Abstract
DNA microarray technology has been a valuable tool to provide a global view of the changes in gene expression that characterize different types of B cell lymphomas, both in relation to clinical parameters but also in comparison with the non-malignant counterparts. The number of transcripts that can be analyzed on an array has dramatically increased, and now most commercially available arrays cover the whole genome, enabling overall analysis of the transcriptome.The backside of collecting this massive amount of information is that even after strict data filtering, it is impossible to do follow-up studies on all findings. Down-stream analysis is time-consuming and when performing confirmatory experiments on the protein level, the experiments are in most cases restricted to proteins recognized by commercially available reagents. Furthermore, since gene expression data is a comparative method not only are the experimental set-up but also the characteristics of both the sample and reference crucial for our ability to answer the questions posed. Thus, initial care must be taken in the design of the experiment and the preparation of the samples.
The aim of this review is to discuss the progress in mantle cell lymphoma research enabled by gene expression analysis and to pinpoint the difficulties in making efficient use of the generated data to provide a fast and accurate clinical diagnosis, efficient stratification of patients into disease sub-groups and improved therapy.
MANTLE CELL LYMPHOMA — HISTORICAL PERSPECTIVES
B cell lymphomas are the malignant counterparts of different developmental stages of normal B lymphocytes and are divided into precursor B cell neoplasms and mature/peripheral B cell neoplasms, according to the WHO classification 1. B cell differentiation is strictly regulated by ho-meostatic controls but even so, malignant transformation occasionally proceeds unimpeded 2. The main groups of B cell neoplasms are precursor B-lymphoblastic leukaemia/lymphoma, Chronic lymphocytic leukaemia/lymphoma (CLL), plasma cell myeloma, extranodal marginal zone (MZ) B cell lymphoma of mucosa-associated lymphoid tissue (MALT) type, follicular lymphoma (FL), mantle cell lymphoma (MCL), diffuse large B cell lymphoma (DLBCL) and Burkitt lymphoma 1.
MCL was originally considered to be derived from naïve B cells and most tumors show unmutated variable heavy chain (VH) genes. However, several studies have shown that 10–20% of MCLs have somatically mutated immunoglobu-lin (Ig) genes, indicating that a sub-group has passed through a differentiation stage involving somatic hypermutation of VH genes [3–8]. Recently, it has been shown that using an alternative method up to 60% of the MCL were shown to have <98% homology with the germline sequence and were thus considered to carry somatically mutated Ig genes 9. In the same study it was further shown that patients with somatically hypermutated VH genes had a better survival and that intraclonal heterogeneity was common in this group. This is in contrast to previous results, where MCL cells, including the VH mutated, has been described to be clonally identical, indicating that the tumor cells are frozen at the developmental stage where the malignant transformation took place [2, 10]. Interestingly, it has been reported that B cells can mutate their VH genes in the absence of a germinal center (GC), but the mechanisms involved are at present not understood [11, 12]. In a previous study, using gene expression analysis of MCL and sub-populations of normal tonsillar B cells we concluded that MCL tumor cells had a transcriptional profile similar to activated B cells and not to na-ïve B cells 13, as previously proposed. This is in accordance with the mutational studies and support the conclusion that MCL is derived from pre-activated B cells and not from naïve B cells [4, 6, 7, 9, 10]. Furthermore, a biased VH usage has been described for MCL, which suggests that the transformation is antigen driven [4, 6, 7, 9, 10]. No antigen has been identified but VH 3–21, which is one of the most frequently used VH genes in MCL, is common in, e.g., rheumatoid factors and may thus define an auto-antigen [2, 14].
The main feature of MCL is over-expression of cyclin D1, which is due to the translocation of BCL1 to the heavy chain locus [15, 16]. This t(11;14)(q13;q32) was identified in the 1970s as a cytogenetic event occurring in rare examples of B cell non-Hodgkin’s lymphoma (NHL). Tsujimoto and colleagues cloned this translocation breakpoint in 1984 17. Cyclin D1 is a positive regulator of the G1/S cell-cycle restriction point and the over-expression induces increased cell cycling. Even if the over-expression of cyclin D1 is the main feature of MCL, it is probably not the sole oncogenic feature, as over-expression of the gene in mice is not lymphomagenic 18.
Rare MCL-like B cell lymphomas lack cyclin D1 expression, but still have the other morphological and immunophe-notypical characteristics of MCL 19. These cyclin D1-negative MCL-like lymphomas have been shown to have a better prognosis and should be considered as an entity separate from MCL [19, 20]. Other phenotypic characteristics of MCL are the expression of CD5, CD19, CD20, CD22, CD79a, CD79b and the lack of CD10 and CD23 21. The lack of CD10 and CD23 expression distinguish MCL from FL and B-CLL, respectively. MCL cells also over-express Bcl-2 but lack t(14;18) (q32:q21), which is characteristic of FL.
MCLs grow in three different patterns in lymph nodes: mantle zone, nodular and diffuse. These growth patterns can be divided further into typical or blastoid variants 21. The characteristics of the different clinical, phenotypic and genotypic features of MCL were agreed on in 1994 22. Morphologically, typical MCLs can have either a nodular or a diffuse growth pattern, with or without residual GCs, and are normally infiltrated with either loose or tight follicular den-dritic cell aggregates 23.
The median age for MCL patients is 60 years (range 18–86) at diagnosis, and the malignancy has a male predominance (3:1) 21. Although the median survival period is short, only 3 to 5 years, and very few patients show long-term survival, a few features indicate a more favorable prognosis. These features include a proliferative index below 10% and arguably, a nodular growth pattern 21. Most patients show disseminated disease at the time of diagnosis, with 80% of patients in stage III or IV, enlarged lymph nodes and frequent (70%) bone-marrow involvement. Involvement of the blood (25%) and gastrointestinal tract is also common 23, while infiltration of the central nervous system is rarely detected and may be related to blastoid transformation 24. Lack of lymph node involvement with isolated massive splenomegaly occurs in some patients, but this is infrequent 21.
MCLs may progress to a blastoid variant, as reviewed by Matolcsy 25 and Muller-Hermelink et al. 26, but this variant may also arise de novo 24. The more aggressive blastoid variant is associated with decreased survival [27, 28] and 26–70% of MCL patients have a morphological progression towards this variant, detected either during life or at autopsy [29, 30]. Identical clonally rearranged Ig genes suggest that blastoid variants arise from the original neoplasm by clonal selection 25. Factors that predict blastoid transformation are leukocytosis, elevated serum lactate dehydro-genase and high proliferative index 24.
