Abstract
Key string algorithm (KSA) could be viewed as robust computational generalization of restriction enzyme method. KSA enables robust and effective identification and structural analyzes of any given genomic sequences, like in the case of NCBI assembly for human genome. We have developed a method, using total frequency distribution of all r-bp key strings in dependence on the fragment length l, to determine the exact size of all repeats within the given genomic sequence, both of monomeric and HOR type. Subsequently, for particular fragment lengths equal to each of these repeat sizes we compute the partial frequency distribution of r-bp key strings; the key string with highest frequency is a dominant key string, optimal for segmentation of a given genomic sequence into repeat units. We illustrate how a wide class of 3-bp key strings leads to a key-string-dependent periodic cell which enables a simple identification and consensus length determinations of HORs, or any other highly convergent repeat of monomeric or HOR type, both tandem or dispersed. We illustrated KSA application for HORs in human genome and determined consensus HORs in the Build 35.1 assembly. In the next step we compute suprachromosomal family classification and CENP-B box / pJα distributions for HORs. In the case of less convergent repeats, like for example monomeric alpha satellite (20-40% divergence), we searched for optimal compact key string using frequency method and developed a concept of composite key string (GAAAC--CTTTG) or flexible relaxation (28 bp key string) which provides both monomeric alpha satellites as well as alpha monomer segmentation of internal HOR structure. This method is convenient also for study of R-strand (direct) / S-strand (reverse complement) alpha monomer alternations. Using KSA we identified 16 alternating regions of R-strand and S-strand monomers in one contig in choromosome 7. Use of CENP-B box and/or pJα motif as key string is suitable both for identification of HORs and monomeric pattern as well as for studies of CENP-B box / pJα distribution. As an example of application of KSA to sequences outside of HOR regions we present our finding of a tandem with highly convergent 3434-bp Long monomer in chromosome 5 (divergence less then 0.3%).
Key Words: Human genome, alpha satellite, alphoid, higher order repeat (HOR), key string algorithm - KSA, frequency distribution of strings, consensus higher order repeat, suprachromosomal families, CENP-B box, pJα motif
INTRODUCTION
Alpha Satellites – Monomers and Higher Order Repeats
Alpha satellites or alphoid DNA consist of fundamental repeat units (monomers) of approximately 171 bp, tandemly arranged in a head-to-tail fashion, where individual monomers diverge by 20-40% [1–8]. Alpha satellite was first discovered in African green monkey [1] and in humans [2]. Subsequently they have been found at the centromeres of human chromosomes and of primates in general [3,4,9–11].
Some stretches of alpha satellites are hierarchically organized into higher-order repeat (HOR) or alphoid arrays, which were studied by restriction endonucleases [1,6,9–14] and reviewed in several publications [7,8,14–19].
HORs are superperiodic pattern superimposed on the ap- proximately periodic tandem of alpha monomers as follows: if an array of n monomers denoted by 1, 2, …n is followed by the next array of monomers denoted by n +1, n+2, … 2n, where the monomer 1 is almost identical (typically 95-100%) to the monomer n+1, the monomer 2 to the monomer n+2, …the monomer n to the monomer 2n, these arrays belong to nmer HOR [7,8,11,15,16].
Stretches of alpha satellites lacking any higher-order periodicity are referred to as monomeric, and their monomers are only ∼ 20-40% identical [13,20,21]. An impressive work was devoted to investigations of monomeric and HOR arrays [1–89].
In addition to their different sequence organization, monomeric and HOR alpha satellite DNA also differ in their functionality [90]: HORs are associated with centromere function on the basis of genomic [21,91], biochemical [92,93] and artificial chromosome assays [21,94,95]. On the other hand, there is no evidence for direct involvement of monomeric alpha satellite DNA in centromere function.
Computational Analysis of NCBI Genome Assembly
HORs and monomeric alpha satellites have been recently studied by computational analysis of the available NCBI human genome assembly. However, due to the incomplete nature of centromeric contigs, i.e., due to centromere gap, most of HOR regions are missing in NCBI genome assembly [90]. Thus, the sequence analysis of NCBI assembly mostly provides alpha satellite content near the centromeric gaps. In some chromosomes genomic assemblies reached into cen-tromeric alpha and in these cases detailed information on HOR structure can be obtained from genome assembly.
Various computational tools have been developed for computational analyses of repetitions in a given genomic sequence (for example, [96–112]), with a goal to achieve a compromise between efficiency and sensitivity requirements. However, there still remain challenges in the case of large scale and/or significantly distorted repetitions.
Analysis of the NCBI assembly was performed recently using two different computational approaches. Rudd and Willard [90] have used standard computational tools. Alpha satellite and other satellites were extracted using Repeat-Masker and characterized as monomeric or HOR using dot matrix program DOTTER. Percent identity among monomeric alpha satellite monomers and among HORs was examined using CLUSTALW. BLAST alignments of all known HORs reported in the literature versus all alpha satellite in the July 2003 assembly was performed in [90], showing that most of HORs reported in the literature were missing in the genome assembly. On the other hand, four new regions of HORs not previously reported in the literature were found (seven HOR copies in chromosome 4, two in chromosome 10, six in chromosome 11, and six in chromosome 19) [90].
Another type of analysis of NCBI assembly on the human genome was based on the use of new computational algorithm Key String Algorithm (KSA) [113–116]. KSA is a simple and robust method to identify HORs and obtain detailed structure of HORs, which was not reported previously.
Key-String Algorithm (KSA) – Robust Computational Generalization of Restriction Enzyme Method
Key String Algorithm (KSA) is based on the use of appropriately chosen short sequence (string) of nucleotides, “key string”, which cuts into fragments a given single-stranded DNA at each location of this string in genomic sequence [113–116]. Each location of a key string sequence could be compared to a restriction site for restriction enzymes. While the restriction enzymes cleave double stranded DNA selectively at specific palindrome sequences, in KSA we have no limitations on the location of action of computational key string. The lengths of ensuing KSA fragments form a length array, which could be compared to an array of lengths of hypothetical restriction fragments resulting from complete digestion, cutting DNA at recognition site corresponding to a chosen key string sequence. Analyzing the KSA fragment length array, we identify and determine a detailed structure of HORs, including precise identification of substitutions, deletions and insertions. In particular, a HOR-specific key string segments a given sequence into HORs. Similarly, for example, in the KSA, a robust monomer-specific key string segments a given sequence into monomers, palindrome-specific key string leads to identification of large palindrome sequences and their substructure etc. KSA provides a straightforward ordering of KSA fragments, regardless of their size (from small fragments of a few bp to as large as tens of kilobases). KSA is characterized by a combination of straightforward computation and visual inspection of computed results, providing a high degree of robustness and requires only a modest scope of computations which can be performed using PC. Due to its robustness, KSA is effective even in the case of significant deletions, insertions and substitutions, enabling a determination of detailed HOR annotation and structure, consensus sequence and exact consensus length in a given genomic sequence, even if it is highly distorted, intertwined and riddled. Using the HOR consensus sequences computed using KSA, in the next step we compute finer characteristics, as for example the suprachromosomal family (SF) classification and CENP-B box / pJα distributions.
KSA is particularly robust in the case of long monomers and higher order repeats, characterized by highly convergent basic structure and sizeable segments of insertions and deletions.
STRAIGHTFORWARD KSA IDENTIFICATION OF HORS - EXAMPLE FOR CHROMOSOME 5 USING COMPACT KEY STRING CCG
The starting point in KSA is to select an appropriate key string (a short sequence of bases). The next point is computational segmentation of a given genomic sequence (for example, a contig from Build assembly) into fragments, each starting with the chosen key string. Then an array of lengths (length array) of fragments is formed, going along the given genomic sequence. The length array is analyzed, searching for regularities and periodicities. If periodicities in length array are found, they reveal the presence of higher order repeats. On the other hand, deviations from periodicity in the length array can be used for an easy and robust identification of insertions, deletions and substitutions with respect to consensus structure. It should be pointed out that the KSA is characterized by a simple way to deal with insertions and deletions of arbitrary complexity.
As an illustration let us consider the KSA segmentation of the contig NT_006713.14 in chromosome 5 (Table 1). We recognize immediately a long-range periodicity in the first part of length array. To this end let us focus to possible repetition of some fragment lengths. In the first part of the length array from Table 1 we consider, for example, the 314 bp length. Each of the repeated 314 bp lengths along the length array is denoted in Table 1 (bold). If the 314-bp length appears at approximately regular distances, as it is the case here, let as determine distances (in bp) between the neighboring 314-bp lengths, i.e., the sum of fragment lengths from the start of a 314-bp length to the end of the length preceding the next 314-bp length. For example, for the first to second 314-bp fragment length we have the distance: 314+37+264+75+62+420+3+500+6+30+304+199 = 2214 bp.
Table 1.
