Abstract
We have demonstrated previously that a wide array of stress signals induces O-GlcNAc transferase (OGT) expression and increases O-GlcNAcylation of many intracellular proteins, a response that is critical for cell survival. Here, we describe a mechanism by which glucose deprivation induces OGT expression and activity in Neuro-2a neuroblastoma cells. Glucose deprivation increases OGT mRNA and protein expression in an AMP-activated protein kinase-dependent manner, whereas OGT enzymatic activity is regulated in a p38 MAPK-dependent manner. OGT is not phosphorylated by p38, but rather it interacts directly with p38 through its C terminus; this interaction increases with p38 activation during glucose deprivation. Surprisingly, the catalytic activity of OGT, as measured toward peptide substrates, is not altered by glucose deprivation. Instead, p38 regulates OGT activity within the cell by recruiting it to specific targets, including neurofilament H. Neurofilament H is O-GlcNAcylated during glucose deprivation in a p38-dependent manner. Interestingly, neurofilament H solubility is increased by glucose deprivation in an O-GlcNAc-dependent manner, suggesting that O-GlcNAcylation of neurofilament H regulates its disassembly from filaments. Not only do these data help to reveal how OGT is regulated by stress, but these findings also describe a possible mechanism by which defective brain glucose metabolism, as found in aging and ischemia, may directly affect axonal structure.
During periods of ischemia, cells experience a severe depletion of extracellular glucose and oxygen, resulting in the activation of numerous stress signaling pathways regulating cell survival (1–4). The brain is extraordinarily sensitive to ischemic conditions, in part because of its large demand for glucose as an energy source (5). Upon ischemia, axonal damage is known to occur, eventually resulting in impairment of brain function (6, 7). Although excito-toxicity and oxidative stress have been shown to contribute to axon injury during ischemia (8), the mechanisms governing the loss of axon structure itself are not completely understood.
We have shown that many stresses increase protein O-GlcNAcylation and promote cell survival by increasing Hsp70 expression via O-GlcNAc transferase (OGT2; EC 2.4.1.94) expression and activity (9). However, the mechanisms by which protein O-GlcNAcylation is regulated under these and other conditions are unclear.
The modification of proteins in the cytosol and nucleus with the monosaccharide O-linked β-N-acetylglucosamine (O-GlcNAc) was described more than 20 years ago (for review see Ref. 10). Protein O-GlcNAcylation is an abundant and dynamic post-translational modification of serine and threonine residues that is essential for life in multicellular organisms (11, 12). Many O-GlcNAcylation sites also are (or adjacent to) known phosphorylation sites, illustrating a complex relationship between O-GlcNAcylation- and phosphorylation-mediated signaling (13–18). More than 500 proteins have now been identified as O-GlcNAcylated, including proteins involved in regulation of the cell cycle, transcription, trafficking, and signaling (19–21).
O-Linked β-N-acetylglucosaminidase (O-GlcNAcase; EC 3.2.1.52), which catalyzes the hydrolysis of O-GlcNAc from proteins, is a nucleocytoplasmic neutral hexosaminidase that also has a domain with high similarity to known acetyltransferases (22, 23). The O-GlcNAcase gene (MGEA5) resides at chromosome 10q24.1, a locus associated with neurodegenerative disease (24, 25). OGT, which catalyzes the addition of O-GlcNAc to proteins using UDP-GlcNAc as the donor nucleotide, resides on the X chromosome at Xq13, a locus also associated with neurological disorders (11, 26). We have demonstrated previously that OGT activity in vitro is responsive linearly to a large range of UDP-GlcNAc concentrations (27). In fact, the ability of OGT to recognize certain peptide substrates seems to be affected by [UDP-GlcNAc] (27). UDP-GlcNAc levels have been shown to change with glucose concentrations (28, 29), consistent with a role for O-GlcNAcylation in nutrient sensing and diabetes (20, 30).
Approximately 2–5% of the glucose that enters the cell is converted into UDP-GlcNAc de novo via the hexosamine biosynthetic pathway (31). Intracellular UDP-GlcNAc concentrations are typically in the 100 micromolar to low millimolar range, similar to ATP (29, 32). During conditions of low glucose, [ATP] and [UDP-GlcNAc] are known to decrease, whereas [AMP] is known to increase, leading to activation of the AMP-activated protein kinase (AMPK) pathway (33). Activation of AMPK has been shown previously to regulate cell survival by phosphorylating tuberin (TSC2), leading to inhibition of mammalian target of rapamycin (mTOR) and inhibition of cell growth (for review see Ref. 34).
Low glucose conditions also activate the p38 MAPK pathway (35, 36). p38 is a member of the stress-activated protein kinase family (for review see Ref. 3), and its activation during energy depletion affects neuronal survival (36). Unfortunately, the mechanisms by which p38 regulates survival in response to glucose deprivation are not clearly understood. Interestingly, a recent high-throughput protein interaction analysis suggested that OGT may interact directly with p38 (37).
We demonstrate here that glucose deprivation indeed decreases UDP-GlcNAc concentrations, yet paradoxically leads to a dramatic increase in protein O-GlcNAcylation, which is in agreement with other reports (38). We further show that this response is mediated by AMPK-dependent OGT expression and p38-dependent OGT activation, consistent with the activation of OGT by cellular stress. We also show that neurofilament H is O-GlcNAcylated during glucose deprivation, altering its function and providing a possible explanation for the loss of axons in ischemia. These findings represent an important advance in our understanding of mechanisms regulating O-GlcNAcylation by OGT, and the findings have important implications for stress signaling relevant to neurodegeneration, aging, and cancer.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments—Neuro-2a murine neuroblastoma cells were obtained from ATCC and maintained in Dulbecco's modified Eagle's medium (5 mm glucose; Mediatech) supplemented with 10% (v/v) fetal bovine serum (Gemini Bio-Products) and penicillin/streptomycin at 37 °C in a humidified incubator with 5% CO2. Cells were seeded 48 h prior to treatments at a density of 1 × 106 cells/100-mm dish. For glucose deprivation treatments, cells were washed twice with glucose-free Dulbecco's modified Eagle's medium (Mediatech) supplemented with 10% (v/v) dialyzed fetal bovine serum (Invitrogen) and penicillin/streptomycin and then incubated in the same medium for the indicated times.
For inhibitor treatments, the protocol was modified as follows. Thirty minutes prior to the start of glucose deprivation, DMSO (Sigma), 20 μm compound C (EMD Biosciences), 5 μg/ml actinomycin D (Sigma), or 10 μm SB203580 (Sigma) was added to the cells. After 30 min, the cells were washed twice with glucose-free media containing the respective inhibitor and then incubated in the same media for the indicated times.
For metformin treatments, 5 mm metformin (Sigma) dissolved in phosphate-buffered saline was added to the plates and incubated for the indicated times.
Plasmids and Transfections—Mammalian expression vectors encoding HA-tagged p38α, MKK3, MKK3 AA, or MKK3 EE were a kind gift of Dr. J. Silvio Gutkind (NIDCR, National Institutes of Health, Bethesda, MD) (39). The mammalian expression vector encoding an untagged form of OGT, pShuttle-OGT, was described previously (9). The prokaryotic expression vector encoding ncOGT was a kind gift of Dr. Suzanne Walker (Harvard Medical School, Boston) (40).
A mammalian expression vector expressing GST-tagged full-length rat OGT (pEF-GST-OGT) was constructed using standard PCR methods using pShuttle-OGT (9) as template. GST-tagged N-terminal OGT truncations were constructed by PCR using the 5′ primers 939, AGA TCT GAG ACT CTT GCC TCT CGA GTT GCA GCT; 959, AGA TCT ATT GCT AAA AGC AGA CAG GAA TAT GAA; 979, AGA TCT TAC CTG AAG AAA ATT CGT GGC AAA GTA; 999, AGA TCT ACC AAA CAA TAC ACA ATG GAA TTA GAG; and 1019, AGA TCT GCT GGC AAC AAA CCC GAC CAC ATG ATT and the 3′ primer GAA TTC TCA GGC TGA CTC AGT GAC TTC AAC AGG, digested with BglII/EcoRI, and subcloned into the BamHI/EcoRI sites of pEF-GST. The GST-tagged C-terminal OGT truncation, GST-OGT-(1–978), was created by PCR using the 5′ primer GGA TCC ATG GCG TCT TCC GTG GGC AAC GTG and the 3′ primer GC GGC CGC TCA TTC TAG ATC GGT CCC CAG TTT, digested with BamHI/NotI, and subcloned into the BamHI/NotI sites of pEF-GST. Neuro-2a cells were transfected using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions.