Most patients with MCL who show response to treatment, or even total remission, soon relapse with new tumors. Thus, more effective treatments must be sought. It is also essential to minimize toxicity, as most patients afflicted by MCL are 60 years of age or more and are likely to experience treatment-related complications 31. There is no defined standard treatment for MCL. The most commonly used regimen is CHOP (cyclophosphamide, doxorubicin, vincris-tine and prednisone), usually in combination with immuno-therapy, such as rituximab [32–34]. Recent results show increased response to this combined therapy, but it does not improve the long-term outcome of MCL patients 35. Consolidation with high-dose chemotherapy and stem cell support has been shown to prolong progression-free survival after conventional chemotherapy, but at present, is not considered to be curative 36. In addition, maintenance therapy with rituximab after a combined treatment with fludarabine, cyclophosphamide, and mitoxantron shows promising results for both MCL and FL 37. Several other regimens are being evaluated, including bortezomib, a proteasome inhibitor 38 along with a number of other biological agents and combinations as reviewed elsewhere [39–41].
Furthermore, MCL cells are highly sensitivity to radiation 40 but since the malignancy most often occurs systemically, this is in most cases not a treatment option. However, in 10–15% of MCL patients, limited stage (I/II) disease is diagnosed. Since these patients only have a few localized tumors, regional radiation therapy has been shown to be a potentially curative treatment 34.
Thus, MCL is far from a homologous entity and even though thumors share many characteristics, they can potentially be divided into sub-groups. Novel technologies, including various microarrays, can be used to identify the molecular differences involved in morphological heterogeneity, clinical behavior and mutational status. These approaches may form the basis to define different therapies tailored to each sub-type of MCL or to find common features useful for improved therapeutic interventions.
GENE EXPRESSION IN LYMPHOMA – DOGMAS AS BASIS FOR ANALYSIS
Highly parallel gene expression analysis has during the last 6–7 years revolutionized the way we look at the different malignant lymphoma entities. It has been clearly shown that the heterogeneity seen in response to treatment and clinical outcome can be correlated to differential gene usage. Global gene expression profiling was also shown early on to be useful for facilitating diagnosis, enabling separation of similar but distinct entities such as acute myeloid leukemia and acute lymphoblastic leukemia (AML-ALL) 42, as well as morphologically similar but clinically different subgroups, such as VH mutated vs. unmutated CLL 43. A recent gene expression study on FL, using whole tumor tissue, concluded that certain genes, such as EphA1 and Smad1, expressed by non-malignant cells in the microenvironment, including granulocytes and vascular structures, affect treatment outcome for patients given Rituximab and CHOP 44. for MCL, different studies have focused on the implications of the overexpression of cyclin D1. Fu et al. 45 used gene expression analysis to show that cyclin D1-negative MCL have similar profiles compared to cyclin D1 positive MCL. In another study, the correlation between a short variant of the cyclin D1 mRNA transcript and increased proliferation in MCL was studied 46 and a third gene expression study demonstrated that cyclin D1 levels are associated with changes in proliferative activity 47. It has further been shown that survival of MCL patients can be predicted, using gene expression analysis. Rosenwald et al. constructed a proliferative signature, based on the Lymphochip, that could predict survival in MCL patients 20. Analyses of MCL have also been performed to define the main features regulating MCL growth, i.e., the apoptotic machinery and genes involved in cell survival [13, 48, 49]. Other studies of MCL have, for example, revealed differences in gene usage, comparing proliferative indexes 50 and difference in morphological subtypes 51, as well as focusing on markers, such as chemokine usage 52 or expression of the cannabinoid receptor 47. It has further been shown that ATM and p53 mutational status can be associated with distinct gene expression signatures, although only p53 mutations could be associated with decreased survival time 53. The leukemic variant of MCL has also been analyzed 54 and MCL cell lines have been assessed as tools for in vitro studies 55. Lately, the technology has also been used in parallel with global genomic analysis to define potential therapeutic targets in MCL 56.
Analysis of B cell lymphomas has centered around the belief that the malignant cell retains most of the characteristics of its benign precursor. Thus, the stage of B cell differentiation at which the malignant transformation took place will influence the behavior of the transformed daughter cells. Consequently, the finding by Alizadeh et al. in 2000 based on gene expression analysis 57, which showed that different subtypes of DLBCL were associated with different normal B cell populations, had a significant impact, not only on the approach to sub-grouping of DLBCL but also on how other B cell lymphomas can be sub-grouped. Knowledge of the normal counterpart will therefore directly facilitate the design of possible methods of intervention, since it pinpoints the events in normal B cell differentiation where the cell is more vulnerable to escaping homeostasis and immune regulation.
Initially, many studies have been performed using focused arrays. Examples are the Lymphochip, used by Staudt and colleagues in analysis of different types of lymphoma [20, 57–60], as well as the Oncochip-CNIO, used by Martinez and colleagues when analyzing MCL 49. In these cases, genes known to be involved in the immune system or related to cancer have been used. In contrast, other arrays, such as the oligonucleotide arrays from Affymetrix, have not been focused towards any groups of genes. Today, when it is possible to scan the whole genome, including uncharacter-ized transcripts, there is no longer any real advantage to focus on a limited number of genes for the initial analysis.
EXPERIMENTAL DESIGN AND ANALYTICAL STRATEGIES – IS THE BEST THERAPEUTIC TARGET A NON-B CELL ANTIGEN
Since gene expression analysis is a comparative technology, the choice of reference material is crucial for the ability to draw strict conclusions. When correlating tumor material to different pathological or clinical parameters, the material is its own reference, as for example when (i) analyzing MCL with and without the blastic variant [51, 61], (ii) correlating gene expression to survival 20 or (iii) comparing gene expression profiles between similar lymphoma entities 62. However, when aiming at identifying potential therapeutic targets, the choice of reference material becomes more difficult. The most important parameter, when selecting a reference, is the composition of the tumor and reference material. The heterogeneity of most archived material makes analysis of the actual gene expression of the malignant cells impossible since it is the sum of the different cell types in the tumor that contributes to the identified signatures. The microenvi-ronment is a key factor for the survival of the tumor and it is clear that both cells and soluble factors influence this milieu and will have a great impact on the analysis, as exemplified by the above gene expression study on FL by Harjunpaa et al. 44. It has already been suggested that the outcome of patients afflicted by FL, in comparison with MCL and DLBCL, is more dependent on the features of the microenvi-ronment than the status of the tumor cells themselves [63, 64]. We have also previously shown that MCL display surface receptors that most likely are involved in cross-talk with neighboring immune cells, such as follicular dendritic cells and that an altered chemokine production may confer survival advantages to the tumor cells 52. Consequently, to be able to make conclusive statements regarding the nature of either the tumor cells or cells in the microenvironment, these players need to be analyzed separately, which today is too seldom performed due to lack of adequate tumor material.