Array of Fragment Lengths, Positions from 1 to 90332 in Contig NT_006713.14 in Chromosome 5
| 134, 92, 391, 199, 314, 37, 264, 75, 62, 420, 3, 500, 6, 30, 304, 199, 314, 37, 264, 75, 62, 420, 170, 333, 6, 334, 199, 314, 37, 264, 75, 62, 416, 503, 6, 30, 304, 199, 314, 37, 264, 75, 62, 420, 170, 333, 6, 533, 314, 37, 264, 75, 62, 420, 503, 6, 30, 304, 199, 314, 37, 264, 75, 62, 420, 503, 6, 334, 199, 314, 37, 264, 75, 62, 420, 503, 6, 30, 304, 199, 314, 37, 264, 75, 62, 420, 503, 6, 334, 199, 314, 37, 264, 75, 62, 420, 503, 6, 30, 304, 199, 314, 37, 264, 75, 62, 420, 503, 6, 334, 199, 314, 37, 264, 75, 482, 170, 333, 6, 334, 199, 314, 340, 37, 264, 557, 503, 6, 334, 199, 314, 37, 304, 37, 264, 75, 62, 293, 127, 503, 6, 334, 199, 314, 37, 303, 37, 264, 75, 62, 46, 374, 503, 6, 334, 199, 314, 37, 303, 37, 264, 75, 62, 46, 125, 420, 171, 333, 6, 533, 143, 171, 37, 506, 86, 1106, 23, 97, 1530, 228, 659, 456, 2006, 2053, 144, 1093, 550, 58, 530, 57, 2336, 307, 214, 444, 1344, 2321, 1305, 2604, 1190, 281, 2031, 367, 1290, 229, 37, 18, 69, 25, 100, 519, 213, 58, 440, 22, 203, 1892, 2390, 910, 504, 682, 349, 536, 1842, 1538, 1733, 660, 682, 82, 1457, 290, 73, 44, 41, 20, 29, 68, 24, 8, 38, 1645, 249, 2007, 554, 881, 1128, 675, 815, 317, 71, 1043, … |
From position 817 to 35555: an approximately long-range periodic sequence revealing an approximate HOR structure with consensus length 2214 bp; from position 35556: irregular sequence.
Similarly, we determine all other distances between the neighboring 314-bp lengths from Table 1, presented in Table 2. Inspection of Table 2 shows a pronounced approximate repeat structure with consensus length 2214 bp. Since this length is an approximate multiple of alpha monomer length 171 bp, this indicates a structure of 13mer HOR. Therefore, this structure will be annotated as 13mer HOR. (This annotation will be directly shown by a later KSA analysis.)
Table 2.
Array of Distances between Neighboring Fragment Lengths 314 (in bp) from Table 1
| 2214, 2214, 2210, 2214, 2214, 2214, 2214, 2214, 2214, 2214, 2214, 2554, 2555, 2554, 2726 |
Distance between the start of each 314-bp fragment length and the start of the next 314-bp fragment length.
In the array in Table 2 there are five deviations from the 2214-bp consensus repeat length. The third length 2210 bp indicates deletion of four bases with respect to consensus. The 12th, 13th and 14th lengths differ from the consensus by additional 340 bp, 341 bp and 340 bp, respectively. Since these are two alpha monomer lengths, a possible interpretation as two alpha monomer insertions with respect to consensus is tempting. Similarly the 15th length 2726 bp has 512 bases in addition to the consensus length, and therefore one should consider a possible interpretation as three alpha monomer insertions with respect to consensus.
The last, 15th HOR copy, ends at the position 35555. The following fragment lengths (after 143, 171, 37, 506, 86, 1106, …) are random numbers. The fragment length 314 does not appear at all in the interval of the next 800000 bases (the 15th fragment length is at position 32830 and the next, 17th, at the position 857353).
Let us now consider more closely the structure of 2214-bp repeats. Therefore, in Table 3 we display alignment of periodic structure composed of fragment lengths from Table 1.
Table 3.
Alignment of Periodic Fragment Lengths from Table 1
| 314, 37, 264, 75, 62, 420, 3, 500, 6, 30, 304, 199, |
| 314, 37, 264, 75, 62, 420, 170, 333, 6, 334, 199, |
| 314, 37, 264, 75, 62, 416, 503, 6, 30, 304, 199, |
| 314, 37, 264, 75, 62, 420, 170, 333, 6, 533, |
| 314, 37, 264, 75, 62, 420, 503, 6, 30, 304, 199, |
| 314, 37, 264, 75, 62, 420, 503, 6, 334, 199, |
| 314, 37, 264, 75, 62, 420, 503, 6, 30, 304, 199, |
| 314, 37, 264, 75, 62, 420, 503, 6, 334, 199, |
| 314, 37, 264, 75, 62, 420, 503, 6, 30, 304, 199, |
| 314, 37, 264, 75, 62, 420, 503, 6, 334, 199, |
| 314, 37, 264, 75, 482, 170, 333, 6, 334, 199, |
| 314, 340, 37, 264, 557, 503, 6, 334, 199, |
| 314, 37, 304, 37, 264, 75, 62, 293, 127, 503, 6, 334, 199, |
| 314, 37, 303, 37, 264, 75, 62, 46, 374, 503, 6, 334, 199, |
| 314, 37, 303, 37, 264, 75, 62, 46, 125, 420, 171, 333, 6, 533, |
For interpretation of alignment we use simple recombination, segmenting or fusing: 500 = 167 + 133, 3 + 167 = 170, 30 + 304 = 334, 170 + 333 = 503, 482 = 62 + 420, 557 = 75 + 482, 62 + 293 + 127 = 482, 62 + 46 + 374 = 482, 46 + 125 = 171, 420 = 46 + 374.
Comparing the first and second HOR copy in Table 3, we see that the corresponding fragment lengths are mostly the same in both repeats (314 bp vs. 314 bp, 37 bp vs. 37 bp, …). Exceptions are the seventh and eighth fragment lengths, 3 bp vs. 170 bp and 500 bp vs. 333 bp, respectively. This can be easily accounted for by substitutions creating a CCG key string within the 500-bp fragment, segmenting the 500-bp fragment into the 167-bp and 333-bp fragments in the first HOR copy. In the next step, the CCG subsequence at the start of 167-bp fragment changes by one-base substitution into CCA, and thus the 3-bp and 167-bp fragments fuse into a single 170-bp fragment. In this way, the 3 bp + 500 bp fragments in the first HOR copy transform into the 170 bp + 333 bp fragments in the second HOR copy. Another difference between the first and the second HOR copy is a trivial fusion of 30-bp and 304-bp fragments in the first HOR copy into a single 334-bp fragment in the second. It can be traced to a one-base substitution in the starting key string CCG at the start of the 304-bp fragment.
Comparing fragment lengths of the second and third HOR copy in Table 3 we see that the sixth fragment length is 416 in comparison to 420. Therefore, the length of the third HOR copy is 2210, i.e., due to four deletions it is by 4 bp smaller than the length 2214 of the second HOR copy. The other fragments in these two HOR copies correspond to each other, taking into account recombinations 503 = 170 + 333 and 30 + 304 = 334.
A more complex situation, including monomer addition, appears for the 12th HOR copy in Table 3. Comparing the 12th to the first HOR copy, segmenting fragments 557 bp = 75 bp + 62 bp + 420 bp and 503 bp = 170 bp + 333 bp we align the fragment lengths to those in the second HOR copy, but there is an additional 340-bp insertion after the fragment 314 bp. (This corresponds to two alpha satellite monomers, 340 bp = 171 bp + 169 bp.) Thus, the length of this HOR copy is 2214 bp + 340 bp = 2554 bp. In the 13th and 14th HOR copy we see insertions 304 bp + 37 bp = 341 bp and 303 bp + 37 bp = 340 bp, respectively. These three HOR copies contain two alpha monomer insertions each.
The 15th HOR copy has insertions at two positions: the 37 bp + 303 bp = 341 bp insertion after the first 314-bp fragment and the 46 bp + 125 bp = 171 bp insertion after the 62-bp HOR-fragment. The 341-bp insertion corresponds to two alpha monomers, of 171 bp and 170 bp, and the 171-bp insertion corresponds to one alpha monomer. Thus, the length of this HOR copy with two insertions is 2214 bp + 341 bp + 171 bp = 2726 bp.
Concluding, from Table 3 we obtain a consensus HOR fragment length-array: 314, 37, 264, 75, 62, 420, 503, 6, 334, 199.
This array represents a periodic cell corresponding to the 2214-bp HOR.
Various key strings will lead to different periodic cells of the length 2214.
It should be noted that a recombination of fragment lengths appears in general once the long-range periodicity is established in the length-array sequence, which makes the use of KSA simple. In such cases, fragment lengths recombinations can generally be used in practice as a phenomenological rule (“rule of thumb”) based on simple straightforward mathematical recombination. Of course, it can also be traced down to substitutions in the corresponding genomic sequences, but in practical KSA use this is not needed once a long-range periodicity of fragment lengths is established.