Adenovirus-mediated Overexpression—Neuro-2a cells were infected with adenovirus encoding green fluorescent protein (Baylor College of Medicine Vector Development Laboratory) or O-GlcNAcase (41) at a multiplicity of infection of 500 for 40 h prior to glucose deprivation.
Protein Analysis—Cells were washed with ice-cold phosphate-buffered saline and collected using cell scrapers, and the resulting pellet was flash-frozen in dry ice and stored at –80 °C until lysis. For most experiments, cell pellets were lysed with 0.5% (v/v) Nonidet P-40 (Sigma) in 20 mm Tris-HCl, pH 7.5, 140 mm NaCl, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and 1 μm PUGNAc with protease and phosphatase inhibitors. Cell lysates were then centrifuged to remove debris. To immunoprecipitate neurofilament H (NF-H), cell pellets were lysed with 1% (w/v) SDS in 20 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and 1 μm PUGNAc with protease and phosphatase inhibitors. The lysate was passed 10 times through a 22-gauge syringe, and 10 volumes of 0.5% Nonidet P-40 lysis buffer (described above) was added to the lysate, homogenized by sonication using a probe sonicator, and centrifuged to remove debris. Protein concentrations were determined by the Bradford assay.
For immunoblot analysis, 20 μg of lysate was added to Laemmli buffer, boiled, and separated on Criterion precast SDS-polyacrylamide gels (Bio-Rad). The gels were subsequently electroblotted to nitrocellulose (Bio-Rad). The membranes were blocked in Tris-buffered saline with 0.1% (v/v) Tween-20 with either 3% (w/v) bovine serum albumin or 5% (w/v) nonfat dry milk. The blocked membranes were then incubated overnight at 4 °C with the appropriate primary antibody against O-GlcNAc (CTD110.6), OGT (DM-17; Sigma), O-GlcNAcase (a kind gift of Dr. Stewart Whiteheart, University of Kentucky, Lexington) (41), actin (Sigma), pAMPK (Thr-172, 40H9; Cell Signaling), AMPK (Cell Signaling), pp38 (Thr-180/Tyr-182, Cell Signaling), p38 (Santa Cruz Biotechnology), HA (HA.11; Covance), GST (Covance), NF-H (RMd0 20; Cell Signaling), or tubulin (Sigma). The blots were then washed, incubated with the appropriate secondary antibody, developed using ECL (GE Healthcare), and exposed to Hyperfilm ECL (GE Healthcare). For densitometric lane profiles, the plot profile function in NIH Image, version 1.63, was used and normalized against actin loading controls.
For co-immunoprecipitation experiments, 1 mg of pre-cleared cell lysate was incubated with 1 μg of antibody against HA (12CA5), pp38, NF-H, or normal mouse IgG overnight at 4 °C. GammaBind G-Sepharose (GE Healthcare) was added and mixed for an additional 2 h at 4°C. The immunoprecipitates were then washed, eluted in Laemmli buffer, and subjected to immunoblot analysis as described above.
mRNA Analysis—RNA was isolated from cells using TRIzol (Invitrogen) according to manufacturer's instructions. The resulting RNAs were used to prepare oligo(dT)-primed cDNA using SuperScript II (Invitrogen) according to manufacturer's instructions. OGT mRNA and 18 S RNA was assayed by qRT-PCR using the primer pairs OGT (TC TCG AGT TGC AGC TTC TCA, CA TGT GGT CAG GTT TGT TGC) and 18 S (AG GAA TTG ACG GAA GGG CAC, GG ACA TCT AAG GGC ATC ACA) with Platinum SYBR Green quantitative PCR SuperMix (Invitrogen) on a MX3000P quantitative PCR machine (Stratagene). The ΔΔCt method was used to quantitate the relative -fold changes in OGT mRNA levels compared with 0 h controls, normalized to 18 S RNA.
OGT and O-GlcNAcase Assays—OGT and O-GlcNAcase activity assays were performed as described previously (9) with minor modifications. Neuro-2a lysates were immunoprecipitated using normal rabbit IgG or the OGT-specific antibody AL-28 and assayed for [3H]GlcNAc incorporation from UDP-[3H]GlcNAc into a substrate peptide derived from CKII (50 μm) for 1 h at room temperature. The reactions were collected onto C18 MacroSpin columns (The Nest Group), washed, eluted with methanol, and counted by scintillation counting. The resulting activity was normalized against the amount of OGT recovered in the immunoprecipitates.
OGT activity was also assayed from whole cell lysates with or without 250 mm CKII peptide using UDP-[3H]GlcNAc for 1 h at room temperature. Reactions were stopped with 50 mm formic acid, collected onto C18 MacroSpin columns, washed, eluted with methanol, and counted by scintillation counting. “Lysate alone” samples represent incorporation into proteins found in the lysate. “With CKII peptide” samples represent incorporation into proteins found in lysate and CKII peptide. The resulting OGT activity was normalized to either the amount of lysate assayed (Fig. 6C) or the amount of OGT in the lysate (Fig. 6D).
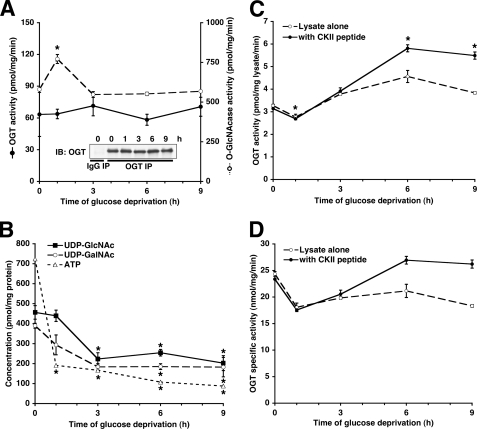
FIGURE 6.
Glucose deprivation-induced protein O-GlcNAcylation cannot be explained by OGT specific catalytic activity or UDP-GlcNAc concentrations. A, OGT immunoprecipitates (IP) from Neuro-2a cells glucose-deprived for the indicated times were assayed for OGT activity (closed circles, solid lines) and normalized to the amount of OGT immunoprecipitated (inset). Lysates from Neuro-2a cells glucose-deprived for the indicated times were assayed for O-GlcNAcase activity (open circles, dashed lines) as described under”Experimental Procedures.“IB, immunoblot. B, lysates from Neuro-2a cells glucose-deprived for the indicated times were assayed for UDP-GlcNAc, UDP-GalNAc, and ATP concentrations as described under”Experimental Procedures.“C, lysates from Neuro-2a cells glucose-deprived for the indicated times were assayed for OGT activity. D, the experiment in C normalized to the amount of OGT in the lysates. *, p < 0.05 compared with 0 h control.
O-GlcNAcase activity was assayed from whole cell lysates using 4 mm p-nitrophenyl-N-acetyl-β-d-glucosaminide as substrate for 1.5 h at 30 °C. Hydrolyzed p-nitrophenol was measured and quantitated by absorbance at 405 nm. The resulting O-GlcNAcase activity was normalized to the amount of lysate assayed.
In Vitro Kinase Assays—Active recombinant GST-tagged p38α (Upstate) was assayed for kinase activity toward recombinant Tau and recombinant ncOGT according to manufacturer's instructions with [γ-32P]ATP. Recombinant Tau was expressed and purified as described previously (42). ncOGT was expressed and purified as described previously (40). Kinase reactions were stopped with Laemmli buffer, separated by SDS-PAGE, stained with Coomassie G250, dried, and exposed to Hyperfilm ECL.
Metabolite Assays—UDP-GlcNAc and UDP-GalNAc concentrations were determined by capillary zone electrophoresis as described previously (43). ATP concentrations in the same samples were determined using the Kinase-Glo ATP-dependent luciferase assay (Promega).
NF-H Solubility Assays—NF-H solubility was determined as described previously with minor modifications (44). Briefly, cell pellets were initially lysed with 0.5% Nonidet P-40 lysis buffer as detailed above. The lysate supernatant was collected as the Nonidet P-40-soluble fraction. The insoluble pellet was washed three times with lysis buffer and then resuspended in 20 mm Tris-HCl, pH 7.5, 8 m urea, 0.1% β-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride, and 1 μm PUGNAc with protease and phosphatase inhibitors. The suspension was homogenized by sonication, and the resulting supernatant was collected as the insoluble fraction.