With this in mind, which is then the best choice of reference material, when aiming to identify therapeutic targets? Most studies use normal B cells, which is a natural choice, they being the ancestor cell. Other examples of different material used for gene expression analysis are (i) hyperplastic lymph nodes, used as reference material in comparison with lymph nodes from patients with MCL 48, (ii) sorted tonsillar B cell subpopulations 13 or (iii) naïve B cells 49. These analyses, comparing malignant and non-malignant B cells, aim at defining differentially expressed gene products. Does it make a difference if the target then is a non-B cell antigen? Rituximab, targeting CD20, has successfully been used in different B cell malignancies, although the effect on MCL patients has been more limited compared to, for example, patients with FL 37. Other examples of Food and Drug Administration (FDA) approved antibodies, targeting both malignant and non-malignant lymphoid cells, include Bexxar (Tositumomab and I-131 Tositumomab), which like Rituxi-mab targets CD20, Campath (Alemtuzumab), targeting CD52 on B cell chronic lymphocytic leukemia and Mylotarg (gemtuzumab ozogamicin), targeting CD33 positive cells in acute myeloid leukemia (www.accessdata.fda.gov/scripts/ cder/drgsatfda/index.cfm). Although effective to different degrees, it is obvious that it is more beneficial for the patient to retain an intact B cell pool. This is especially crucial for immunodeficient patients. B cell malignancies, like other types of cancer, frequently affect individuals with an impaired immune system, such as patients suffering from acquired immunodeficiency syndrome (AIDS) or patients treated with immunosuppressive agents. The role of Epstein Barr virus infection for development of NHL in immunosup-pressed individuals, for example after transplantation, and in individuals infected by human immunodeficiency virus (HIV), is particularly well documented 65. Other biological regimens like bortezomib, a proteasome inhibitor, are not specific for B cells but still show promising results 38.
Consequently, it remains to be seen whether B cell lym-phoma-associated antigens, not present on non-malignant B cells, will be more efficient as targets for new antibody-mediated therapies. However, a larger variety of reference material and strategies must be considered to enable a focused analysis for the identification of suitable targets on the malignant cells or on surrounding cells that facilitate tumor growth and survival.
FROM GENOMIC TO PROTEOMIC ANALYSIS IN MANTLE CELL LYMPHOMA – HOW FAR ARE WE FROM DEFINING NEW DIAGNOSTIC OR THERAPEUTIC TARGETS
Conventional chromosomal comparative genomic hybridization (CGH) has been used to identify common MCL-associated cytogenetic abnormalities [66–69]. Recently, high-resolution microarray-based CGH assays have been performed on a number of both primary MCL primary samples, as well as MCL cell lines [70–74]. Similar to gene expression analysis, genetic abnormalities were shown to be associated with the clinical outcome of MCL patients [20, 72] and were correlated to clinical sub-groups and parameters. Only a few genomic studies of MCL have been correlated with gene expression data [56, 73] and conversely, gene expression studies most often lack parallel analysis of the underlying genomic alterations. However, for gene expression analysis aimed at defining new therapeutic antigens, correlation at the genomic level may be of less importance than correlation at the proteome level. Despite this, the bulk of MCL data generated on expression arrays remains to be converted into information on the corresponding proteins.
The standard methods for analyzing proteins which correspond to known differentially regulated genes are immu-nohistochemistry (IHC) and Western blot, using commercially available antibodies. Since paraffin-embedded tumor tissues are often readily available in biobanks IHC analysis is thus the most frequently used method. However, IHC requires that the antigen is resistant to the harsh fixation and treatments used in histotechnology and consequently some antigens are difficult or impossible to analyze using this method 75. Western blot, on the other hand, suffers from other limitations. In Western blot analysis, separation of the different cell types in the tumor tissue needs initially to be performed, to be able to pinpoint the cell type carrying the protein of interest. In most cases, such fresh tumor samples are difficult to collect. Furthermore, both techniques are limited by the availability of suitable commercial antibodies. In a recent study, we have used tailor-made antibodies generated within the Human Proteome Resource (HPR) project [76, 77] to transfer our gene expression data to the proteome level 78. In short, gene transcripts upregulated in MCL tumors were used to generate antibodies. In subsequent IHC analysis, a large fraction of these antibodies recognized MCL tissue but not benign B cells. Several such antibodies are being further characterized using large scale tissue mi-coarrays with an aim to define antigens useful for improved diagnostics or therapy (Ek et al., manuscript in preparation).
A few de novo parallel proteome analyses of MCL have been performed, for example using commercial antibody microarrays 79 or two-dimensional gel electrophoresis, in MCL cell lines (GRANTA-519 and MAVER-1) or excised tissue, respectively [80, 81]. However, the number of proteins that can be analyzed is limited compared to parallel gene expression profiling. The antibody microarray analyzed 512 antibodies, whereas the 2D gel electrophoresis could identify 750 spots in the MCL tumor material. High-density antibody-based microarrays may offer a way to efficiently transfer gene expression data into proteomics, but this technique like IHC and Western blot requires the availability of suitable antibodies. The generation of tailor-made antibodies designed for microarray applications is on-going and uses large recombinant antibody libraries, which will eventually offer an unbiased analysis of novel gene products 82. A summary of the process where gene expression data is used to focus interest on antigens involved in a specific malignancy and how these targets can be validated at the protein level and in functional analysis is shown in Fig. (1). As discussed above, there is a clear need for specific tailor-made reagents capable of validating all findings without discrimination.
Fig. (1).
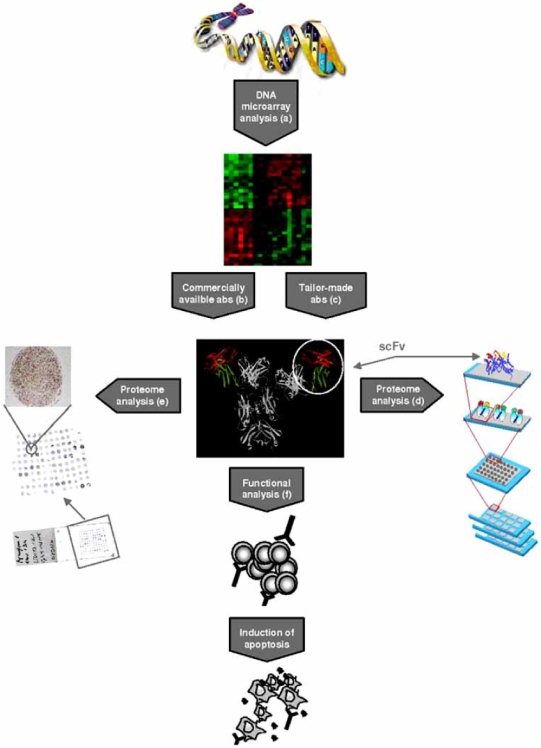
Bioinformatic analysis of DNA* microarray data from malignant and normal cells identifies differentially expressed target genes, as illustrated by (a) the heat map, where genes upregulated in malignant cells are shown in red and downregulated genes are shown in green. (b) Downstream analysis is often focused on a limited number of targets for which commercially antibodies (abs) are available. (c) Tailor-made antibodies allow downstream analysis of all identified gene products. Antibodies or antibody-fragments (scFvs) can be used in various applications, for example (d) antibody microarrays, where different materials such as plasma, whole cells or tissue extracts can be analyzed, or on (e) tissue microarrays, where a large number of different tissues is analyzed on a single slide or (f) for functional analysis, where the therapeutic potential can be assessed.
*The DNA illustration is adapted from http://genomics.energy.gov with permission from the U.S. Department of Energy Genome Program. The antibody illustration was created by Mats Ohlin from the B12 antibody (1HZH, www.pdb.org). The antibody microarray illustration was kindly provided by Christer Wingren.