The periodic cell with highest frequency of appearance for HOR in a given genomic sequence and a chosen key string corresponds to consensus HOR. Partial deviations from exact length array reveal locations of violations of consensus periodicity (deletions and/or insertions with respect to consensus, and/or substitutions within the key string). This enables a precise identification and location of deletions and insertions within HORs.
KSA IDENTIFICATION OF HORS IN CHROMOSOME 5 USING DIFFERENT 3-bp KEY STRINGS CONSISTING OF C AND G BASES
Table 4 presents the results of straightforward KSA segmentation of contig NT_006713.14 in chromosome 5 by using eight different 3-bp key strings consisting of C and G bases. In all cases the same HOR with consensus length 2214 bp was identified by segmentation into fragments. Each key string is associated with a specific periodic cell of 2214 bp and they all correspond to the same HOR (with different start base).
Table 4.
2214-bp Periodic Cells of the Same 13mer HOR Identified in Contig NT_006713.14 in Chromosome 5 (Build 36.1) Using Strings CCG, CCC, CGC, CGG, GCC, GCG, GGC and GGG
| Key string | Periodic cells (fragment lengths in bp) | Start position |
|---|---|---|
| CCG | (314, 37, 264, 75, 62, 420, 503, 6, 334, 199) | 11883 |
| CCC | (511, 97, 58, 30, 141, 185, 326, 49, 137, 137, 89, 267, 187) | 13744 |
| CGC | (99, 341, 185, 72, 85, 88, 81, 13, 482, 184, 171, 413) | 11610 |
| CGG | (1237, 221, 756) | 11685 |
| GCC | (46, 170, 170, 97, 88, 326, 506, 6, 168, 58, 113, 131, 40, 170, 125) | 11529 |
| GCG | (169, 361, 489, 512, 341, 171, 171) | 10177 |
| GGC | (162, 179, 170, 162, 36, 276, 39, 94, 102, 144, 508, 146, 25, 171) | 11358 |
| GGG | (285, 226, 196, 10, 77, 909, 511) | 12014 |
KSA IDENTIFICATION OF HORS IN DIFFERENT CHROMOSOMES USING A SINGLE 3-bp KEY STRING CCG
Using a single key string, we can determine HORs in different chromosomes. This will be shown here for the Build 36.1 genome assembly by performing a straightforward CCG-key-string KSA segmentation for chromosomes 1, 4, 5, 7, 8, 10, 11, 17, 19 and X, which were previously investigated for HORs in KSA using ColorHOR [115,116]. In the present straightforward KSA segmentation, for each chromosome we compute the array of fragment lengths using the CCG key string. By an easy visual inspection we look for periodicity in this length array. Any periodicity directly reveals a periodic cell corresponding to highly convergent tandem repeats. These results are shown in Table 5 for ten chromosomes. A single choice of key string, CCG, enables a simple precise identification of all HORs in genome assembly for these chromosomes.
Table 5.
HORs and the Corresponding Periodic Cells Identified in Build 36.1 Assembly for Different Human Chromosomes by Straightforward KSA Segmentation Using Key String CCG
| Chr. | Contig | nmer | Consensus length (bp) | Periodic cell (fragment lengths in bp) |
|---|---|---|---|---|
| 1 | NT_077389.3 | 11mer | 1866 | (180, 156, 338, 486, 63, 16, 46, 82,162, 171, 166) |
| 4 | NT_022853.14 | 13mer | 2210 | (182, 156, 36, 306, 26, 169, 169, 338, 167, 98, 252, 311) |
| 5 | NT_006713.14 | 13mer | 2214 | (314, 37, 264, 75, 62, 420, 503, 6, 334,199) |
| 7 | NT_023603.5 | 16mer | 2734 | (2213, 323, 110, 88) |
| 8 | NT_023678.15 | 11mer | 1868 | (246, 1622) |
| 10 | NT_079540.1 | 18mer | 3058 | (98, 242, 340, 339, 340, 340, 98, 278, 304,340, 269, 70) |
| 11 | NT_035158.2 | 12mer | 2047 | (34, 66, 28, 143, 143, 24, 28, 152, 15, 82, 74, 89, 73, 171, 142, 584, 199) |
| 17 | NT_024862.13 | 14mer | 2379 | (134, 204, 2041) |
| 19 | NT_011295.10 | 17mer | 2896 | (734, 19, 313, 305, 215, 70, 314, 926) |
| 19 | NT_113948.1 | 13mer | 2214 | (437, 925, 340, 131, 67, 314) |
| X | NT_011630.14 | 12mer | 2057 | (39, 314, 24, 144, 223, 117, 27, 357, 171, 167, 144, 36, 294) |
In general, an arbitrary key string, like CCG, will not reveal alpha satellite monomers, since these monomers diverge from each other by about 20-40 % and therefore periodic position of a key string has a small probability. (Only specific strings, presenting robust segments of alpha satellites, can provide segmentation into alpha monomers as will be discussed later.)
Positions of periodic cells in the corresponding contigs: chromosome 1 - 278067 , chromosome 4 - 906, chromosome 5 – 11883, chromosome 7 - 107592, chromosome 8 – 2076, chromosome 10 – 184305, chromosome 11 – 495035, chromosome 17 – 562619, chromosome 19 (17mer) - 15797329, chromosome 19 (13mer) – 77025, and chromosome X – 6120763. In contigs in chromosomes 4, 5 and 8 the identified HOR arrays are positioned at the beginning of each contig, with cut off at the start of contig. In chromosome 7 the identified HOR array is embedded within the region of monomeric alpha satellite. In all other contigs from Table 1 the identified HOR array are positioned at the beginning of each contig, with cut off at the start of each contig.
KSA ANALYSIS OF MONOMERIC ALPHA SATELLITES AND HORS CHOOSING PHENOMENOLOGICAL ROBUST KEY STRING (APPROXIMATELY 4-6 BP)
Initially, we have used in KSA a three-to-six-bp key string from a large class of strings, to identify HORs which provides an easily detectable periodic pattern in fragment length array [113,114]. The method is simpler and more effective if repeat copies are more convergent and repeat sequence longer, while the size of insertions and deletions is not relevant.
For KSA identification of alpha monomers a practical recommendation was given for a choice of key string [115]: to choose a short (4-6 bp) subsequence from the known human alpha satellite consensus sequence [39,61,117]. For example, a particularly robust 6-bp subsequence is GAAACA and only slightly less robust are AGAAAC, GAGCAG, AAACAC and AGAGAA.
In [116] we used key strings convenient for segmentation into alpha monomers given in Table 6.
Table 6.
Convenient Key Strings for Segmentation of Build 35.1 Assembly for Some Human Chromosomes into Alpha Monomers [116]
| Key string | HOR | Chr. |
|---|---|---|
| GTTTCC | 11mer (1866 bp) | 1 |
| GTTTCG | 13mer (2211 bp) | 4 |
| ACACAC | 13mer (2214 bp) | 5 |
| AGAAAC | 16mer (2734 bp) | 7 |
| CCCC | 11mer (1869 bp) | 8 |
| AAAGCA | 18mer (3058 bp) | 10 |
| AAGGTGC | 12mer (2047 bp) | 11 |
| TTGGCCT | 14mer (2379 bp) | 17 |
| AAGTGG | 13mer (2214 bp) | 19 |
| AACTACC | 17mer (2896 bp) | 19 |
| GTTTCGAAAC | 12mer (2057 bp) | X |
Using a key string with higher frequency of appearance, the KSA provides segmentation into shorter fragments, on the average. In order to identify certain periodicity or higher order periodicity, we need a key string segmenting a given sequence into fragments which are, on the average, sizably shorter than the length of periodic pattern to be identified. Therefore, for example, we cannot identify alpha satellite monomers by using a randomly chosen key string which segments the given genomic sequence into fragments of the average length comparable to the length of alpha satellite monomer.
However, it is possible to find a key string corresponding to a unique robust subsequence within the periodic pattern (monomer or HOR), appearing only once per periodic pattern, which segments a given sequence directly into alpha monomers.
Subsequently, the presence of a HOR can be recognized from periodicity in the monomer length array.
For example, using the 5-bp key string GAAAC for KSA analysis of the contig NT_023603.5 in chromosome 7 (Build 36.1), we obtain the array of fragment lengths, which reveals the periodic cell:
170, 66, 105, 66, 59, 45, 39, 27, 105, 66, 105, 65, 60, 22, 23, 126, 44, 66, 60, 45, 66, 106, 66, 105, 171, 65, 61, 45, 67, 105, 39, 27, 105, 66, 105, 66, 105, 65, 105
with the corresponding the HOR length
170 bp + 66 bp + 105 bp + 66 bp + … + 105 bp = 2734 bp.
Such periodic cell appears, for example, at positions 73843 to 76576. Going along this array we can easily combine fragment lengths into approximately 171 bp segments:
170, 66 + 105 = 171, 66 + 59 + 45 = 170, 39 + 27 + 105 = 171, …. 65 + 105 = 170
In this way we obtain an array of 16 alpha monomers (in bp):
170, 171, 170, 171, 171, 170, 170, 171, 172, 171, 171, 171, 172, 171, 171, 171
giving a segmentation of the 2734 16mer HOR into alpha monomers.