Statistical Measurements—All experiments were performed on at least three separate occasions (n ≥ 3). In all charts, error bars represent S.E. The p values were calculated using the paired two-tailed Student's t test.
RESULTS
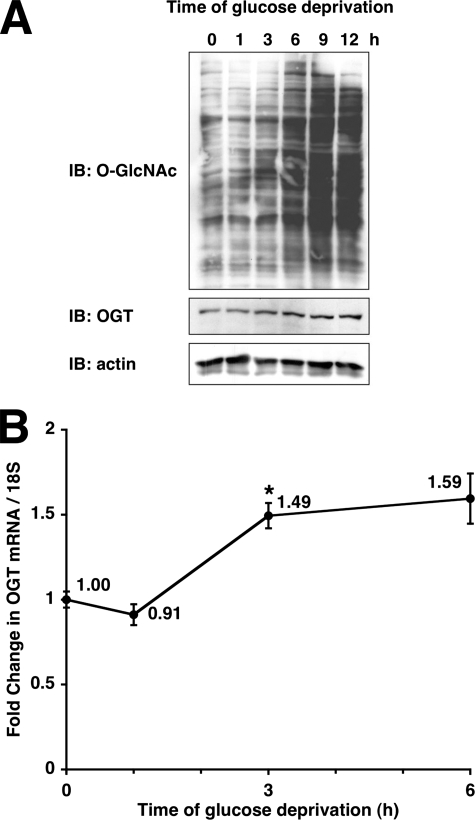
Glucose Deprivation Induces Protein O-GlcNAcylation and OGT Expression in an AMPK-dependent Manner—To clarify the role of nutrient sensing in the regulation of O-GlcNAc, we investigated Neuro-2a neuroblastoma cells in the context of glucose deprivation. As expected, there was a slight decrease in protein O-GlcNAcylation at the early (1–3 h) time points because of decreased flux through the hexosamine biosynthetic pathway. Interestingly, protein O-GlcNAcylation increased significantly and dramatically in the later (6–12 h) time points in response to glucose deprivation (Fig. 1A). Competition of these protein O-GlcNAcylation immunoblots using GlcNAc shows that the vast majority of these immunoreactive bands are indeed specific for O-GlcNAcylation (supplemental Fig. S1).
FIGURE 1.
Glucose deprivation induces protein O-GlcNAcylation and OGT expression. A, lysates from Neuro-2a cells glucose-deprived for the indicated times were immunoblotted (IB) for O-GlcNAc, OGT, and actin. B, mRNA prepared from Neuro-2a cells glucose deprived for the indicated times were subjected to qRT-PCR using primers specific for OGT and 18 S RNA. *, p < 0.05 compared with 0 h control.
We also observed a slight increase in OGT protein and mRNA levels (Fig. 1). These observations are consistent with our previous work showing that many forms of cellular stress induce O-GlcNAcylation, a response that is required for cell survival (9). Also consistent with our previous work, we demonstrated that this increase in protein O-GlcNAcylation is independent of protein degradation (supplemental Fig. S2) and represents an actual increase in OGT activity in vivo. These data are in agreement with a recent report showing that glucose deprivation stimulates O-GlcNAcylation in HepG2 human hepatoma cells (38).
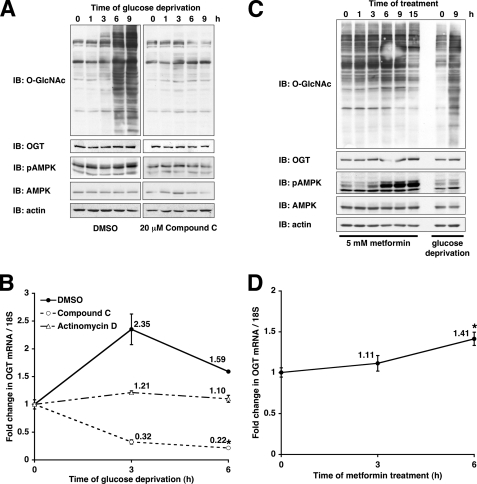
Because the AMPK signaling pathway is known to be activated under conditions of glucose deprivation (4, 33), we next investigated the possibility that AMPK could be regulating this increase in protein O-GlcNAcylation. Pretreatment of cells with the specific AMPK inhibitor compound C (45) for the most part prevented the glucose deprivation-induced increase in protein O-GlcNAcylation (Fig. 2A). Compound C also prevented the mild increase in OGT protein and mRNA expression induced by glucose deprivation (Fig. 2, A and B), suggesting that AMPK regulates OGT expression during glucose deprivation.
FIGURE 2.
AMPK is necessary but not sufficient for glucose deprivation-induced protein O-GlcNAcylation. A, lysates from Neuro-2a cells glucose-deprived for the indicated times (following 30-min pretreatment with DMSO or 20 μm compound C) were immunoblotted (IB) for O-GlcNAc, OGT, pAMPK, AMPK, and actin. B, mRNA prepared from Neuro-2a cells glucose-deprived for the indicated times (following 30-min pretreatment with DMSO, 20 μm compound C, or 5 μg/ml actinomycin D) were subjected to qRT-PCR using primers specific for OGT and 18 S RNA. C, lysates from Neuro-2a cells treated for the indicated times with 5 mm metformin or glucose deprivation were immunoblotted for O-GlcNAc, OGT, pAMPK, AMPK, and actin. D, mRNA prepared from Neuro-2a cells treated with 5 mm metformin for the indicated times were subjected to qRT-PCR using primers specific for OGT and 18 S RNA. *, p < 0.05 compared with 0 h control.
To determine whether AMPK activation was sufficient for the increase in protein O-GlcNAcylation as seen in glucose deprivation, we treated cells with metformin, a known activator of AMPK signaling (45). Indeed, metformin treatment increased OGT mRNA and protein expression (Fig. 2, C and D). Treatment of cells with metformin did result in a slight increase in protein O-GlcNAcylation. However, the magnitude of the increase was much lower when compared with glucose deprivation (Fig. 2C), suggesting that other stress signaling pathways are required for the maximal induction of protein O-GlcNAcylation as seen with glucose deprivation.
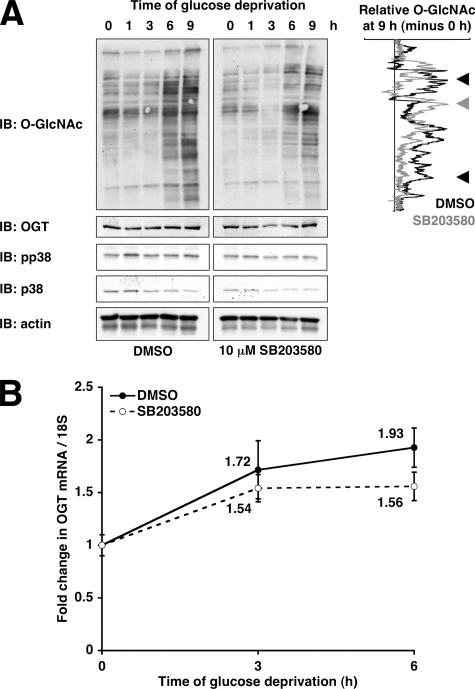
p38 Inhibition Partially Prevents Glucose Deprivation-induced Protein O-GlcNAcylation—The p38 MAPK pathway is activated by many different types of cellular stress (46), and OGT has been found to be a putative interactor for p38 in a high-throughput yeast two-hybrid screen (37), making p38 an attractive potential regulator of protein O-GlcNAcylation. Pretreatment of cells with the specific p38 inhibitor SB203580 (47) partially prevented the increase in protein O-GlcNAcylation induced by glucose deprivation (Fig. 3A, black arrowheads) without preventing the increase in OGT protein and mRNA expression (Fig. 3).
FIGURE 3.
Inhibition of p38 partially prevents glucose deprivation-induced protein O-GlcNAcylation. A, lysates from Neuro-2a cells glucose-deprived for the indicated times (following 30-min pretreatment with DMSO or 10 μm SB203580) were immunoblotted (IB) for O-GlcNAc, OGT, pp38, p38, and actin. Densitometric lane profiles for the 9-h time points (subtracted by the 0 h time points and normalized to actin loading) are displayed for the O-GlcNAc immunoblot. The black arrowheads mark bands decreased by SB203580, and the gray arrowhead marks bands increased by SB203580. B, mRNA prepared from Neuro-2a cells glucose-deprived for the indicated times (following 30-min pretreatment with DMSO or 10 μm SB203580) were subjected to qRT-PCR using primers specific for OGT and 18 S RNA.