To finally make sense of gene and protein expression data, relevant in vivo models for functional analysis of the identified potential targets are needed. Murine models have until recently been lacking, except for a xenograft using the UPN1 cell line 83, but in 2006 two separate models were presented in which MCL-like tumors could be induced using mitogenic stimulation in Eμ-cyclin D1 carrying transgenic mice 84. In a second study, a mutant form of cyclin D1 prevented export from the nucleus, thus showing that constitutively nuclear cyclin D1 induces B-cell lymphomas in transgenic mice 85. Further evaluation of these models and there utility in MCL research is needed.
In summary, we conclude that although extensive research on MCL using gene expression analysis has been performed, parallel, unbiased tools to transfer the data to the protein level needs to be implemented to facilitate the transfer of the new findings into the clinic. These tools include combinations of high-throughput affinity proteomics, such as antibody and tissue microarrays, which recently have shown great promise in defining new and improved approaches to facilitate diagnosis, treatment decisions and in the long term may potentially contribute to improved treatment outcome for patients.
ACKNOWLEDGEMENTS
This review is partly based on studies supported by grants from the Leukemia and Lymphoma Society (USA, grant no. 6085-06) and from CREATE Health, a Strategic Center for Translational Cancer Research (www.createhealth. lth.se). We would also like to thank Dr. Michael Dictor and Dr. Mats Jerkeman for valuable comments on the manuscript.
ABBREVIATIONS
- AIDS
Acquired immunodeficiency syndrome
- ALL
Acute lymphoblastic leukemia
- AML
Acute myeloid leukemia
- CHOP
Cyclophosphamide, doxorubicin, vincristine and prednisone
- CLL
Chronic Lymphocytic Leukemia
- CGH
Comparative genomic hybridization
- DLBCL
Diffuse large B cell lymphoma
- FDA
Food and Drug Administration
- FL
Follicular lymphoma
- GC
Germinal center
- HIV
Human immunodeficiency virus
- HPR
Human Proteome Resource
- Ig
Immunoglobulin
- IHC
Immunohistochemistry
- MCL
Mantle cell lymphoma
- MZ
Marginal zone
- MALT
Mucosa associated lymphoid tissue
- NHL
Non Hodgkin’s lymphoma
- VH
Variable heavy chain
REFERENCES
- 1.Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD. The World Health Organization classification of hematological malignancies report of the Clinical Advisory Committee Meeting, Airlie House, Virginia, November 1997. Mod Pathol. 2000;13:193–207. doi: 10.1038/modpathol.3880035. [DOI] [PubMed] [Google Scholar]
- 2.Shaffer AL, Rosenwald A, Staudt LM. Lymphoid malignancies: the dark side of B-cell differentiation. Nat Rev Immunol. 2002;2:920–32. doi: 10.1038/nri953. [DOI] [PubMed] [Google Scholar]
- 3.Welzel N, Le T, Marculescu R, Mitterbauer G, Chott A, Pott C, Kneba M, Du MQ, Kusec R, Drach J, Raderer M, Mannhalter C, Lechner K, Nadel B, Jaeger U. Templated nucleo-tide addition and immunoglobulin JH-gene utilization in t(11;14) junctions: implications for the mechanism of translocation and the origin of mantle cell lymphoma. Cancer Res. 2001;61:1629–36. [PubMed] [Google Scholar]
- 4.Pittaluga S, Tierens A, Pinyol M, Campo E, Delabie J, De Wolf-Peeters C. Blastic variant of mantle cell lymphoma shows a heterogenous pattern of somatic mutations of the rearranged immunoglobulin heavy chain variable genes. Br J Haematol. 1998;102:1301–6. doi: 10.1046/j.1365-2141.1998.00907.x. [DOI] [PubMed] [Google Scholar]
- 5.Laszlo T, Nagy M, Kelenyi G, Matolcsy A. Immunoglobulin V(H) gene mutational analysis suggests that blastic variant of mantle cell lymphoma derives from different stages of B-cell maturation. Leuk Res. 2000;24:27–31. doi: 10.1016/s0145-2126(99)00156-3. [DOI] [PubMed] [Google Scholar]
- 6.Thorselius M, Walsh S, Eriksson I, Thunberg U, Johnson A, Backlin C, Enblad G, Sundstrom C, Roos G, Rosenquist R. Somatic hypermutation and V(H) gene usage in mantle cell lymphoma. Eur J Haematol. 2002;68:217–24. doi: 10.1034/j.1600-0609.2002.01662.x. [DOI] [PubMed] [Google Scholar]
- 7.Camacho FI, Algara P, Rodriguez A, Ruiz-Ballesteros E, Mollejo M, Martinez N, Martinez-Climent JA, Gonzalez M, Mateo M, Caleo A, Sanchez-Beato M, Menarguez J, Garcia-Conde J, Sole F, Campo E, Piris MA. Molecular heterogeneity in MCL, defined by the use of specific VH genes and the frequency of somatic mutations. Blood. 2003;101:4042–46. doi: 10.1182/blood-2002-11-3456. [DOI] [PubMed] [Google Scholar]
- 8.Kienle D, Krober A, Katzenberger T, Ott G, Leupolt E, Barth TF, Moller P, Benner A, Habermann A, Muller-Hermelink HK, Bentz M, Lichter P, Dohner H, Stilgenbauer S. VH mutation status and VDJ rearrangement structure in mantle cell lymphoma: correlation with genomic aberrations, clinical characteristics, and outcome. Blood. 2003;102:3003–9. doi: 10.1182/blood-2003-05-1383. [DOI] [PubMed] [Google Scholar]
- 9.Lai R, Lefresne SV, Franko B, Hui D, Mirza I, Mansoor A, Amin HM, Ma Y. Immunoglobulin VH somatic hypermutation in mantle cell lymphoma: mutated genotype correlates with better clinical outcome. Mod Pathol. 2006;19:1498–505. doi: 10.1038/modpathol.3800677. [DOI] [PubMed] [Google Scholar]
- 10.Walsh SH, Thorselius M, Johnson A, Soderberg O, Jerkeman M, Bjorck E, Eriksson I, Thunberg U, Landgren O, Ehinger M, Lofvenberg E, Wallman K, Enblad G, Sander B, Porwit-MacDonald A, Dictor M, Olofsson T, Sundstrom C, Roos G, Rosenquist R. Mutated VH genes and preferential VH3-21 use define new subsets of mantle cell lymphoma. Blood. 2003;101:4047–54. doi: 10.1182/blood-2002-11-3479. [DOI] [PubMed] [Google Scholar]