(Alpha monomers in Build 36.1 sequences have the same strand convention as [117], R-strand (direct), while in [113] the genomic sequence from the clone AC017075.8 in chromosome 7 was with strand convention which is reverse complement to [117], S-strand).
Concluding, a key string convenient to detect alpha monomers directly simultaneously detects a periodic cell for each HOR copy, i.e., it gives at the same time segmentation both alpha monomers and identification of HORs. If no periodicity is present in the array of monomer lengths, the sequence of monomers is of monomeric type.
A shortcoming of using very short key strings (4-6 bp) to identify alpha monomers is that not a single key string is convenient for different chromosomes, i.e., the key string is chromosome-dependent and may also be dependent on region within a chromosome sequence.
The lengths of constituent alpha monomers in HOR depend on the choice of key string and of the starting monomer in HOR. Therefore, different sets of monomer lengths are obtained by KSA decomposition of the same HOR using different choices of the key string. In the later sections we develop a systematic method for the choice of key strings.
By an appropriate choice of the key string, KSA can provide straightforward segmentation of genomic sequence into HOR copies, without internal fragments within HOR (i.e., without constituent alpha monomers). For example, such HOR-segmenting key string for the NCBI assembly for chromosome 7 is an almost palindrome-like key string TTTTTTAAAAA. This string appears only once in each HOR copy and always at the same position within each HOR copy. It was referred to as a “beautiful” string [114]. This key string exhibits the highest degree of robustness. Segmentation of the clone AC017075.8 using this key string reveals the presence of 55 HOR copies [114].
FREQUENCY DISTRIBUTION OF STRINGS VERSUS FRAGMENT LENGTH IN GENOMIC SEQUENCE FOR IDENTIFICATION OF ALL REPEATS AND FOR DETERMINATION OF OPTIMAL KEY STRING
A key-string frequency distribution for a given genomic sequence can be described by considering a set of all r-bp key strings, where the number 4r is comparable to a repeat length (as for example the alpha monomer length or HOR length).
In hypothetical situation of equal probability of appearance of each of the r-bp strings within a given genomic sequence, the average length of a KSA fragment should be 4r bp. For example, in such case the average length of KSA fragment for a particular 3-bp key string (for example, CCG) should be 43 = 64 bp; for a particular 6-bp key string (for example, GTTTCC) the average length should be 46 = 4096 bp.
In the realistic situation of genomic sequence, we compute for a particular key-string of length r the total frequency of appearance of all KSA fragments with all possible 4r key strings of the fragment length l bp. Their superposition displays the total length-frequency distribution (fr vs. l) for a set of all 4r r-bp key strings.
As an example, we calculate the total length–frequency distribution for the contig NT_011295.10 in chromosome 19 using all possible 6-bp key strings. Fig. (1) shows graphical presentation of frequency (f6) vs. fragment length (l). There are two most pronounced peaks: for the frequency of fragment length l = 171 bp and of fragment length l = 2896 bp. Less pronounced peaks appear at approximate multiples of 171: l = 341 bp, 512 bp …
Fig. (1).
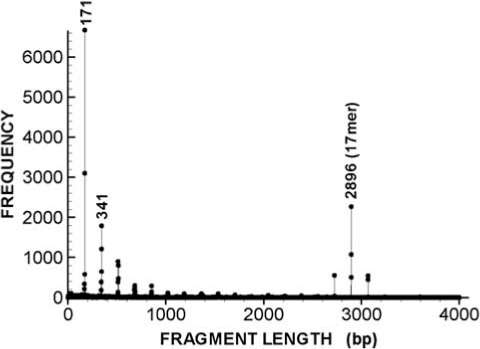
Total frequency f6 as fraction of the fragment length computed for contig NT_011295.10 in chromosome 19 using all possible 6-bp strings (For description see the text).
Table 7 displays frequencies of fragment lengths around most pronounced peaks in the frequency-length diagram from Fig. (1). Table 8 presents the 6-bp and 8-bp strings with highest frequency for fragment lengths l = 171 bp and l = 2896 bp. Table 9 displays a section of array of fragment lengths obtained by using the dominant key string (i.e., having highest frequency f8 for fragment length l = 2896 bp). Table 10 displays a section of array of fragment lengths obtained by using the dominant key string (i.e., having highest frequency f6 for fragment length l = 171 bp). Therefrom we obtain the periodic cell of length 2896:
Table 7.
Frequencies f6 Around Fragment Lengths l = 171 bp and l =2896 bp with Most Pronounced Peaks in Fig. (1)
| Monomer | 17mer | ||
|---|---|---|---|
| l(bp) | f6 | l(bp) | f6 |
| 165 | 10 | 2890 | 1 |
| 166 | 338 | 2891 | 0 |
| 167 | 205 | 2892 | 1 |
| 168 | 49 | 2893 | 0 |
| 169 | 574 | 2894 | 502 |
| 170 | 3095 | 2895 | 1072 |
| 171 | 6668 | 2896 | 2262 |
| 172 | 73 | 2897 | 4 |
| 173 | 50 | 2898 | 8 |
| 174 | 11 | 2899 | 4 |
| 175 | 3 | 2900 | 0 |
Table 8.
Strings with Highest Frequency for Fragment Lengths l = 171 bp and l = 2896 bp in Contig NT_011295.10 in Chromosome 19
| l = 171 bp (r = 6): AGTTGA, GTTGAA, TTGTGA, CTTTGT, TTTGTG, TGTGAT, AGTTTT, CATTCA, TGGATA, TTTGAA, AGCAGT |
| l = 2896 bp (r = 6): ACCAGA, ACTACC, AGGAGC, ATATCA, ATCAGG, CAGGAG, CATGTG, CTGAGA, GAGAAA, GCATGT, GTGTAG, TACCAG |
| l = 2896 bp (r = 8): ATCAGGAG, CTCTTTGT, TCAGGAGC, AAAAAGAA, AAAGAAAT, AACTACCA, AAGAAATA, AATATCTG, ACAGAAGG, ACCAGAGT, ACGGAGTT, ACTACCAG |
Table 9.
Segmentation of Contig NT_011295.10 in Chromosome 19 (Build 36.1) Using the Dominant Key String ATCAGGAG
| 15380, 101979, 71804, 71055, 74442, 68120, 26761, 2896, 2896, 2896, 2896, 2724, 2894, 2895, 2895, 171, 2896, 171, 1628 |
Dominant key string has the highest frequency f8 for fragment length l = 2896 bp. Fragment lengths are shown only for a section of contig from position 153368026 to the end. Underlined: region of 17mer HOR. The last fragment length of 1628 bp corresponds to a truncated HOR copy. The 2724-bp sequence has one-monomer deletion with respect to the 2896-bp consensus. Two 171-bp sequences represent insertion to consensus HOR: 2895 bp + 171 bp = 3066 bp, 2896 bp + 171 bp = 3067 bp. Two HOR copies have one-base and two-base deletions with respect to the 2896-bp consensus.
Table 10.
Aligned Array of Fragment Lengths for the Region of 2896-bp 17mer HOR in Contig NT_011295.10 in Chromosome 19 (Build 36.1)
| 166, 171, 170, 171, 340, 171, 171, 511, 170, 171, 342, 171, 171, |
| 166, 171, 170, 171, 340, 171, 171, 511, 170, 171, 342, 171, 171, |
| 166, 171, 170, 171, 340, 171, 171, 511, 170, 171, 342, 171, 171, |
| 166, 171, 170, 171, 340, 171, 171, 511, 170, 171, 342, 171, 171, |
| 166, 171, 170, 171, 339, 171, 171, 681 171, 171, 171, 171 |
| 166, 171, 170, 171, 339, 171, 171, 511, 170, 170, 342, 171, 171, |
| 166, 171, 170, 171, 339, 171, 171, 511, 170, 171, 342, 171, 171, |
| 166, 171, 170, 171, 339, 171, 171, 511, 170, 171, 342, 171, 171, 171, |
| 166, 171, 170, 171, 339, 171, 682, 171, 171, 342, 171, 171, 171 |
| 166, 171, 170, 171, 339, 171, 162 (end of contig) |
This is obtained by segmentation using dominant r = 6 key string, AGTTGA, having highest frequency of fragment length l = 171 bp (see Table 8).
166, 171, 170, 171, 340, 171, 171, 511, 170, 171, 342,171, 171.
The first four HOR copies are equal to this consensus periodic cell. The fifth HOR copy has one 171-bp monomer deletion, one base deletion and a fusion 511 + 170 = 681 and segmentation 342 = 171 + 171 (due to a base substitution in key strings). Therefrom, its length is 2896 bp – 171 bp – 1 bp = 2724 bp. The 8th and 9th HOR copies have insertion of one 171-bp monomer each.