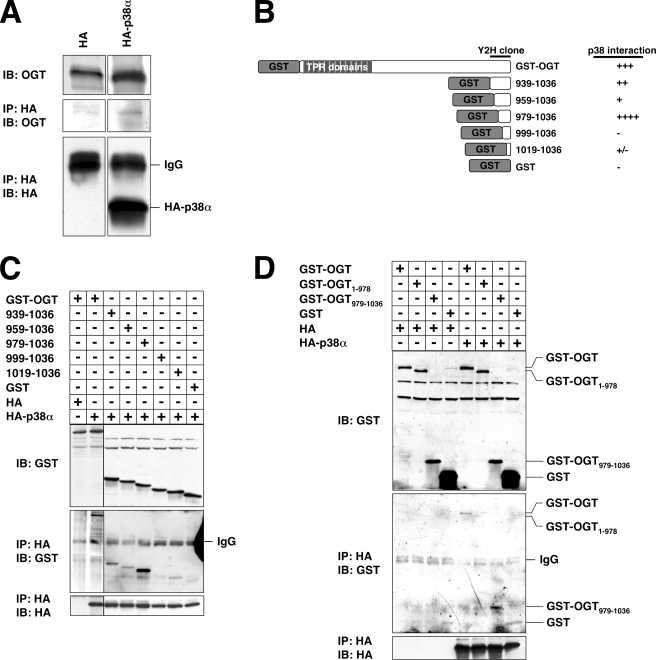
p38 Interacts with the C Terminus of OGT—To verify that p38 and OGT physically interact, we performed co-immunoprecipitation assays using HA-tagged p38α with endogenously expressed OGT. Indeed, OGT was found to co-immunoprecipitate with p38 (Fig. 4A).
FIGURE 4.
p38 interacts with the C terminus of OGT. A, lysates from Neuro-2a cells transfected with HA or HA-p38α were immunoprecipitated (IP) for HA and immunoblotted (IB) for OGT and HA. B, diagram of GST-tagged OGT N-terminal truncations created and tested for the ability to interact with p38. C and D, lysates from Neuro-2a cells transfected with the indicated constructs were immunoprecipitated for HA and immunoblotted for GST and HA.
Upon scrutiny of the yeast two-hybrid data, we discovered that the OGT clone found to interact with p38 contained residues 939 to the stop codon (37). Based on these data, we created N-terminal truncations of OGT to confirm this interaction and to further identify the p38-binding site on OGT (Fig. 4B). Residues 939–1036 of OGT were in fact sufficient for binding p38 in this assay (Fig. 4C). When OGT was truncated to residues 979–1036, its binding to p38 was maximal (Fig. 4C), suggesting that residues 939–978 may actually be inhibitory to p38 binding. Further truncation down to residues 999–1036 completely abolished binding to p38 (Fig. 4C), suggesting that residues between 979 and 998 are necessary for binding p38. In fact, residues 979–1036 seemed to bind p38 better than full-length OGT (Fig. 4D), in agreement with the hypothesis that other regions of OGT may be inhibitory to p38 binding.
To confirm that the C terminus of OGT was necessary for binding p38, we created a C-terminal truncation of OGT containing residues 1–978. As expected, OGT-(1–978) was not able to interact with p38 (Fig. 4D). Altogether, these experiments demonstrate that residues 979–1036 are necessary and sufficient for binding p38.
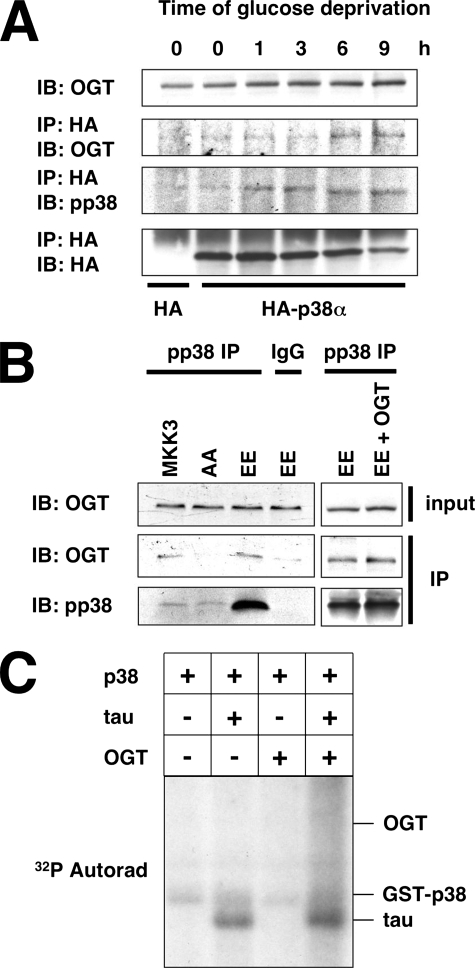
OGT Interacts with Activated p38 during Glucose Deprivation—To understand how p38 could regulate OGT during glucose deprivation, we sought to determine whether its interaction with OGT was altered by glucose deprivation. Interestingly, the interaction of OGT with p38 increased during glucose deprivation and corresponded with maximal p38 activation at the 9-h time point (Fig. 5A).
FIGURE 5.
OGT interacts with the activated form of p38. A, lysates from Neuro-2a cells transfected with HA or HA-p38α and glucose-deprived for the indicated times were immunoprecipitated (IP) for HA and immunoblotted (IB) for OGT, pp38, and HA. B, lysates from Neuro-2a cells transfected with MKK3, MKK3 AA, MKK3 EE, or MKK3 EE with OGT were immunoprecipitated for pp38 or normal mouse IgG and immunoblotted for OGT and pp38. C, 50 ng of activated recombinant GST-p38 was assayed for in vitro kinase activity toward recombinant Tau (2 μg), recombinant OGT (8 μg), or recombinant Tau in the presence of OGT.
We next determined whether p38 activation was necessary for binding to OGT. Toward this end, we transfected Neuro-2a cells with plasmids expressing different forms of MKK3, the upstream kinase of p38. When cells were transfected with the wild type (MKK3) or constitutive active form of MKK3 (EE), OGT was found to interact with pp38 (Fig. 5B). However, when cells were transfected with the dominant negative form of MKK3 (AA), OGT did not interact with p38 (Fig. 5B), suggesting that activation of p38 is necessary for its interaction with OGT. Interestingly, the amount of OGT bound to p38 was not increased in the cells expressing MKK3 EE when compared with wild type MKK3 transfected cells, implying that the amount of OGT was a limiting factor. Next, we overexpressed OGT along with MKK3 EE and found an increase in the amount of OGT bound to p38 (Fig. 5B), confirming that the induction of OGT protein expression is necessary for its increased interaction with p38.
Because activated p38 interacts with OGT, it stood to reason that it might be able to phosphorylate OGT. To examine this possibility, we performed an in vitro kinase assay using recombinant activated p38 and recombinant Tau and OGT as substrates. As reported previously, p38 was able to phosphorylate Tau protein (Fig. 5C, Ref. 48) as well as ATF2 (data not shown; Ref. 49). However, p38 was not able to phosphorylate recombinant OGT, suggesting that OGT is not a direct substrate of p38 (Fig. 5C). In addition, we sought to determine whether OGT would block the phosphorylation of Tau by p38 in vitro. However, co-incubation of Tau with OGT seemed to have little negative effect on its phosphorylation by p38 and even appeared to slightly increase its phosphorylation (Fig. 5C).
OGT Specific Catalytic Activity and [UDP-GlcNAc] Are Not Increased during Glucose Deprivation—In order to determine whether the interaction of p38 with OGT could have an effect on its activity, we next examined the specific activity of OGT by immunoprecipitating OGT and assaying its activity toward the CKII peptide substrate during glucose deprivation. Surprisingly, the OGT specific catalytic activity was not increased (when corrected for the amount of OGT immunoprecipitated) during glucose deprivation, despite the large increase in protein O-GlcNAcylation (Fig. 6A). O-GlcNAcase activity in these same lysates was also not decreased during glucose deprivation (Fig. 6A). In fact, there was a small but significant increase in O-GlcNAcase activity at the 1-h time point.