- 11.Weller S, Faili A, Garcia C, Braun MC, Le Deist FF, de Saint Basile GG, Hermine O, Fischer A, Reynaud CA, Weill JC. CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc Natl Acad Sci USA. 2001;98:1166–70. doi: 10.1073/pnas.98.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto M, Lo SF, Carruthers CJ, Min J, Mariathasan S, Huang G, Plas DR, Martin SM, Geha RS, Nahm MH, Chaplin DD. Affinity maturation without germinal centres in lymphotoxin-alpha-deficient mice. Nature. 1996;382:462–6. doi: 10.1038/382462a0. [DOI] [PubMed] [Google Scholar]
- 13.Ek S, Hogerkorp CM, Dictor M, Ehinger M, Borrebaeck CAK. Mantle cell lymphomas express a distinct genetic signature affecting lymphocyte trafficking and growth regulation as compared with subpopulations of normal human B cells. Cancer Res. 2002;62:4398–405. [PubMed] [Google Scholar]
- 14.He X, Goronzy JJ, Zhong W, Xie C, Weyand CM. VH3-21 B cells escape from a state of tolerance in rheumatoid arthritis and secrete rheumatoid factor. Mol Med. 1995;1:768–80. [PMC free article] [PubMed] [Google Scholar]
- 15.Stacey DW. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol. 2003;15:158–63. doi: 10.1016/s0955-0674(03)00008-5. [DOI] [PubMed] [Google Scholar]
- 16.Donnellan R, Chetty R. Cyclin D1 and human neoplasia. Mol Pathol. 1998;51:1–7. doi: 10.1136/mp.51.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsujimoto Y, Yunis J, Onorato-Showe L, Erikson J, Nowell PC, Croce CM. Molecular cloning of the chromosomal breakpoint of B-cell lymphomas and leukemias with the t(11;14) chromosome translocation. Science. 1984;224:1403–6. doi: 10.1126/science.6610211. [DOI] [PubMed] [Google Scholar]
- 18.Lovec H, Grzeschiczek A, Kowalski MB, Moroy T. Cyclin D1/bcl-1 cooperates with myc genes in the generation of B-cell lymphoma in transgenic mice. EMBO J. 1994;13:3487–95. doi: 10.1002/j.1460-2075.1994.tb06655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yatabe Y, Suzuki R, Tobinai K, Matsuno Y, Ichinohasama R, Okamoto M, Yamaguchi M, Tamaru J, Uike N, Hashimoto Y, Morishima Y, Suchi T, Seto M, Nakamura S. Significance of cyclin D1 overexpression for the diagnosis of mantle cell lymphoma: a clinicopathologic comparison of cyclin D1-positive MCL and cyclin D1-negative MCL-like B-cell lymphoma. Blood. 2000;95:2253–61. [PubMed] [Google Scholar]
- 20.Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink HK, Smeland EB, Chiorazzi M, Giltnane JM, Hurt EM, Zhao H, Averett L, Henrickson S, Yang L, Powell J, Wilson WH, Jaffe ES, Simon R, Klausner RD, Montserrat E, Bosch F, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Fisher RI, Miller TP, LeBlanc M, Ott G, Kvaloy S, Holte H, Delabie J, Staudt LM. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–97. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 21.Kurtin PJ. Mantle cell lymphoma. Adv Anat Pathol. 1998;5:376–98. doi: 10.1097/00125480-199811000-00011. [DOI] [PubMed] [Google Scholar]
- 22.Zucca E, Stein H, Coiffier B. European Lymphoma Task Force (ELTF): Report of the workshop on Mantle Cell Lymphoma (MCL) Ann Oncol. 1994;5:507–11. doi: 10.1093/oxfordjournals.annonc.a058904. [DOI] [PubMed] [Google Scholar]
- 23.Swerdlow SH, Williams ME. From centrocytic to mantle cell lymphoma: a clinicopathologic and molecular review of 3 decades. Hum Pathol. 2002;33:7–20. doi: 10.1053/hupa.2002.30221. [DOI] [PubMed] [Google Scholar]
- 24.Raty R, Franssila K, Jansson SE, Joensuu H, Wartiovaara-Kautto U, Elonen E. Predictive factors for blastoid transformation in the common variant of mantle cell lymphoma. Eur J Cancer. 2003;39:321–9. doi: 10.1016/s0959-8049(02)00456-2. [DOI] [PubMed] [Google Scholar]
- 25.Matolcsy A. High-grade transformation of low-grade non-Hodgkin’s lymphomas: mechanisms of tumor progression. Leuk Lymphoma. 1999;34:251–9. doi: 10.3109/10428199909050950. [DOI] [PubMed] [Google Scholar]
- 26.Muller-Hermelink HK, Zettl A, Pfeifer W, Ott G. Pathology of lymphoma progression. Histopathology. 2001;38:285–306. doi: 10.1046/j.1365-2559.2001.01120.x. [DOI] [PubMed] [Google Scholar]
- 27.Lardelli P, Bookman MA, Sundeen J, Longo DL, Jaffe ES. Lymphocytic lymphoma of intermediate differentiation. Morphologic and immunophenotypic spectrum and clinical correlations. Am J Surg Pathol. 1990;14:752–63. doi: 10.1097/00000478-199008000-00007. [DOI] [PubMed] [Google Scholar]
- 28.Raty R, Franssila K, Joensuu H, Teerenhovi L, Elonen E. Ki-67 expression level, histological subtype, and the International Prognostic Index as outcome predictors in mantle cell lymphoma. Eur J Haematol. 2002;69:11–20. doi: 10.1034/j.1600-0609.2002.01677.x. [DOI] [PubMed] [Google Scholar]
- 29.Norton AJ, Matthews J, Pappa V, Shamash J, Love S, Rohatiner AZ, Lister TA. Mantle cell lymphoma: natural history defined in a serially biopsied population over a 20-year period. Ann Oncol. 1995;6:249–56. doi: 10.1093/oxfordjournals.annonc.a059154. [DOI] [PubMed] [Google Scholar]
- 30.Weisenburger DD, Vose JM, Greiner TC, Lynch JC, Chan WC, Bierman PJ, Dave BJ, Sanger WG, Armitage JO. Mantle cell lymphoma. A clinicopathologic study of 68 cases from the Nebraska Lymphoma Study Group. Am J Hematol. 2000;64:190–6. doi: 10.1002/1096-8652(200007)64:3<190::aid-ajh9>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 31.Porcu P, Nichols CR. Evaluation and management of the “new” lymphoma entities: mantle cell lymphoma, lymphoma of mucosa-associated lymphoid tissue, anaplastic large-cell lymphoma, and primary mediastinal B-cell lymphoma. Curr Probl Cancer. 1998;22:283–368. doi: 10.1016/s0147-0272(98)90003-4. [DOI] [PubMed] [Google Scholar]
- 32.Howard OM, Gribben JG, Neuberg DS, Grossbard M, Poor C, Janicek MJ, Shipp MA. Rituximab and CHOP induction therapy for newly diagnosed mantle-cell lymphoma: molecular complete responses are not predictive of progression-free survival. J Clin Oncol. 2002;20:1288–94. doi: 10.1200/JCO.2002.20.5.1288. [DOI] [PubMed] [Google Scholar]