Concluding, the optimal key string for segmentation into alpha monomers, and subsequently also into HORs, is computed as a string with highest frequency of the fragment length 171 bp in a given genomic sequence. On the other hand, the optimal key string for direct segmentation into HORs (without internal alpha monomer structure, i.e., without segmenting into constituent alpha monomers) is computed as a string with highest frequency of the fragment length determined by the long-range peak from the total frequency distribution.
This method can be generalized to any type of tandem repeats, monomeric or HOR, as well as to dispersed repeats (with the use of fragments within each repeat unit).
This involves three steps of computation:
first, by computation of total frequency distribution for all key strings of a size r the lengths of repeat structures in a given sequence are determined;
second, computation of frequency vs. fragment length distribution for each repeat length determined in the preceding step provides the dominant key string;
third, segmentation using dominant key string leads to determination of consensus of repeat structure and insertions as well as insertions, deletions and substitutions with respect to consensus.
COLORHOR ALGORITHM FOR SCAN OF HORs
ColorHOR is a graphical user interface method based on KSA [115]. It enables a fast computational identification of HORs in a given genomic sequence, without requiring a priori information on the composition of genomic sequence. ColorHOR provides a color representation of HORs, giving a direct visual identification of HORs. In this way we determined the HOR annotation of Build 35.1 assembly for human genome. New HORs have been found in chromosomes 4, 8, 9, 10, 11 and 19 and exact consensus lengths have been determined for all HORs present in Build 35.1 assembly [115].
ColorHOR procedure involves the following steps: computational construction of the length-frequency distribution, computational construction of alpha staircase and computational construction of colored bands and color-motif [115]. The first step displays diagrammatically the frequency N versus fragment length Δ. The second step computes the cumulative frequency Nc of the fragment length Δ = 171 bp, up to a base position n along genomic sequence and displays diagrammatically Nc versus n diagram (referred to as alpha staircase). Any local clustering of the 171 bp fragment lengths along genomic sequence results in a sharp increase (stair) in this diagram. The location of each alpha monomer containing section within the sequence is associated with a stair in the alpha staircase, providing a fast graphical identification of segments containing alpha monomers. The third step provides the length-frequency (N versus Δ) diagram for the alpha monomer containing section. Identifying highest peaks in this diagram, the coloring rule is defined: to each length corresponding to pronounced peaks a particular color is assigned. Accordingly, the stripes displaying the corresponding key-string fragments along the band presenting genomic sequence are colored. In this way, a colored band with repetitive color-motif is obtained at the location of each HOR-containing section of genomic sequence [115]. The ColorHOR method was applied to Build 35.1 assembly for all chromosomes and more closely demonstrated for chromosome 1 [115].
KSA CONSENSUS HORS
Using KSA we have determined consensus HORs for Build 35.1 assembly for chromosomes 1, 4, 5, 7, 8, 10, 11, 17, 19, and X. Aligned monomers contained in consensus nmer HOR are denoted
t01, t02, … t0n.
This array is equal to consensus HOR if the monomer sequences correspond to the convention of [117] (will be referred to as R-strand (direct) monomers); this is the case for 16mer in chromosome 7, 11mer in chromosome 8, 14mer in chromosome 17, and 17mer in chromosome 19 deduced from Build 35.1 assembly. If the consensus HOR contains alpha monomers which are reverse complement to convention of [117] (will be referred to as S-strand monomers), then the array t01, t02, … t0n is reversed complement to consensus HOR; this is the case for 11mer in chromosome 1, 13mer in chromosome 4, 13mer in chromosome 5, 18mer in chromosome 10, 12mer in chromosome 11, 13mer in chromosome 19 and 12mer in chromosome X deduced from Build 35.1 assembly [115]. In Table 11 we display consensus HOR for the 13mer HOR which was recently found using KSA [116]. Table 12 shows divergence among alpha monomers in consensus HORs from chromosomes 5 and 1; Table 13 displays some minimal divergences between constituent monomers. Table 14 displays average divergence among monomers in consensus HORs. Table 15 discusses the use of composite GAAAC--CTTTG semi-palindromic key string for identification of alpha monomer sequences.
Table 11.
Consensus 13mer HOR in Human Chromosome 5
| t01=171 |
| GTCTGCAAGCGGATAATGGGCTTCGCTTTGTGTCCTTTGGTGGAAACGGGAATATCTTCTAATAAAAACTAGACAGAAATATTCTCACAATCGTCTTTGTGATG |
| TGGGCATTCAACTAACACAGTTGAACATTTCTTCTCACAGAGCAGTTTTGAAACACTCTTTTGCTAG |
| t02=170 |
| AATTGC-AGGTGAATCTTTGGAGCGCTTTGAAGCCTTTGTTGGAAATGGGAATATCTTCACACACAAACTAGCCAGAAGCATTCTCAGAAACTTCTTTGTGATG |
| TGTGCGTTGAACCCAGAGAGATGAACCTTTCCTTTGATAGAGCAGTTTTGAAACGTGTTTTTGTAAG |
| t03=170 |
| AATCTGCCAGCGGACACTTGGAGCGCTTTGAGGGCTATGGTGGAGAAGGAAACATCTTCCCATAAAAACTAGAAAGAAGCATTCTCGGAAACATTTATGTGAAG |
| CGTGCCTTCAACTCACAGAGTTGAACCTTCCTTTTGATAGAACAGTTTTGAAACACTCTTTTG-AAC |
| t04=171 |
| AATCTACAATTGGATAATTGGAACCCTTTGATGCCCATGGTAGAAAAGGAAATATCCTCATATAAAAACTAGACAGAAGGATTCACAGAAAATGCTTTGTGATG |
| TGTGCATTCAAATCACGGAGTTGAATCTTTCTTTTGTTAGAGCAGTTTTGAAACACTGTTTCTGTGG |
| t05=171 |
| GATCCGCAAGTGGATATTTGGACAGCTTTGAGATCTTTGCTGGAAATGGGAATATCTTCACATATAAACTAGACAGAAGCATTCTCAGAAACTTCTTCGTGATG |
| TGTGCATTCTACTCCCAAATTTGAATCTTCCTTCTCATGAAGCAGTTTTGAAACACTCTATTTGTGC |
| t06=170 |
| AATCTGAAAGTGGATATTTGGAGCTCTTTGAGGGCTATGGCGGAAAAGAAAATATATTCACAT-TAAACT AGACAGCAGCATTCTCAGAAACTTCTTTAGGAT |
| GTCTGCAGTAAACTCACAGAGTTGAACATACCTTTCCGTAGAGCAGTTTTGAAACACTCTGTTTGTGG |
| t07=167 |
| AATCTGCATGTGGATATCTGGAGCGATTTGAGGCCTATGGTCAAAAAGGAAATATCTTCCTGGGAAAAATAGACGAAAGCATTCTCAGAAACTGCTTTGTGATA |
| TGTGCATTCGACTCACCGAGTTGAAACTTTTTTTTGATAGAGCAGTTTTGAAACACTCTGTAG |
| t08=171 |
| AATCTGCAAGTGGATATTTGGAGCGCTTTGAGGCCTATGGTAGAAAAAGAAATATCTGCCTCTAAAAACTAGACAGAAGCATTCTGAGAAACTTCTTTGTGATG |
| TTTGCATTCAACTACCAGAGTTGAATCTTCCTTTTGATAGGGCAGTTTGGAAACACTCTTTTTGTAG |
| t09=171 |
| AATCTGCAAGTGGATATTTGGACTGCTTTGAGGCCTTCATCGGAGACGGGAATATCTTCACATAAACACTAGGCAGAAGCATTCTCAGAAACTACTTTGTGATC |
| TGTCCATTCAACTCACAGAGTTGAACCTTCCTTTTTATGGAGCAGTTTTGAAACACTGTTTTTGGAG |
| t10=171 |
| AATCTGCAAGTAGACATTTGGAGTGCTTTGAGGGCTGTGGTGCCAAAGGAAATGTCTTCCCATGGAAACTAGACTGAAGCATTCTCAGCAACTTCTTTGTGACG |
| TTTGCATTCATCTCACAGTGTTGAACATACCTTTCCATAGAGTAGTTTTGAGACACTATTTTTGTAG |
| t11=170 |
| AATCTGCAAGTAGATATTTGGAGCGCTTTGAGGCCTTCGTTGGAAACCGGAATATCTTCACAGAAAAAGTAGATAGAGGCATTCTCAGAAACTTTTTTGTGATA |
| TGTTGATTCATCTGACAGCGTTGAACCTTTCTTTTGATAGAGCAGTTTTGAAAAACTC-TTTTGTCG |
| t12=170 |
| AATCTGCAAGTGGATATTCGGACCACTTTGAGGCCTTCATAGGAAACAGTAATACCTTCACATAAAAACTAGATAGAAGCATTGTCAGAAAGTTCTTTGTGATG |
| TGTGAATTCAACTCACAGAGTTGAACC-TTCCTTTAATAGAGCAGTTTTGAAACACTCTTCTTCTAG |
| t13=171 |
| AATCTGCCAGTGGATACTTGGAGCGCTTTGAGGGCTATTGTGCCAATGGAGATATCTTCCCCTAAAAACTAGACAGAAGCATTCTCAGAAACTACTTTGTGATG |
| TTTGCATTCAACTCACAGAGTTGAACATACCTCTTCATAGAGCAGTTTTGAAAACCTCTTTTTGTAG |
Sequence corresponds to reverse complement of convention according to [117]. For description see the text.
