167-4838(02)00303-5. [DOI] [PubMed] [Google Scholar]
- 37.Metallo SJ, Schepartz A. Distribution of labor among bZIP segments in the control of DNA affinity and specificity. Chem. Biol. 1994;1:143–151. doi: 10.1016/1074-5521(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 38.Hollenbeck JJ, Oakley MG. GCN4 binds with high affinity to DNA sequences containing a single consensus half-site. Biochemistry. 2000;39:6380–6389. doi: 10.1021/bi992705n. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y-H, Yang JT, Chau KH. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 40.Hammarström M, Hellgren N, Van den Berg S, Berglund H, Härd T. Rapid screening for improved solubility to small human proteins produced as fusion proteins in Escherichia coli. Protein Sci. 2002;11:313–321. doi: 10.1110/ps.22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park C, Campbell JL, Goddard WA., III Can the monomer of the leucine zipper proteins recognize the dimer binding site without dimerization? J. Am. Chem. Soc. 1996;118:4235–4239. [Google Scholar]
- 42.Kohler JJ, Metallo SJ, Schneider TL, Schepartz A. DNA specificity enhanced by sequential binding of protein monomers. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11735–11739. doi: 10.1073/pnas.96.21.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cranz S, Berger C, Baici A, Jelesarov I, Bosshard HR. Monomeric and dimeric bZIP transcription factor GCN4 binds at the same rate to their target DNA site. Biochemistry. 2004;43:718–727. doi: 10.1021/bi0355793. [DOI] [PubMed] [Google Scholar]
- 44.O’Neil KT, Shuman JD, Ampe C, DeGrado WF. DNA-induced increase in the α-helical content of C/EBP and GCN4. Biochemistry. 1991;30:9030–9034. doi: 10.1021/bi00101a017. [DOI] [PubMed] [Google Scholar]
- 45.Weiss MA, Ellenberger TE, Wobbe CR, Lee JP, Harrison SC, Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990;347:575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- 46.Johnson NP, Lindstrom J, Baase WA, von Hippel PH. Double-stranded DNA templates can induce α-helical conformation in peptides containing lysine and alanine: Functional implications for leucine zipper and helix-loop-helix transcription factors. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4840–4844. doi: 10.1073/pnas.91.11.4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Padmanabhan S, Zhang W, Capp MW, Anderson CF, Record JMT. Binding of cationic (+4) alanine- and glycine-containing oligopeptides to double-stranded DNA: Thermodynamic analysis of effects of coulombic interactions and α-helix induction. Biochemistry. 1997;36:5193–5206. doi: 10.1021/bi962927a. [DOI] [PubMed] [Google Scholar]
- 48.Bracken C, Carr PA, Cavanagh J, Palmer AG., III Temperature dependence of intramolecular dynamics of the basic leucine zipper of GCN4: Implications for the entropy of association with DNA. J. Mol. Biol. 1999;285:2133–2146. doi: 10.1006/jmbi.1998.2429. [DOI] [PubMed] [Google Scholar]
- 49.Berger C, Piubelli L, Haditsch U, Bosshard HR. Diffusion-controlled DNA recognition by an unfolded, monomeric bZIP transcription factor. FEBS Lett. 1998;425:14–18. doi: 10.1016/s0014-5793(98)00156-2. [DOI] [PubMed] [Google Scholar]
- 50.Kohler JJ, Schepartz A. Kinetic studies of Fos-Jun-DNA complex formation: DNA binding prior to dimerization. Biochemistry. 2001;40:130–142. doi: 10.1021/bi001881p. [DOI] [PubMed] [Google Scholar]
- 51.Rentzeperis D, Jonsson T, Sauer RT. Acceleration of the refolding of Arc repressor by nucleic acids and other polyanions. Nat. Struct. Biol. 1999;6:569–573. doi: 10.1038/9353. [DOI] [PubMed] [Google Scholar]
- 52.Locker J, Ghosh D, Luc P-V, Zheng J. Definition and prediction of the full range of transcription factor binding sites-The hepatocyte nuclear factor 1 dimeric site. Nucleic Acids Res. 2002;30:3809–3817. doi: 10.1093/nar/gkf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Hippel PH, Berg OG. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 54.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]