- 33.Boye J, Elter T, Engert A. An overview of the current clinical use of the anti-CD20 monoclonal antibody rituximab. Ann Oncol. 2003;14:520–35. doi: 10.1093/annonc/mdg175. [DOI] [PubMed] [Google Scholar]
- 34.Hiddemann W, Dreyling M. Mantle cell lymphoma: therapeutic strategies are different from CLL. Curr Treat Options Oncol. 2003;4:219–26. doi: 10.1007/s11864-003-0023-x. [DOI] [PubMed] [Google Scholar]
- 35.Lenz G, Dreyling M, Hoster E, Wormann B, Duhrsen U, Metzner B, Eimermacher H, Neubauer A, Wandt H, Steinhauer H, Martin S, Heidemann E, Aldaoud A, Parwaresch R, Hasford J, Unterhalt M, Hiddemann W. Immunochemotherapy with rituximab and cyclophosphamide, doxorubicin, vincristine, and prednisone significantly improves response and time to treatment failure, but not long-term outcome in patients with previously untreated mantle cell lymphoma: results of a prospective randomized trial of the German Low Grade Lymphoma Study Group (GLSG) J Clin Oncol. 2005;23:1984–92. doi: 10.1200/JCO.2005.08.133. [DOI] [PubMed] [Google Scholar]
- 36.Dreyling M, Lenz G, Hoster E, Van Hoof A, Gisselbrecht C, Schmits R, Metzner B, Truemper L, Reiser M, Steinhauer H, Boiron JM, Boogaerts MA, Aldaoud A, Silingardi V, Kluin-Nelemans HC, Hasford J, Parwaresch R, Unterhalt M, Hiddemann W. Early consolidation by myeloablative radiochemotherapy followed by autologous stem cell transplantation in first remission significantly prolongs progression-free survival in mantle-cell lymphoma: results of a prospective randomized trial of the European MCL Network. Blood. 2005;105:2677–84. doi: 10.1182/blood-2004-10-3883. [DOI] [PubMed] [Google Scholar]
- 37.Forstpointner R, Unterhalt M, Dreyling M, Bock HP, Repp R, Wandt H, Pott C, Seymour JF, Metzner B, Hanel A, Lehmann T, Hartmann F, Einsele H, Hiddemann W. Maintenance therapy with rituximab leads to a significant prolongation of response duration after salvage therapy with a combination of rituximab, fludarabine, cyclophosphamide, and mitoxantrone (R-FCM) in patients with recurring and refractory follicular and mantle cell lymphomas: Results of a prospective randomized study of the German Low Grade Lymphoma Study Group (GLSG) Blood. 2006;108:4003–8. doi: 10.1182/blood-2006-04-016725. [DOI] [PubMed] [Google Scholar]
- 38.Fisher RI, Bernstein SH, Kahl BS, Djulbegovic B, Robertson MJ, de Vos S, Epner E, Krishnan A, Leonard JP, Lonial S, Stadtmauer EA, O’Connor OA, Shi H, Boral AL, Goy A. Multicenter phase II study of bortezomib in patients with relapsed or refractory mantle cell lymphoma. J Clin Oncol. 2006;24:4867–74. doi: 10.1200/JCO.2006.07.9665. [DOI] [PubMed] [Google Scholar]
- 39.Brody J, Advani R. Treatment of mantle cell lymphoma: current approach and future directions. Crit Rev Oncol Hematol. 2006;58:257–65. doi: 10.1016/j.critrevonc.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Zelenetz AD. Mantle cell lymphoma: an update on management. Ann Oncol. 2006;17(4):iv12–4. doi: 10.1093/annonc/mdj992. [DOI] [PubMed] [Google Scholar]
- 41.Goy A. New directions in the treatment of mantle cell lymphoma: an overview. Clin Lymphoma Myeloma. 2006;7(Suppl 1):S24–32. doi: 10.3816/clm.2006.s.005. [DOI] [PubMed] [Google Scholar]
- 42.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 43.Rosenwald A, Alizadeh AA, Widhopf G, Simon R, Davis RE, Yu X, Yang L, Pickeral OK, Rassenti LZ, Powell J, Botstein D, Byrd JC, Grever MR, Cheson BD, Chiorazzi N, Wilson WH, Kipps TJ, Brown PO, Staudt LM. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639–47. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harjunpaa A, Taskinen M, Nykter M, Karjalainen-Lindsberg ML, Nyman H, Monni O, Hemmer S, Yli-Harja O, Hautaniemi S, Meri S, Leppa S. Differential gene expression in non-malignant tumour microenvironment is associated with out-come in follicular lymphoma patients treated with rituximab and CHOP. Br J Haematol. 2006;135:33–42. doi: 10.1111/j.1365-2141.2006.06255.x. [DOI] [PubMed] [Google Scholar]
- 45.Fu K, Weisenburger DD, Greiner TC, Dave S, Wright G, Rosenwald A, Chiorazzi M, Iqbal J, Gesk S, Siebert R, De Jong D, Jaffe ES, Wilson WH, Delabie J, Ott G, Dave BJ, Sanger WG, Smith LM, Rimsza L, Braziel RM, Muller-Hermelink HK, Campo E, Gascoyne RD, Staudt LM, Chan WC. Cyclin D1-negative mantle cell lymphoma: a clinico-pathologic study based on gene expression profiling. Blood. 2005;106:4315–21. doi: 10.1182/blood-2005-04-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sander B, Flygare J, Porwit-Macdonald A, Smith CI, Emanuelsson E, Kimby E, Liden J, Christensson B. Mantle cell lymphomas with low levels of cyclin D1 long mRNA transcripts are highly proliferative and can be discriminated by elevated cyclin A2 and cyclin B1. Int J Cancer. 2005;117:418–30. doi: 10.1002/ijc.21166. [DOI] [PubMed] [Google Scholar]
- 47.Islam TC, Asplund AC, Lindvall JM, Nygren L, Liden J, Kimby E, Christensson B, Smith CI, Sander B. High level of cannabinoid receptor 1, absence of regulator of G protein signalling 13 and differential expression of Cyclin D1 in mantle cell lymphoma. Leukemia. 2003;17:1880–90. doi: 10.1038/sj.leu.2403057. [DOI] [PubMed] [Google Scholar]
- 48.Hofmann WK, de Vos S, Tsukasaki K, Wachsman W, Pinkus GS, Said JW, Koeffler HP. Altered apoptosis pathways in mantle cell lymphoma detected by oligonucleotide microarray. Blood. 2001;98:787–94. doi: 10.1182/blood.v98.3.787. [DOI] [PubMed] [Google Scholar]
- 49.Martinez N, Camacho FI, Algara P, Rodriguez A, Dopazo A, Ruiz-Ballesteros E, Martin P, Martinez-Climent JA, Garcia-Conde J, Menarguez J, Solano F, Mollejo M, Piris MA. The molecular signature of mantle cell lymphoma reveals multiple signals favoring cell survival. Cancer Res. 2003;63:8226–32. [PubMed] [Google Scholar]