Table 12.
Table of Divergence (%) between Monomers from HOR Consensus of 11mers in Chromosome 1 (Columns) and 13mers from Chromosome 5 (Rows)
| t01 | t02 | t03 | t04 | t05 | t06 | t07 | t08 | t09 | t10 | t11 | |
| 171 | 171 | 171 | 171 | 171 | 166 | 171 | 165 | 171 | 167 | 171 | |
| t01=171 | 32.2 | 32.7 | 29.8 | 32.7 | 26.3 | 42.1 | 34.5 | 39.8 | 36.8 | 40.4 | 36.3 |
| t02=170 | 29.2 | 28.1 | 26.3 | 27.5 | 24.0 | 37.4 | 28.7 | 33.9 | 30.4 | 35.1 | 32.2 |
| t03=170 | 23.4 | 25.1 | 21.6 | 27.5 | 22.2 | 36.3 | 24.0 | 35.7 | 27.5 | 32.7 | 26.9 |
| t04=171 | 26.3 | 28.1 | 21.1 | 26.3 | 24.6 | 35.7 | 21.6 | 37.4 | 24.6 | 33.9 | 26.3 |
| t05=171 | 26.9 | 25.1 | 22.2 | 25.7 | 21.6 | 33.9 | 26.3 | 32.7 | 29.2 | 32.7 | 29.8 |
| t06=170 | 25.7 | 25.7 | 22.2 | 26.9 | 22.8 | 35.1 | 22.2 | 33.9 | 24.6 | 33.9 | 25.1 |
| t07=167 | 25.1 | 27.5 | 25.1 | 29.8 | 25.7 | 38.6 | 25.1 | 38.6 | 27.5 | 33.3 | 30.4 |
| t08=171 | 19.3 | 22.2 | 17.5 | 24.0 | 18.7 | 29.2 | 18.1 | 31.6 | 19.3 | 28.1 | 21.6 |
| t09=171 | 23.4 | 22.8 | 22.2 | 22.2 | 17.5 | 30.4 | 21.6 | 29.2 | 25.7 | 28.7 | 24.6 |
| t10=171 | 22.8 | 25.7 | 22.2 | 28.1 | 24.0 | 36.8 | 24.6 | 35.1 | 26.9 | 32.2 | 26.3 |
| t11=170 | 26.3 | 25.7 | 24.0 | 24.0 | 20.5 | 34.5 | 25.7 | 31.6 | 28.1 | 28.7 | 30.4 |
| t12=170 | 25.1 | 23.4 | 23.4 | 23.4 | 18.1 | 33.9 | 23.4 | 30.4 | 24.6 | 30.4 | 26.9 |
| t13=171 | 22.8 | 22.8 | 21.1 | 22.2 | 19.3 | 35.1 | 20.5 | 34.5 | 22.2 | 31.0 | 24.0 |
Table 13.
Minimal Divergence between Monomers in Some Pairs of Monomers from Consensus HORs (Suprachromosomal Family Assignment (SF) is Given for each HOR)
| Div. (%) | |
|---|---|
| mon.t04 in 13mer from chr.5 (SF5) / mon.t05 in 13mer from chr.19 (SF5) | 1 |
| mon.t11 in 12mer from chr.11 (SF3) / mon.t10 in 12mer from chr.X (SF3) | 4 |
| mon.t08 in 13mer from chr.5 (SF5) / mon.t17 in 17mer from chr.19 (SF5) | 5 |
| mon.t06 in 13mer from chr.19 (SF5) / mon.t03 in 17mer from chr.19 (SF5) | 6 |
| mon.t03 in 11mer from chr.1 (SF3) / mon.t02 in 12mer from chr.11 (SF3) | 10 |
| mon.t03 in 11mer from chr.1 (SF3) / mon.t01 in 12mer from chr.X (SF3) | 10 |
| mon.t08 in 13mer from chr.5 (SF5) / mon.t11 in 12mer from chr.X (SF3) | 15 |
| mon.t05 in 11mer from chr.1 (SF3) / mon.t03 in 16mer from chr.7 (SF5) | 18 |
| mon.t07 in 11mer from chr.8 (SF2) / mon.t04 in 14mer from chr.17 (SF3) | 18 |
| mon.t05 in 11mer from chr.1 (SF3) / mon.t09 in 13mer from chr.5 (SF5) | 18 |
| mon.t05 in 11mer from chr.1 (SF3) / mon.t05 in 11mer from chr.8 (SF2) | 21 |
| mon.t05 in 11mer from chr.1 (SF3) / mon.t03 in 18mer from chr.10 (SF1) | 21 |
| mon.t03 in 11mer from chr.1 (SF3) / mon.t10 in 13mer from chr.4 (SF5) | 21 |
| mon.t03 in 18mer from chr.10 (SF1) / mon.t08 in 12mer from chr.11 (SF3) | 23 |
Table 14.
Average Divergence (%) Among Monomers in Consensus HORs
| 11mer Chr.1 | 13mer Chr.4 | 13mer Chr.5 | 16mer Chr.7 | 11mer Chr.8 | 18mer Chr.10 | 12mer Chr.11 | 14mer Chr.17 | 13mer Chr.19 | 17mer Chr.19 | 12mer Chr.X | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 11mer/Chr.1 | 28 | 32 | 28 | 28 | 31 | 30 | 26 | 26 | 28 | 27 | 26 |
| 13mer/Chr.4 | 28 | 27 | 28 | 32 | 31 | 31 | 31 | 28 | 27 | 30 | |
| 13mer/Chr.5 | 23 | 24 | 27 | 27 | 26 | 26 | 22 | 22 | 26 | ||
| 16mer/Chr.7 | 24 | 28 | 27 | 26 | 27 | 24 | 23 | 26 | |||
| 11mer/Chr.8 | 28 | 30 | 29 | 29 | 28 | 27 | 29 | ||||
| 18mer/Chr.10 | 23 | 29 | 29 | 27 | 26 | 29 | |||||
| 12mer/Chr.11 | 25 | 25 | 26 | 25 | 24 | ||||||
| 14mer/Chr.17 | 25 | 26 | 26 | 25 | |||||||
| 13mer/Chr.19 | 24 | 23 | 26 | ||||||||
| 17mer/Chr.19 | 23 | 25 | |||||||||
| 12mer/Chr.X | 25 |
Table 15.
Key String GAAAC--CTTTG Fragment Array for a Section from Position 5787 to 20911 in Contig NT_007758.11 in Chromosome 7
| a) 340, 679, 340, 340, 340, 340, 1355, 679, 171, 168, 335, 342, 169, 171, 169, 509, 171, 169, 341, 169, 340, 171, 169, 340, 513, 169, 511, 171, 169, 171, 511, 509, 171, 335, 171, 171, 511, 681, 169, 681, 682, 171, 170, 171 |
| b) 169, 171, 169, 171, 168, 171, 169, 171, 169, 171, 169, 171, 169, 171, 164, 171, 169, 171, 169, 171, 169, 171, 168, 171, 169, 171, 171, 168, 170, 165, 171, 171, 169, 171, 169, 171, 170, 168, 171, 169, 170, 171, 169, 171, 169,171, 169, 171, 169, 171, 171, 171, 169, 171, 169, 171, 171, 169, 171, 169, 171, 171, 169, 171, 169, 171, 169, 166, 171, 171, 171, 170, 170, 169, 171, 170, 171, 169, 171, 171, 170, 169, 171, 171, 170, 170, 171, 170, 171 |
(a) and the corresponding fragment array with subsequent segmentation of alpha monomer multiples (b).
KSA consensuses for HORs in other chromosomes are presented in Supplementary data.
KSA IDENTIFICATION OF ALPHA MONOMERS USING COMPOSITE SEMI-PALINDROMIC KEY STRING GAAAC--CTTTG
In a search for a single key string which will be convenient for identification of alpha monomers in all chromosomes we found for R-strand (direct) sequences (convention like in [117]) the complex key string GAAAC--CTTTG (-- denote any two bases). For S-strand (reverse complement) sequences the key string is reverse complement, i.e., CAAAG--GTTTC.
For example, let us analyze the sequence of contig NT_007758.11 in chromosome 7.
A sizeable number of monomers start with a sequence which differs by one or more substitutions from the GAAAC--CTTTG key string. Any such monomer is in the key string segmentation fused with a monomer preceding it, leading to multiple monomer lengths (see Table 15a). For example, the first 340-bp fragment in Table 15a) starts with the key string GAAAC--CTTTG and the first 169 bases form a 169-bp monomer. The following twelve bases, GTAAC TTATTTG contain two substitutions (A → T at the second and C → A at the eighth position in the key string) and thus the remaining bases, which form a 171-bp alpha monomer, are fused with the preceding 169-bp monomer into a single 340-bp fragment. In this way, the first 340-bp fragment in Table 15a) is segmented into 169-bp and 171-bp monomers in Table 15b).