To be sure that UDP-GlcNAc concentrations were not increasing and thus driving the increased protein O-GlcNAcylation, we measured intracellular UDP-GlcNAc, UDP-GalNAc, and ATP concentrations during glucose deprivation. As expected, the concentrations of these metabolites decreased during glucose deprivation by more than 2-fold (Fig. 6B).
Next, we set out to confirm that the total OGT activity in the lysate was actually increased by glucose deprivation. We were able to assay OGT activity in whole cell lysate with or without the addition of the CKII peptide substrate and found that total OGT activity did indeed increase during glucose deprivation (Fig. 6C). However, these increases disappeared when normalized to the amount of OGT in the lysate (Fig. 6D), confirming our earlier experiments showing that OGT specific catalytic activity is not increased during glucose deprivation.
p38 Recruits OGT to Specific Targets Including Neurofilament H during Glucose Deprivation—Because OGT specific catalytic activity was not increased, whereas the interaction of OGT with p38 was increased during glucose deprivation, we put forth the hypothesis that p38 may interact with OGT and target it to specific substrates, thus increasing protein O-GlcNAcylation during glucose deprivation. In an attempt to test this hypothesis, we performed a large scale co-immunoprecipitation to identify proteins that interact with OGT during glucose deprivation (data not shown). One potential target identified by this screen was NF-H, a known substrate of both OGT and p38 (50, 51).
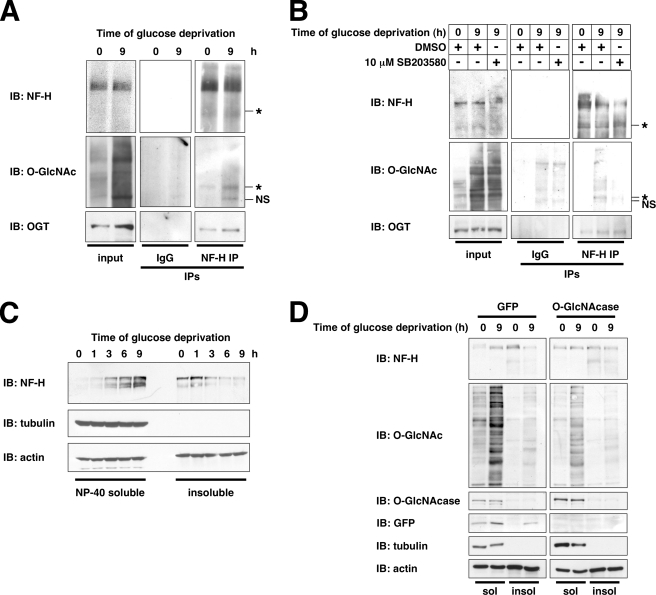
To confirm that NF-H interacted with OGT during glucose deprivation, we performed co-immunoprecipitation assays using endogenously expressed NF-H and OGT. As expected, NF-H interacted with OGT during glucose deprivation (Fig. 7A). More importantly, O-GlcNAcylation of a faster migrating form (110,000, apparent molecular weight) of NF-H increased during glucose deprivation (Fig. 7A).
FIGURE 7.
During glucose deprivation, NF-H is O-GlcNAcylated in a p38-dependent manner, and its solubility is increased. A, lysates from Neuro-2a cells glucose-deprived for the indicated times were immunoprecipitated (IP) for NF-H or normal mouse IgG and immunoblotted (IB) for NF-H, O-GlcNAc, and OGT. An asterisk marks the O-GlcNAcylated NF-H species. NS, nonspecific bands appearing in normal IgG immunoprecipitates. B, lysates from Neuro-2a cells glucose-deprived for the indicated times (following 30-min pretreatment with DMSO or 10 μm SB203580) were immunoprecipitated for NF-H or normal mouse IgG and immunoblotted for NF-H, O-GlcNAc, and OGT. An asterisk marks O-GlcNAcylated NF-H species. C, Nonidet P-40-soluble and -insoluble fractions prepared from Neuro-2a cells glucose-deprived for the indicated times were immunoblotted for NF-H, tubulin, and actin. D, Nonidet P-40-soluble (sol) and -insoluble (insol) fractions prepared from Neuro-2a cells infected with green fluorescent protein (GFP) or O-GlcNAcase adenovirus and glucose-deprived for the indicated times were immunoblotted for NF-H, O-GlcNAc, O-GlcNAcase, green fluorescent protein, tubulin, and actin.
To demonstrate that NF-H O-GlcNAcylation during glucose deprivation requires p38, we pretreated cells with SB203580. Indeed, inhibition of p38 abolished the glucose deprivation-induced O-GlcNAcylation of NF-H (Fig. 7B), suggesting that p38 activity is required for OGT to O-GlcNAcylate NF-H. Surprisingly, inhibition of p38 did not seem to affect the interaction of OGT with NF-H during glucose deprivation (Fig. 7B), suggesting that the interaction of OGT with NF-H may not be necessarily p38-dependent, whereas its activity toward NF-H requires p38 activation.
Neurofilament H Solubility Is Increased by O-GlcNAcylation during Glucose Deprivation—To examine what effect NF-H O-GlcNAcylation may have on its function, we determined its solubility during glucose deprivation. NF-H solubility increased dramatically during glucose deprivation (Fig. 7C), indicating a shift in the pool of NF-H from the insoluble, filamentous form to a more soluble form (44).
Next, we determined whether NF-H O-GlcNAcylation was required for this increase in solubility during glucose deprivation by using adenoviral-mediated overexpression of O-GlcNAcase. Overexpression of O-GlcNAcase completely prevented the increase in NF-H solubility during glucose deprivation (Fig. 7D), suggesting that O-GlcNAcylation of NF-H is necessary for an increase in solubility.
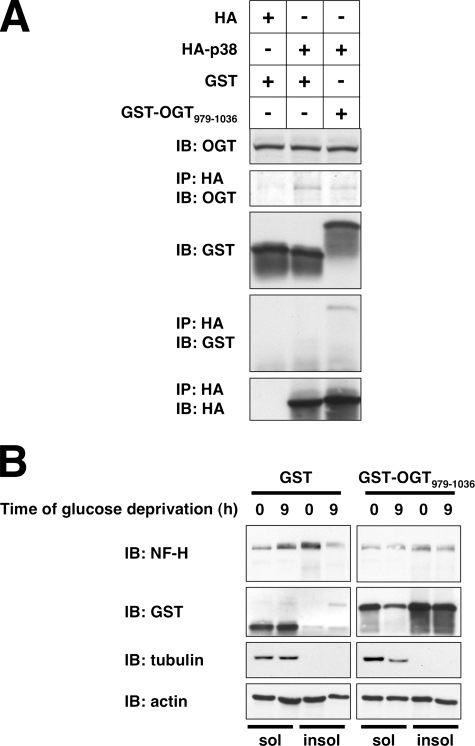
To further confirm that O-GlcNAcylation affects NF-H solubility, we took advantage of the observation that GST-OGT-(979–1036) appears to bind p38 better than full length OGT. Because this short fragment lacks the catalytic portion of OGT, overexpression of this protein would not be expected to increase O-GlcNAcylation and should effectively compete with endogenous OGT for binding to p38. To demonstrate that GST-OGT-(979–1036) competes the OGT-p38 interaction, we performed co-immunoprecipitation assays and found that overexpression of this fragment decreases the amount of endogenous OGT bound to p38 (Fig. 8A).
FIGURE 8.
GST-OGT-(979–1036) overexpression disrupts the OGT-p38 interaction and prevents the glucose deprivation-induced NF-H solubility change. A, lysates from Neuro-2a cells transfected with the indicated constructs were immunoprecipitated (IP) for HA and immunoblotted (IB) for OGT, GST, and HA. B, Nonidet P-40-soluble (sol) and -insoluble (insol) fractions prepared from Neuro-2a cells transfected with GST or GST-OGT-(979–1036) and glucose-deprived for the indicated times were immunoblotted for NF-H, GST, tubulin, and actin.
Next, we determined whether GST-OGT-(979–1036) could prevent the increase in NF-H solubility seen during glucose deprivation. As expected, overexpression of GST-OGT-(979–1036) prevented the glucose deprivation-induced increase in NF-H solubility (Fig. 8B), suggesting that the interaction of OGT with p38 is necessary for NF-H solubility.