- 50.Ek S, Bjorck E, Porwit-MacDonald A, Nordenskjold M, Borrebaeck CAK. Increased expression of Ki-67 in mantle cell lymphoma is associated with de-regulation of several cell cycle regulatory components, as identified by global gene expression analysis. Haematologica. 2004;89:686–95. [PubMed] [Google Scholar]
- 51.Zhu Y, Hollmen J, Raty R, Aalto Y, Nagy B, Elonen E, Kere J, Mannila H, Franssila K, Knuutila S. Investigatory and analytical approaches to differential gene expression profiling in mantle cell lymphoma. Br J Haematol. 2002;119:905–15. doi: 10.1046/j.1365-2141.2002.03931.x. [DOI] [PubMed] [Google Scholar]
- 52.Ek S, Bjorck E, Hogerkorp CM, Nordenskjold M, Porwit-MacDonald A, Borrebaeck CAK. Mantle cell lymphomas acquire increased expression of CCL4, CCL5 and 4-1BB-L implicated in cell survival. Int J Cancer. 2006;118:2092–7. doi: 10.1002/ijc.21579. [DOI] [PubMed] [Google Scholar]
- 53.Greiner TC, Dasgupta C, Ho VV, Weisenburger DD, Smith LM, Lynch JC, Vose JM, Fu K, Armitage JO, Braziel RM, Campo E, Delabie J, Gascoyne RD, Jaffe ES, Muller-Hermelink HK, Ott G, Rosenwald A, Staudt LM, Im MY, Karaman MW, Pike BL, Chan WC, Hacia JG. Mutation and genomic deletion status of ataxia telangiectasia mutated (ATM) and p53 confer specific gene expression profiles in mantle cell lymphoma. Proc Natl Acad Sci USA. 2006;103:2352–7. doi: 10.1073/pnas.0510441103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rizzatti EG, Falcao RP, Panepucci RA, Proto-Siqueira R, Anselmo-Lima WT, Okamoto OK, Zago MA. Gene expression profiling of mantle cell lymphoma cells reveals aberrant expression of genes from the PI3K-AKT, WNT and TGFbeta signalling pathways. Br J Haematol. 2005;130:516–26. doi: 10.1111/j.1365-2141.2005.05630.x. [DOI] [PubMed] [Google Scholar]
- 55.Ek S, Ortega E, Borrebaeck CAK. Transcriptional profiling and assessment of cell lines as in vitro models for mantle cell lymphoma. Leuk Res. 2005;29:205–13. doi: 10.1016/j.leukres.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 56.Rinaldi A, Kwee I, Taborelli M, Largo C, Uccella S, Martin V, Poretti G, Gaidano G, Calabrese G, Martinelli G, Baldini L, Pruneri G, Capella C, Zucca E, Cotter FE, Cigudosa JC, Catapano CV, Tibiletti MG, Bertoni F. Genomic and expression profiling identifies the B-cell associated tyrosine kinase Syk as a possible therapeutic target in mantle cell lymphoma. Br J Haematol. 2006;132:303–16. doi: 10.1111/j.1365-2141.2005.05883.x. [DOI] [PubMed] [Google Scholar]
- 57.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Staudt LM, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 58.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, Lopez-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–47. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 59.Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, Chan WC, Zhao T, Haioun C, Greiner TC, Weisenburger DD, Lynch JC, Vose J, Armitage JO, Smeland EB, Kvaloy S, Holte H, Delabie J, Campo E, Montserrat E, Lopez-Guillermo A, Ott G, Muller-Hermelink HK, Connors JM, Braziel R, Grogan TM, Fisher RI, Miller TP, LeBlanc M, Chiorazzi M, Zhao H, Yang L, Powell J, Wilson WH, Jaffe ES, Simon R, Klausner RD, Staudt LM. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med. 2003;198:851–62. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2003;100:9991–6. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Vos S, Krug U, Hofmann WK, Pinkus GS, Swerdlow SH, Wachsman W, Grogan TM, Said JW, Koeffler HP. Cell Cycle Alterations in the Blastoid Variant of Mantle Cell Lymphoma (MCL-BV) as Detected by Gene Expression Profiling of Mantle Cell Lymphoma (MCL) and MCL-BV. Diagn Mol Pathol. 2003;12:35–43. doi: 10.1097/00019606-200303000-00005. [DOI] [PubMed] [Google Scholar]
- 62.Thieblemont C, Nasser V, Felman P, Leroy K, Gazzo S, Callet-Bauchu E, Loriod B, Granjeaud S, Gaulard P, Haioun C, Traverse-Glehen A, Baseggio L, Bertucci F, Birnbaum D, Magrangeas F, Minvielle S, Avet-Loiseau H, Salles G, Coiffier B, Berger F, Houlgatte R. Small lymphocytic lymphoma, marginal zone B-cell lymphoma, and mantle cell lymphoma exhibit distinct gene-expression profiles allowing molecular diagnosis. Blood. 2004;103:2727–37. doi: 10.1182/blood-2003-06-2160. [DOI] [PubMed] [Google Scholar]
- 63.Kuppers R. Prognosis in follicular lymphoma--it’s in the microenvironment. N Engl J Med. 2004;351:2152–3. doi: 10.1056/NEJMp048257. [DOI] [PubMed] [Google Scholar]
- 64.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, Fisher RI, Braziel RM, Rimsza LM, Grogan TM, Miller TP, LeBlanc M, Greiner TC, Weisenburger DD, Lynch JC, Vose J, Armitage JO, Smeland EB, Kvaloy S, Holte H, Delabie J, Connors JM, Lansdorp PM, Ouyang Q, Lister TA, Davies AJ, Norton AJ, Muller-Hermelink HK, Ott G, Campo E, Montserrat E, Wilson WH, Jaffe ES, Simon R, Yang L, Powell J, Zhao H, Goldschmidt N, Chiorazzi M, Staudt LM. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–69. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 65.Grossbard ML. Atlas of clinical oncology. London: BC Decker Inc; 2002. Malignant lymphomas; pp. 135–151. [Google Scholar]
- 66.Bea S, Ribas M, Hernandez JM, Bosch F, Pinyol M, Hernandez L, Garcia JL, Flores T, Gonzalez M, Lopez-Guillermo A, Piris MA, Cardesa A, Montserrat E, Miro R, Campo E. Increased number of chromosomal imbalances and high-level DNA amplifications in mantle cell lymphoma are associated with blastoid variants. Blood. 1999;93:4365–74. [PubMed] [Google Scholar]
- 67.Martinez-Climent JA, Vizcarra E, Sanchez D, Blesa D, Marugan I, Benet I, Sole F, Rubio-Moscardo F, Terol MJ, Climent J, Sarsotti E, Tormo M, Andreu E, Salido M, Ruiz MA, Prosper F, Siebert R, Dyer MJ, Garcia-Conde J. Loss of a novel tumor suppressor gene locus at chromosome 8p is associated with leukemic mantle cell lymphoma. Blood. 2001;98:3479–82. doi: 10.1182/blood.v98.12.3479. [DOI] [PubMed] [Google Scholar]