Using this semi-palindromic key string we identify the R-strand (direct) sequences, and using its reverse complements the S-strand (reverse complement) sequences. In this way we have performed identification of monomers in complete Build 36.1 assembly for all human chromosomes.
KSA ANALYSIS OF MONOMERIC ALPHA SATELLITES AND HORS USING COMPOSITE KSA – 28-bp (KS28) ALPHA MONOMER KEY STRING
Although the GAAAC--CTTTG key string is a convenient single key string for analysis of alpha monomers in complete Build 36.1 assembly for all human chromosomes, it requires the second step with subsequent segmentation of alpha monomer multiples into single alpha monomers which makes the analysis more complex. Therefore we have searched for a single key string which will segment any sequence of alpha monomers (HOR or monomeric) directly into alpha monomers, without need for additional recombination of fragment lengths.
Here we propose the 28-bp sequence
TGAGAAACTG CTTTGTGATG TGTGCATT
for R-strand sequences, and its reverse complement for S-strand sequences. Here, the sequence is cut at positions where at least 19 bases out of 28 are found in a given genomic sequence after alignment of this flexible key string with genomic sequence under study. Additionally, we allow in alignment for one base insertion or deletion in the key string. For convenience, in the R-strand monomers this key string is placed at the end of monomeric sequence and in the S-strand monomers at the beginning.
For example, analyzing the contig NT_023603.5 in chromosome 7, the first two alpha monomers of R-strand identified in this way are given in Table 16. In aligning key string to the first monomer we insert one base G between the fifth and sixth base in the 28-bp key string, thus extending it to 29 bp. In this way, we obtain that 20 bases from the key string are aligned with corresponding bases from genomic sequence being analyzed. This provides segmentation of the first 169-bp monomer (Table 16). The next subsequences of genomic sequence aligning with the key string are the last 28 bases of the 171-bp monomer starting at position 29737 within the contig (21 bases out of 28 in the key string are aligned with genomic sequence) (Table 16).
Table 16.
First Two Alpha Monomers of R-Strand in the Contig NT_023603.5 Identified by KSA Using KS28
| 170-bp monomer, start at position 24767 |
| CAACTAACAGAATTGAACCTTTCTTTTGTTAGAGCAGTTTTGAAACACTCTTTTTGTAGAATCTGCAAGTTGATATTTGGATAGATTTGAGGA |
| TTTCATTGGAAACGGGAATATCTTCATATAAAAAGTAGACAGAAGCATTCTGAGAAACTTTTTGTGATGTTTGCATT |
| 171-bp monomer, start at position 24937 |
| CAAGTCACAGAGTTCAACATTCTCTTTCATAGAGCACGTTTGAAACACTCCCTTTGTAGTATCTGGAAGTTGACATTTGGAGCGCTTTGAGG |
| TCTATGGTGAAAAAGGAAATCTCTTCCCATAAAAACTAGACAGAAGCATTCTCAGAATCTTGTTTGTGATGTGTGTGCT |
Bases aligned with the key string are bold.
R-STRAND (DIRECT) – S-STRAND (REVERSE COMPLEMENT) ALPHA MONOMER ALTERNATION
Performing KSA analysis using the key string KS28 we have found alternating regions (islands) R-strand (direct) and S-strand (reverse complement) alpha monomers. As an illustration, we present our analysis of R-strand and S-strand regions in the contig NT_007758.11 in chromosome 7, where we found sixteen alternating regions of alpha monomers, eight of R-strand and eight of S-strand (Table 17). In this contig R-strand regions contain 1395 alpha monomers of the lengths 166-176 bp and S-strand regions 880 alpha monomers. (In total, we identified and classified 4500 alpha monomers of lengths 166-176 bp in Build 36.1 assembly for chromosome 7.) In addition, we found 147 other alpha monomers or their segments outside of the 166-176 length intervals. In this way we identified altogether 2422 alpha monomers in the contig NT_007758.11.
Table 17.
R (Direct) - and S (Reverse Complement)-Strand Regions of Alpha Monomers in Contig NT_007758.11 in Chromosome 7
| Start position | R- or S-monomers |
|---|---|
| 1 | R |
| 39657 | S |
| 166275 | R |
| 187460 | S |
| 194693 | R |
| 199558 | S |
| 207568 | R |
| 252906 | S |
| 268826 | R |
| 296455 | S |
| 312212 | R |
| 387671 | S |
| 398809 | R |
| 421029 | S |
| 435498 | R |
| 485642 | Q (2326593) |
| 2985969 | S |
| 2986655 | Q |
Insertions into alpha monomer regions (position/length in bp): 42717/4756, 62854/1304, 80670/1529, 87037/1833, 92299/1780, 95622/4238, 100713/1387, 109990/523, 112397/1178, 128138/262, 132310/4004, 137241/376, 160098/6177, 188851/1884, 208421/477, 214283/998, 221715/932, 223158/477, 224314/304, 229237/7046, 239533/695, 278055/283, 304255/1116, 307597/1187, 310500/682, 314737/482, 348427/481, 351665/7023, 392184/430, 393472/5337, 402048/6320, 418939/484, 434972/526, 442287/3524, 463933/1498, 469667/1163, and 479414/1248. These lengths are typical of SINE/LINE. Q denotes broad regions without alpha monomers.
We have investigated divergence between R- and S-strand alpha monomers by aligning S-strand monomers to reverse complement of R-strand monomers. They are almost identical (divergence less than 0.5 %).
SUPRACHROMOSOMAL FAMILY ASSIGNMENT OF HORS AND ALPHA SATELLITE MONOMERS
Sequence comparison of alpha satellite monomers revealed 12 types of alphoid monomers, which form five su-prachromosomal families (SFs) [7,18,62,117]. They all descend from two basic types of monomers, A and B.
In subtypes of alpha satellite DNA consisting of dimers which belong to SF1 and SF2 (-J1J2- and -D1D2-, respectively) [118], majority of CENP-B boxes are regularly distributed in every other monomer unit leading to the “every other monomer scheme” [95,119]. On the other hand, in HORs which belong to SF3, the CENP-B boxes are distributed apparently irregularly and specifically to each chromosome [7,118,120]. As for pJα motif distribution, no systematic investigation was reported so far.
In the case of HORs we calculate divergence for pairwise comparison of all monomers from consensus HOR and SF monomers. To each monomer from consensus HOR the corresponding SF monomer with the lowest mutual divergence is assigned [116]. As an example, we present here the divergence matrix for 12mer consensus HOR in chromosome 11, revealing the SF3 classification (Table 18). Therefrom, the suprachromosomal classification of this HOR is W3 W4 W3 R1(W2a) W1 W5 W4 W3 W2 W1 W5 W4 (athe second lowest divergence for t04).
Table 18.
Comparison of Aligned Monomers in 12mer Consensus HOR in Chromosome 11 to SF Monomers (Divergence (%))
| t01 | t02 | t03 | t04 | t05 | t06 | t07 | t08 | t09 | t10 | t11 | t12 | ||
| 171 | 171 | 171 | 171 | 167 | 171 | 171 | 171 | 171 | 166 | 171 | 175 | ||
| J1* | 171 | 28.7 | 21.6 | 28.1 | 25.7 | 28.1 | 25.1 | 22.2 | 26.3 | 27.5 | 30.4 | 22.2 | 25.1 |
| D2* | 171 | 24.0 | 16.4 | 25.7 | 22.8 | 24.0 | 21.1 | 19.3 | 22.8 | 22.8 | 26.9 | 21.6 | 22.2 |
| W4* | 171 | 21.1 | 9.9 | 24.6 | 17.0 | 22.2 | 15.2 | 8.8 | 19.9 | 19.3 | 24.6 | 17.0 | 13.5 |
| W5* | 171 | 25.1 | 18.1 | 28.7 | 21.6 | 26.9 | 9.9 | 18.7 | 24.0 | 22.2 | 27.5 | 7.6 | 24.6 |
| M1* | 171 | 20.5 | 15.8 | 22.8 | 17.5 | 21.6 | 19.9 | 17.5 | 21.1 | 19.9 | 25.1 | 19.9 | 21.1 |
| R2* | 171 | 19.3 | 11.7 | 22.2 | 17.5 | 20.5 | 17.5 | 14.0 | 19.3 | 18.7 | 22.8 | 17.0 | 18.1 |
| J2* | 169 | 24.0 | 24.0 | 26.3 | 25.1 | 23.4 | 28.1 | 26.3 | 21.6 | 22.2 | 27.5 | 27.5 | 28.1 |
| D1* | 171 | 17.0 | 19.3 | 19.9 | 18.1 | 18.7 | 23.4 | 19.9 | 18.1 | 19.3 | 21.1 | 22.2 | 21.1 |
| W1* | 167 | 14.0 | 21.6 | 21.6 | 17.5 | 11.7 | 23.4 | 21.6 | 15.2 | 21.6 | 14.0 | 23.4 | 24.0 |
| W2* | 171 | 23.4 | 21.1 | 28.7 | 16.4 | 26.3 | 24.6 | 22.8 | 21.6 | 4.1 | 28.1 | 24.0 | 22.2 |
| W3* | 171 | 13.5 | 20.5 | 17.5 | 19.3 | 18.7 | 24.6 | 22.8 | 5.8 | 22.2 | 21.1 | 24.0 | 23.4 |
| R1* | 171 | 17.0 | 15.8 | 19.3 | 14.0 | 17.5 | 20.5 | 16.4 | 16.4 | 15.2 | 21.1 | 19.9 | 18.1 |
Underlined: SF monomer having lowest divergence to the monomer tn (nth monomer in consensus HOR).