DISCUSSION
Physiologically, glucose deprivation can occur in a number of instances, including during ischemic events and as a result of aging in the brain (1, 52). During cerebral ischemia, the brain is deprived of oxygen as well as glucose (1). It has been observed that ischemia results directly in selective injury and loss of axons in the brain (6, 7). However, the precise mechanisms underlying ischemic-induced axonal damage remain unclear.
Glucose metabolism in the brain has been shown to decline with age (52, 53). Although the brain is able to use ketone bodies as a minor source of fuel, glucose remains the primary source of energy for the brain (54). Even though the direct effects of limiting glucose usage on brain function are not clearly understood, studies showing that glucose deprivation leads to axonal loss imply that it has negative consequences for brain function (6, 7). In fact, it has been proposed that the age-related decline in glucose metabolism, along with the resulting reduced O-GlcNAcylation of neuronal proteins, could be a key mediator in the development of late-onset Alzheimer disease (55–59). It is worthy to note that the incidence of dementia and Alzheimer disease is significantly increased in patients following cerebral ischemia (60–62), suggesting a direct association between defective glucose metabolism and brain function.
Here, we report a potential mechanism by which glucose deprivation may influence axonal structural stability (Fig. 9). We present evidence that glucose metabolism activates AMPK, which mediates an increase in OGT mRNA and protein expression. During glucose deprivation, p38 is also activated, an activation that is required for its interaction with OGT. This p38-OGT interaction seems to alter the function of OGT in a way such that specific proteins, including NF-H, are now better substrates for OGT, possibly by directly recruiting OGT to target sequences or sites. One effect of this p38-mediated activation of OGT is the O-GlcNAcylation of NF-H, resulting in its depolymerization from neurofilaments or other intermediate filament structures. NF-H is a major structural component of mature axons, the loss of which in neurofilaments has negative consequences on axonal structure and caliber (63).
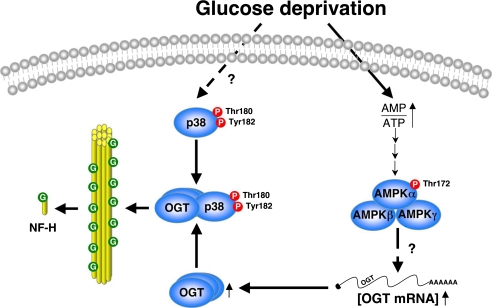
FIGURE 9.
Model for OGT regulation by AMPK and p38 during glucose deprivation. Glucose deprivation activates AMPK via an increase in the AMP/ATP ratio, resulting in increased OGT expression. Glucose deprivation activates p38 by phosphorylation, which interacts with OGT, recruiting it to specific targets, including neurofilament H, to increase its solubility.
NF-H is a type IV intermediate filament protein with a head, rod, and tail domain. The central coiled-coil α-helical rod domain allows NF-H to form heterotetramers with NF-L or NF-M, but this domain is not known to be phosphorylated or O-GlcNAcylated (50, 64). The C-terminal tail domain of NF-H, which protrudes and forms a side arm from the neurofilaments, contains more than 30 Lys-Ser-Pro (KSP) repeat phosphorylation sites (64). Recently, Ackerley et al. (51) demonstrated that p38α can phosphorylate several of these KSP repeats on NF-H and can associate with phosphorylated neurofilament aggregations, as seen in amyotrophic lateral sclerosis. Our laboratory has shown previously that many of these KSP repeats on NF-H can also be O-GlcNAcylated; however, the functional significance of this finding remains unclear (50). Interestingly, Ludemann et al. (65) have shown that O-GlcNAcylation of the KSP repeats in NF-M, which has a much shorter tail domain, seems decreased in amyotrophic lateral sclerosis, whereas its phosphorylation is increased, suggesting a direct reciprocal relationship between O-GlcNAcylation and phosphorylation of the tail domain of neurofilaments. Nonetheless, it is not known how NF-H tail domain phosphorylation/O-GlcNAcylation might affect its solubility. Instead, NF-H tail phosphorylation has been shown to affect axonal transport of neurofilaments (66, 67).
In general, phosphorylation of the head domain of intermediate filament proteins is thought to control its polymerization into filaments (67). Only one phosphorylation site at Ser-61 in the head domain of NF-H has been identified (68). Whether phosphorylation of NF-H at this site affects its assembly has not been investigated at this time. In contrast, there are three known sites of O-GlcNAcylation in the head domain of NF-H: at Thr-53, Ser-54, and Ser-56 (50). However, the precise function of O-GlcNAcylation on these residues is also not known. Further studies regarding the phosphorylation and O-GlcNAcylation of the head domain of NF-H will help to elucidate their roles in regulating the assembly and solubility of neurofilaments. Also, the significance of the increase in solubility of NF-H in terms of cell survival is not completely understood. Although loss of neurofilaments has been shown to be associated with axonal damage (69), whether this is actually beneficial to the overall survival of the cell remains unclear.
The fact that p38 is a positive regulator of NF-H O-GlcNAcylation during glucose deprivation suggests that this is part of a stress response pathway. Surprisingly, instead of interacting with OGT through its N-terminal TPR motifs, p38 interacts with the OGT C terminus, a region proximal to its catalytic domain. It is possible that binding of p38 to OGT may induce a conformational change in OGT, altering its ability to recognize certain proteins or sequences. Clearly, the role of p38 in mediating O-GlcNAcylation during glucose deprivation and other forms of cellular stress warrants further investigation. In fact, recent studies suggest that the p38 pathway itself may be regulated by the hexosamine biosynthetic pathway and O-GlcNAcylation (70, 71).
Furthermore, although p38 does not directly phosphorylate OGT in vitro, our data cannot rule out the possibility that OGT may be phosphorylated by another kinase(s) under these conditions. However, our observation that OGT specific catalytic activity is not changed during glucose deprivation suggests that phosphorylation of OGT is unlikely to affect its catalytic activity directly (Fig. 6, A, C, and D). Instead, phosphorylation of OGT may affect its ability to recognize its substrates or its interaction with other targeting proteins. It should be noted that p38 inhibition only partially prevented glucose deprivation-induced protein O-GlcNAcylation (Fig. 3A), implying that there are many other unknown OGT targeting proteins. The identification of other proteins that are O-GlcNAcylated during glucose deprivation may be a promising avenue for revealing some of these OGT-regulating proteins. Furthermore, this mode of OGT regulation through recruitment may also extend to other forms of cellular stimulation.
Regardless of how OGT is being targeted to its substrates during glucose deprivation, its increased expression by AMPK seems to be a critical prerequisite, because inhibition of AMPK almost completely blocks glucose deprivation-induced O-GlcNAcylation (Fig. 2A). Our data indicate that AMPK is a positive regulator of OGT mRNA expression. The transcription factors that are responsible for regulating transcription of OGT are unknown. In actuality, our data imply that AMPK might regulate OGT mRNA stability, because inhibition of transcription merely prevented the glucose deprivation-induced increase in OGT mRNA, whereas inhibition of AMPK caused a decrease in OGT mRNA during glucose deprivation (Fig. 2B). In accordance with this hypothesis, Yun et al. (72) have shown that during glucose deprivation of DU145 prostate carcinoma cells, vascular endothelial growth factor mRNA stability is increased in an AMPK-dependent manner. However, our observations cannot rule out the possibility that OGT expression may also be regulated transcriptionally as well as translationally.