- 68.Monni O, Oinonen R, Elonen E, Franssila K, Teerenhovi L, Joensuu H, Knuutila S. Gain of 3q and deletion of 11q22 are frequent aberrations in mantle cell lymphoma. Genes Chromosomes Cancer. 1998;21:298–307. doi: 10.1002/(sici)1098-2264(199804)21:4<298::aid-gcc3>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 69.Bentz M, Plesch A, Bullinger L, Stilgenbauer S, Ott G, Muller-Hermelink HK, Baudis M, Barth TF, Moller P, Lichter P, Dohner H. t(11;14)-positive mantle cell lymphomas exhibit complex karyotypes and share similarities with B-cell chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2000;27:285–94. [PubMed] [Google Scholar]
- 70.Schraders M, Pfundt R, Straatman HM, Janssen IM, van Kessel AG, Schoenmakers EF, van Krieken JH, Groenen PJ. Novel chromosomal imbalances in mantle cell lymphoma detected by genome-wide array-based comparative genomic hybridization. Blood. 2005;105:1686–93. doi: 10.1182/blood-2004-07-2730. [DOI] [PubMed] [Google Scholar]
- 71.Tagawa H, Karnan S, Suzuki R, Matsuo K, Zhang X, Ota A, Morishima Y, Nakamura S, Seto M. Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene. 2005;24:1348–58. doi: 10.1038/sj.onc.1208300. [DOI] [PubMed] [Google Scholar]
- 72.Rubio-Moscardo F, Climent J, Siebert R, Piris MA, Martin-Subero JI, Nielander I, Garcia-Conde J, Dyer MJ, Terol MJ, Pinkel D, Martinez-Climent JA. Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood. 2005;105:4445–54. doi: 10.1182/blood-2004-10-3907. [DOI] [PubMed] [Google Scholar]
- 73.de Leeuw RJ, Davies JJ, Rosenwald A, Bebb G, Gascoyne RD, Dyer MJ, Staudt LM, Martinez-Climent JA, Lam WL. Comprehensive whole genome array CGH profiling of mantle cell lymphoma model genomes. Hum Mol Genet. 2004;13:1827–37. doi: 10.1093/hmg/ddh195. [DOI] [PubMed] [Google Scholar]
- 74.Kohlhammer H, Schwaenen C, Wessendorf S, Holzmann K, Kestler HA, Kienle D, Barth TF, Moller P, Ott G, Kalla J, Radlwimmer B, Pscherer A, Stilgenbauer S, Dohner H, Lichter P, Bentz M. Genomic DNA-chip hybridization in t(11;14)-positive mantle cell lymphomas shows a high frequency of aberrations and allows a refined characterization of consensus regions. Blood. 2004;104:795–801. doi: 10.1182/blood-2003-12-4175. [DOI] [PubMed] [Google Scholar]
- 75.Wester K, Asplund A, Backvall H, Micke P, Derveniece A, Hartmane I, Malmstrom PU, Ponten F. Zinc-based fixative improves preservation of genomic DNA and proteins in histoprocessing of human tissues. Lab Invest. 2003;83:889–99. doi: 10.1097/01.lab.0000074892.53211.a5. [DOI] [PubMed] [Google Scholar]
- 76.Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, Berglund L, Bergstrom K, Brumer H, Cerjan D, Ekstrom M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J, Forsberg M, Bjorklund MG, Gumbel K, Halimi A, Hallin I, Hamsten C, Hansson M, Hedhammar M, Hercules G, Kampf C, Larsson K, Lindskog M, Lodewyckx W, Lund J, Lundeberg J, Magnusson K, Malm E, Nilsson P, Odling J, Oksvold P, Olsson I, Oster E, Ottosson J, Paavilainen L, Persson A, Rimini R, Rockberg J, Runeson M, Sivertsson A, Skollermo A, Steen J, Stenvall M, Sterky F, Stromberg S, Sundberg M, Tegel H, Tourle S, Wahlund E, Walden A, Wan J, Wernerus H, Westberg J, Wester K, Wrethagen U, Xu LL, Hober S, Ponten F. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–32. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 77.Uhlen M, Ponten F. Antibody-based proteomics for human tissue profiling. Mol Cell Proteomics. 2005;4:384–93. doi: 10.1074/mcp.R500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 78.Ek S, Andreasson U, Hober S, Kampf C, Ponten F, Uhlen M, Merz H, Borrebaeck CAK. From gene expression analysis to tissue microarrays: a rational approach to identify therapeutic and diagnostic targets in lymphoid malignancies. Mol Cell Proteomics. 2006;5:1072–81. doi: 10.1074/mcp.M600077-MCP200. [DOI] [PubMed] [Google Scholar]
- 79.Ghobrial IM, McCormick DJ, Kaufmann SH, Leontovich AA, Loegering DA, Dai NT, Krajnik KL, Stenson MJ, Melhem MF, Novak AJ, Ansell SM, Witzig TE. Proteomic analysis of mantle-cell lymphoma by protein microarray. Blood. 2005;105:3722–30. doi: 10.1182/blood-2004-10-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marengo E, Robotti E, Bobba M, Liparota MC, Rustichelli C, Zamo A, Chilosi M, Righetti PG. Multivariate statistical tools applied to the characterization of the proteomic profiles of two human lymphoma cell lines by two-dimensional gel electro-phoresis. Electrophoresis. 2006;27:484–94. doi: 10.1002/elps.200500323. [DOI] [PubMed] [Google Scholar]
- 81.Antonucci F, Chilosi M, Parolini C, Hamdan M, Astner H, Righetti PG. Two-dimensional molecular profiling of mantle cell lymphoma. Electrophoresis. 2003;24:2376–85. doi: 10.1002/elps.200305457. [DOI] [PubMed] [Google Scholar]
- 82.Wingren C, Borrebaeck CAK. Antibody microarrays: current status and key technological advances. Omics. 2006;10:411–27. doi: 10.1089/omi.2006.10.411. [DOI] [PubMed] [Google Scholar]
- 83.M’Kacher R, Farace F, Bennaceur-Griscelli A, Violot D, Clausse B, Dossou J, Valent A, Parmentier C, Ribrag V, Bosq J, Carde P, Turhan AG, Bernheim A. Blastoid mantle cell lymphoma: evidence for nonrandom cytogenetic abnormalities additional to t(11;14) and generation of a mouse model. Cancer Genet Cytogenet. 2003;143:32–8. doi: 10.1016/s0165-4608(02)00823-3. [DOI] [PubMed] [Google Scholar]
- 84.Smith MR, Joshi I, Jin F, Al-Saleem T. Murine model for mantle cell lymphoma. Leukemia. 2006;20:891–3. doi: 10.1038/sj.leu.2404177. [DOI] [PubMed] [Google Scholar]
- 85.Gladden AB, Woolery R, Aggarwal P, Wasik MA, Diehl JA. Expression of constitutively nuclear cyclin D1 in murine lymphocytes induces B-cell lymphoma. Oncogene. 2006;25:998–1007. doi: 10.1038/sj.onc.1209147. [DOI] [PMC free article] [PubMed] [Google Scholar]