Recently, Taylor et al. (38) showed that glucose deprivation induced protein O-GlcNAcylation and OGT expression in HepG2 hepatoma cells. Although our data showing that glucose deprivation induces protein O-GlcNAcylation are largely in agreement with that report, here we are able to outline several means by which OGT and protein O-GlcNAcylation are regulated during glucose deprivation by AMP-activated protein kinase and p38 MAPK signaling pathways, resulting in NF-H O-GlcNAcylation and alteration of its solubility. These findings represent a possible mechanism by which defective glucose metabolism may influence axonal structural stability. A greater understanding of how protein O-GlcNAcylation is regulated during glucose deprivation may lead to better insights into the mechanisms of cerebral ischemia, aging, neurodegeneration, and cancer.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of mammalian expression plasmids encoding p38 and MKK3 from Dr. J. Silvio Gutkind (NIDCR, National Institutes of Health, Bethesda, MD) and the prokaryotic expression plasmid encoding ncOGT from Dr. Suzanne Walker (Harvard Medical School, Boston, MA). We also appreciate the generous gift of O-GlcNAcase antibodies from Dr. Stewart Whiteheart (University of Kentucky, Lexington). We thank Kaoru Sakabe for creating the GST-tagged full-length OGT mammalian expression plasmid. We also thank Tonia Vassilowitch, Michael Housley, Dr. Wagner Dias, Dr. Chad Slawson, and other members of our laboratory for technical assistance and critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HD13563 and CA42486 (to G. W. H.). This work was also supported by a National Science Foundation graduate research fellowship (to W. D. C.). Under a licensing agreement between The Johns Hopkins University, Covance Research Products, Sigma-Aldrich, and Santa Cruz Biotechnology, G. W. H. receives royalties from the sale of the CTD110.6 O-GlcNAc antibody. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: OGT, O-linked β-N-acetylglucosaminyltransferase; ncOGT, nucleocytoplasmic OGT; O-GlcNAc, O-linked β-N-acetylglucosamine; O-GlcNAcase, O-linked β-N-acetylglucosaminidase; AMPK, AMP-activated protein kinase; DMSO, dimethyl sulfoxide; NF-H, neurofilament H; HA, hemagglutinin; GST, glutathione S-transferase; MAPK, mitogen-activated protein kinase; qRT, quantitative reverse transcription; MKK. mitogen-activated protein kinase kinase; PUGNAc, O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino N-phenyl carbamate.
References
- 1.Schurr, A. (2002) Neurochem. Int. 41 1–8 [DOI] [PubMed] [Google Scholar]
- 2.Tabakman, R., Jiang, H., Shahar, I., Arien-Zakay, H., Levine, R. A., and Lazarovici, P. (2005) Ann. N. Y. Acad. Sci. 1053 84–96 [DOI] [PubMed] [Google Scholar]
- 3.Irving, E. A., and Bamford, M. (2002) J. Cereb. Blood Flow Metab. 22 631–647 [DOI] [PubMed] [Google Scholar]
- 4.McCullough, L. D., Zeng, Z., Li, H., Landree, L. E., McFadden, J., and Ronnett, G. V. (2005) J. Biol. Chem. 280 20493–20502 [DOI] [PubMed] [Google Scholar]
- 5.LaManna, J. C., and Lust, W. D. (1997) Neurosurg. Clin. N. Am. 8 145–163 [PubMed] [Google Scholar]
- 6.Lyeth, B. G., Jenkins, L. W., Hamm, R. J., Dixon, C. E., Phillips, L. L., Clifton, G. L., Young, H. F., and Hayes, R. L. (1990) Brain Res. 526 249–258 [DOI] [PubMed] [Google Scholar]
- 7.Cervos-Navarro, J., and Lafuente, J. V. (1991) J. Neurol. Sci. 103, (Suppl.) S3–S14 [DOI] [PubMed] [Google Scholar]
- 8.Bramlett, H. M., and Dietrich, W. D. (2004) J. Cereb. Blood Flow Metab. 24 133–150 [DOI] [PubMed] [Google Scholar]
- 9.Zachara, N. E., O'Donnell, N., Cheung, W. D., Mercer, J. J., Marth, J. D., and Hart, G. W. (2004) J. Biol. Chem. 279 30133–30142 [DOI] [PubMed] [Google Scholar]
- 10.Hart, G. W., Housley, M. P., and Slawson, C. (2007) Nature 446 1017–1022 [DOI] [PubMed] [Google Scholar]
- 11.Shafi, R., Iyer, S. P., Ellies, L. G., O'Donnell, N., Marek, K. W., Chui, D., Hart, G. W., and Marth, J. D. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 5735–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Donnell, N., Zachara, N. E., Hart, G. W., and Marth, J. D. (2004) Mol. Cell. Biol. 24 1680–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamemura, K., Hayes, B. K., Comer, F. I., and Hart, G. W. (2002) J. Biol. Chem. 277 19229–19235 [DOI] [PubMed] [Google Scholar]
- 14.Cheng, X., Cole, R. N., Zaia, J., and Hart, G. W. (2000) Biochemistry 39 11609–11620 [DOI] [PubMed] [Google Scholar]
- 15.Medina, L., Grove, K., and Haltiwanger, R. S. (1998) Glycobiology 8 383–391 [DOI] [PubMed] [Google Scholar]
- 16.Du, X. L., Edelstein, D., Dimmeler, S., Ju, Q., Sui, C., and Brownlee, M. (2001) J. Clin. Investig. 108 1341–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang, W. H., Kim, J. E., Nam, H. W., Ju, J. W., Kim, H. S., Kim, Y. S., and Cho, J. W. (2006) Nat. Cell Biol. 8 1074–1083 [DOI] [PubMed] [Google Scholar]
- 18.Comer, F. I., and Hart, G. W. (2001) Biochemistry 40 7845–7852 [DOI] [PubMed] [Google Scholar]
- 19.Wells, L., Vosseller, K., and Hart, G. W. (2001) Science 291 2376–2378 [DOI] [PubMed] [Google Scholar]
- 20.Love, D. C., and Hanover, J. A. (2005) Sci. STKE 2005, RE13. [DOI] [PubMed]
- 21.Zachara, N. E., and Hart, G. W. (2006) Biochim. Biophys. Acta 1761 599–617 [DOI] [PubMed] [Google Scholar]
- 22.Braidman, I., Carroll, M., Dance, N., Robinson, D., Poenaru, L., Weber, A., Dreyfus, J. C., Overdijk, B., and Hooghwinkel, G. J. (1974) FEBS Lett. 41 181–184 [DOI] [PubMed] [Google Scholar]
- 23.Toleman, C., Paterson, A. J., Whisenhunt, T. R., and Kudlow, J. E. (2004) J. Biol. Chem. 279 53665–53673 [DOI] [PubMed] [Google Scholar]
- 24.Heckel, D., Comtesse, N., Brass, N., Blin, N., Zang, K. D., and Meese, E. (1998) Hum. Mol. Genet. 7 1859–1872 [DOI] [PubMed] [Google Scholar]
- 25.Bertram, L., Blacker, D., Mullin, K., Keeney, D., Jones, J., Basu, S., Yhu, S., McInnis, M. G., Go, R. C., Vekrellis, K., Selkoe, D. J., Saunders, A. J., and Tanzi, R. E. (2000) Science 290 2302–2303 [DOI] [PubMed] [Google Scholar]
- 26.Haltiwanger, R. S., Blomberg, M. A., and Hart, G. W. (1992) J. Biol. Chem. 267 9005–9013 [PubMed] [Google Scholar]
- 27.Kreppel, L. K., and Hart, G. W. (1999) J. Biol. Chem. 274 32015–32022 [DOI] [PubMed] [Google Scholar]
- 28.Wang, J., Liu, R., Hawkins, M., Barzilai, N., and Rossetti, L. (1998) Nature 393 684–688 [DOI] [PubMed] [Google Scholar]
- 29.Marshall, S., Nadeau, O., and Yamasaki, K. (2004) J. Biol. Chem. 279 35313–35319 [DOI] [PubMed] [Google Scholar]
- 30.Akimoto, Y., Hart, G. W., Hirano, H., and Kawakami, H. (2005) Med. Mol. Morphol. 38 84–91 [DOI] [PubMed] [Google Scholar]
- 31.McClain, D. A., and Crook, E. D. (1996) Diabetes 45 1003–1009 [DOI] [PubMed] [Google Scholar]
- 32.Tomiya, N., Ailor, E., Lawrence, S. M., Betenbaugh, M. J., and Lee, Y. C. (2001) Anal. Biochem. 293 129–137 [DOI] [PubMed] [Google Scholar]
- 33.Culmsee, C., Monnig, J., Kemp, B. E., and Mattson, M. P. (2001) J. Mol. Neurosci. 17 45–58 [DOI] [PubMed] [Google Scholar]
- 34.Towler, M. C., and Hardie, D. G. (2007) Circ. Res. 100 328–341 [DOI] [PubMed] [Google Scholar]
- 35.Barone, F. C., Irving, E. A., Ray, A. M., Lee, J. C., Kassis, S., Kumar, S., Badger, A. M., White, R. F., McVey, M. J., Legos, J. J., Erhardt, J. A., Nelson, A. H., Ohlstein, E. H., Hunter, A. J., Ward, K., Smith, B. R., Adams, J. L., and Parsons, A. A. (2001) J. Pharmacol. Exp. Ther. 296 312–321 [PubMed] [Google Scholar]
- 36.Takman, R., Jiang, H., Schaefer, E., Levine, R. A., and Lazarovici, P. (2004) J. Mol. Neurosci. 22 237–250 [DOI] [PubMed] [Google Scholar]
- 37.Li, S., Armstrong, C. M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P. O., Han, J. D., Chesneau, A., Hao, T., Goldberg, D. S., Li, N., Martinez, M., Rual, J. F., Lamesch, P., Xu, L., Tewari, M., Wong, S. L., Zhang, L. V., Berriz, G. F., Jacotot, L., Vaglio, P., Reboul, J., Hirozane-Kishikawa, T., Li, Q., Gabel, H. W., Elewa, A., Baumgartner, B., Rose, D. J., Yu, H., Bosak, S., Sequerra, R., Fraser, A., Mango, S. E., Saxton, W. M., Strome, S., Van Den Heuvel, S., Piano, F., Vandenhaute, J., Sardet, C., Gerstein, M., Doucette-Stamm, L., Gunsalus, K. C., Harper, J. W., Cusick, M. E., Roth, F. P., Hill, D. E., and Vidal, M. (2004) Science 303 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor, R. P., Parker, G. J., Hazel, M. W., Soesanto, Y., Fuller, W., Yazzie, M. J., and McClain, D. A. (2008) J. Biol. Chem. 283 6050–6057 [DOI] [PubMed] [Google Scholar]
- 39.Marinissen, M. J., Chiariello, M., Pallante, M., and Gutkind, J. S. (1999) Mol. Cell. Biol. 19 4289–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gross, B. J., Kraybill, B. C., and Walker, S. (2005) J. Am. Chem. Soc. 127 14588–14589 [DOI] [PubMed] [Google Scholar]
- 41.Slawson, C., Zachara, N. E., Vosseller, K., Cheung, W. D., Lane, M. D., and Hart, G. W. (2005) J. Biol. Chem. 280 32944–32956 [DOI] [PubMed] [Google Scholar]
- 42.Scott, C. W., Blowers, D. P., Barth, P. T., Lo, M. M., Salama, A. I., and Caputo, C. B. (1991) J. Neurosci. Res. 30 154–162 [DOI] [PubMed] [Google Scholar]
- 43.Lehmann, R., Huber, M., Beck, A., Schindera, T., Rinkler, T., Houdali, B., Weigert, C., Haring, H. U., Voelter, W., and Schleicher, E. D. (2000) Electrophoresis 21 3010–3015 [DOI] [PubMed] [Google Scholar]
- 44.Shea, T. B. (1994) FEBS Lett. 343 131–136 [DOI] [PubMed] [Google Scholar]
- 45.Zhou, G., Myers, R., Li, Y., Chen, Y., Shen, X., Fenyk-Melody, J., Wu, M., Ventre, J., Doebber, T., Fujii, N., Musi, N., Hirshman, M. F., Goodyear, L. J., and Moller, D. E. (2001) J. Clin. Investig. 108 1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cowan, K. J., and Storey, K. B. (2003) J. Exp. Biol. 206 1107–1115 [DOI] [PubMed] [Google Scholar]
- 47.Cuenda, A., Rouse, J., Doza, Y. N., Meier, R., Cohen, P., Gallagher, T. F., Young, P. R., and Lee, J. C. (1995) FEBS Lett. 364 229–233 [DOI] [PubMed] [Google Scholar]
- 48.Goedert, M., Hasegawa, M., Jakes, R., Lawler, S., Cuenda, A., and Cohen, P. (1997) FEBS Lett. 409 57–62 [DOI] [PubMed] [Google Scholar]
- 49.Derijard, B., Raingeaud, J., Barrett, T., Wu, I. H., Han, J., Ulevitch, R. J., and Davis, R. J. (1995) Science 267 682–685 [DOI] [PubMed] [Google Scholar]
- 50.Dong, D. L., Xu, Z. S., Hart, G. W., and Cleveland, D. W. (1996) J. Biol. Chem. 271 20845–20852 [DOI] [PubMed] [Google Scholar]
- 51.Ackerley, S., Grierson, A. J., Banner, S., Perkinton, M. S., Brownlees, J., Byers, H. L., Ward, M., Thornhill, P., Hussain, K., Waby, J. S., Anderton, B. H., Cooper, J. D., Dingwall, C., Leigh, P. N., Shaw, C. E., and Miller, C. C. (2004) Mol. Cell. Neurosci. 26 354–364 [DOI] [PubMed] [Google Scholar]
- 52.Gold, P. E. (2005) Neurobiol. Aging 26 Suppl. 1, 60–64 [DOI] [PubMed] [Google Scholar]
- 53.Hoyer, S. (1998) J. Neural. Transm. Suppl. 54 187–194 [DOI] [PubMed] [Google Scholar]
- 54.Fehm, H. L., Kern, W., and Peters, A. (2006) Prog. Brain Res. 153 129–140 [DOI] [PubMed] [Google Scholar]
- 55.Fukuyama, H., Ogawa, M., Yamauchi, H., Yamaguchi, S., Kimura, J., Yonekura, Y., and Konishi, J. (1994) J. Nucl. Med. 35 1–6 [PubMed] [Google Scholar]
- 56.Hoyer, S. (2004) Eur. J. Pharmacol. 490 115–125 [DOI] [PubMed] [Google Scholar]
- 57.Arnold, C. S., Johnson, G. V., Cole, R. N., Dong, D. L., Lee, M., and Hart, G. W. (1996) J. Biol. Chem. 271 28741–28744 [DOI] [PubMed] [Google Scholar]
- 58.Liu, F., Iqbal, K., Grundke-Iqbal, I., Hart, G. W., and Gong, C. X. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10804–10809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dias, W. B., and Hart, G. W. (2007) Mol. Biosyst. 3 766–772 [DOI] [PubMed] [Google Scholar]
- 60.Leys, D., and Pasquier, F. (2000) J. Neural. Transm. Suppl. 59 31–36 [DOI] [PubMed] [Google Scholar]
- 61.White, L., and Launer, L. (2006) Alzheimer Dis. Assoc. Disord. 20 Suppl. 2, S79–S83 [DOI] [PubMed] [Google Scholar]
- 62.Vermeer, S. E., Prins, N. D., den Heijer, T., Hofman, A., Koudstaal, P. J., and Breteler, M. M. (2003) N. Engl. J. Med. 348 1215–1222 [DOI] [PubMed] [Google Scholar]
- 63.Elder, G. A., Friedrich, V. L., Jr., Kang, C., Bosco, P., Gourov, A., Tu, P. H., Zhang, B., Lee, V. M., and Lazzarini, R. A. (1998) J. Cell Biol. 143 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Omary, M. B., Ku, N. O., Tao, G. Z., Toivola, D. M., and Liao, J. (2006) Trends Biochem. Sci 31 383–394 [DOI] [PubMed] [Google Scholar]
- 65.Ludemann, N., Clement, A., Hans, V. H., Leschik, J., Behl, C., and Brandt, R. (2005) J. Biol. Chem. 280 31648–31658 [DOI] [PubMed] [Google Scholar]
- 66.Watson, D. F., Griffin, J. W., Fittro, K. P., and Hoffman, P. N. (1989) J. Neurochem. 53 1818–1829 [DOI] [PubMed] [Google Scholar]
- 67.Nixon, R. A., and Sihag, R. K. (1991) Trends Neurosci. 14 501–506 [DOI] [PubMed] [Google Scholar]
- 68.Trinidad, J. C., Specht, C. G., Thalhammer, A., Schoepfer, R., and Burlingame, A. L. (2006) Mol. Cell. Proteomics 5 914–922 [DOI] [PubMed] [Google Scholar]
- 69.King, C. E., Adlard, P. A., Dickson, T. C., and Vickers, J. C. (2000) Clin. Exp. Pharmacol. Physiol. 27 548–552 [DOI] [PubMed] [Google Scholar]
- 70.Kneass, Z. T., and Marchase, R. B. (2005) J. Biol. Chem. 280 14579–14585 [DOI] [PubMed] [Google Scholar]
- 71.Fulop, N., Zhang, Z., Marchase, R. B., and Chatham, J. C. (2007) Am. J. Physiol. 292 H2227–H2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yun, H., Lee, M., Kim, S. S., and Ha, J. (2005) J. Biol. Chem. 280 9963–9972 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.