Average divergence of consensus HOR with respect to all S-strand SF monomers reverse complement monomers of [117]) is 21%, while for SF monomers with lowest divergence it is 11%.
Using KSA we determined some new SF assignments: SF5 for 13mer (2211 bp), SF5 for 13mer (2214 bp), SF2 for 11mer (1869 bp), SF1 for 18mer (3058 bp), SF3 for 12mer (2047 bp), SF3 for 14mer (2379 bp), and SF5 for 17mer (2896 bp) in chromosomes 4, 5, 8, 10, 11, 17, and 19, respectively [116].
CENP-B BOX AND pJα DISTRIBUTIONS IN HORS AND ALPHA MONOMERS
In the 17-bp canonical CENP-B box motif 5’-PyTTCG TTGGAAPuCGGGA-3’ (R-strand sequence) only the underlined nucleotides (core recognition sequence) are essential for CENP-B box to bind with CENP-B proteins [117,120–125]. In de novo assembly of human centromeres the role of human centromeres was investigated using various synthetic repetitive sequences; only the combination of both the CENP-B box and HOR provided successful binding [119, 126]. CENP-B box appears only in alpha satellite HORs [18,119,120,124,126] while no CENP-B boxes were detected in monomeric alpha satellites [78,81].
Within the same region of monomeric unit, in some monomers a sequence motif was found, recognized by alpha satellite binding protein pJα [117]. The 17-bp pJα motif 5’-TTCCTTTTPyCACCPuTAG-3’ reflects some of nucleotides derived from alpha satellite monomer which were shown to be effective in binding experiments. A shorter pJα core sequence CCTTTTPyC [117], presenting an essential part of the pJα motif, was effective when dimerized, while a number of mutations outside of this core did not abolish binding.
Using KSA method we have identified CENP-B box and pJα motif distributions in alpha monomers (with more than two monomers in each HOR copy) contained in Build 35.1 assembly, after performing the KSA identification and determination of detailed alpha monomer structure [116]. Then the consensus distribution of CENP-B box and pJα motif was determined for each HOR [116].
In chromosome 5 we identified SF5 13mer which is the only HOR without any CENP-B box and pJα motif. This 13mer is highly homologous (96%) to the 13mer in chromosome 19. In chromosome 19 a new SF5 17mer has one CENP-B box and one pJα motif. Deleting four monomers in this 17mer, we get good alignment with 13mer in the same chromosome. In chromosome 10 a new SF1 18mer has eight CENP-B boxes in every other monomer except one. In chromosome 4 a new SF5 13mer has CENP-B box in three consecutive monomers. We found four exceptions to the rule that a CENP-B box belongs to the type B and pJα motif to type A monomers. Such cases are, for example, 16mers in chromosome 7 and 17mers in chromosome 19 [116].
The KSA study of the CENP-B box and pJα motif distributions is performed for monomeric and dimeric alpha satellites too.
As an illustration, let us consider the CENP-B box / pJα distribution in the sequence of alpha monomers in the first R-strand monomer region from Table 19. We see in the first section of array the appearance of CENP-B box forming an approximately every other monomer scheme, with monomers of length 169 bp. This reflects an approximate basic dimeric structure of alternating 169-bp (with CENP-B box) and 171-bp (without CENP-B box) monomers. In the last section of array dimeric structure is dissolved and there appears more irregularly distributed pJα motif mostly in 171-bp monomers. Such irregular distribution of pJα motif prevails in the remaining part of this contig and the density of pJα motif decreases, for example close to the end of contig the distribution is:
Table 19.
CENP-B box / pJα Distribution (Essential Part) in the Sequence of Alpha Monomers
| R3(169) – C(120), R5(169) – C(120), R7(169) – C(120), R8(169) – C(120), R10(169) – |
| C(120), R12(169) – C(120), R14(169) – C(120), R16(169) – C(120), R18(169) – C(120), |
| R20(169) – C(120), R22(164) – C(115), R32(169) – P(126), R34(169) – C(120), |
| R36(169) – C(120), R40(168) – C(119), R44(169) – C(120), R50(164) – C(115), |
| R52(169) – C(120), R53(171) – P(126), R54(169) – C(120), R56(169) – C(126), |
| R60(169) – C(120), R63(168) – C(119), R65(165) – C(116), R68(169) – C(120), |
| R73(168) – C(119), R78(169) – C(120), R80(169) – C(120), R82(169) – C(120), |
| R93(169) – C(120), R95(169) – C(120), R98(169) – C(120), R100(169) – C(120), |
| R102(169) – P(126), R106(171) – C(120), R113(169) – C(120), R119(171) – P(126), |
| R122(171) – P(126), R127(171) – P(126), R130(171) – P(126), R131(169) – P(124), |
| R134(171) – P(126), R135(171) – P(126), R137(170) – P(125), R141(169) – C(120), |
| R144(170) – P(125), R146(172) – P(127), R158(171) – P(126), R161(171) – P(126), |
| R163(171) – P(126), R166(171) – C(120), R176(171) – P(126), R177(171) – P(126), |
| R181(171) – P(126), R184(171) – P(126), R187(171) – P(126), R188(171) – P(126), |
| R189(171) – P(126), R191(171) – P(126), R192(171) – P(126), R193(171) – P(126) |
Section from position 1 to 32686 of R-strand monomer region from Table 15 (contig NT_007758.11 in chromosome 7).
Essential part of CENP-B box: -TTCG----A--CGGG-. Essential part of pJα motif: S-strand-------CPuAAAAGG--. Rn(l) denotes the nth monomer (in order of appearance), l is the monomer length (in bp). C(nc) denotes a CENP-B box starting at position nc within the monomer. P(np) denotes a pJα motif starting at position np within the monomer. For example R3(169) – C(120): the third monomer in the array is of length 169 bp and contains a CENP-B box starting at position 120 within the monomer. Table displays a list of monomers in order of appearance, deleting all those without CENP-B box and pJα motif.
R2390(172)–P(127), R2396(172)–P(127), R2399(166)–P(121), R2406(172)–P(127), …
The SF classification of alpha monomers or HORs is used as a basis for discussion of CENP-B box and pJα motif distributions in alpha monomers.
KSA IDENTIFICATION OF ALPHA MONOMERS AND HORS USING ESSENTIAL CENP-B BOX OR α AS KEY STRING
CENP-B box or pJα lie within every alpha monomer at the same location and therefore fragment lengths obtained by segmenting array of alpha monomers using CENP-B box or pJα are approximately multiples of 171, while outside of alpha monomer regions fragment lengths are irregular and much larger. If the monomer array forms a HOR pattern, a periodic pattern can be seen in the fragment length array, i.e., a periodic cell appears.
For a given genomic sequence we perform four KSA computations, using CENP-B box (R-strand), CENP-B box (S-strand), pJα motif (R-strand) and pJα motif (R-strand). Combining islands of alpha monomer containing sections in these four KSA segmentations we obtain alpha monomer and HOR identification in genomic sequence. (This choice of key string is not effective only in exceptional cases of HORs without any CENP-B box or pJα motif; the only known case of this type is the 13mer in chromosome 5 [116]).
Let as illustrate this method for whole FASTA file of chromosome X (Build 35.1). In this case we find two HOR islands for CENP-B box key string (S-strand), none island for CENP-B box key string (R-strand), five monomeric and two HOR islands for pJα key string (S-string), and seven monomeric islands for pJα key string (R-string). In Table 20 we present some sections of KSA fragment length arrays using pJα key string (S-strand) and in Table 21 using CENP-B box (S-strand). At the beginning or end of HORs a transitional region towards monomeric pattern can be seen.
Table 20.
Fragment Lengths for a Section with four Alpha Monomeric and one HOR Islands (Underlined) in Chromosome X
| … 41993, 576201, 529156, 747, 824, 687, 344, 681, 696, 686, 172, 687, 27895, 1302, 689, 683, 343, 346, 343, 506, 866, 859, 681, 344, 328, 349, 688, 344, 1350, 344, … 515, 339, 344, 339, 20542, 1022, 340, 1881, 686, 1888, 1201, 688, 1039, 1539, 686, 341, 344, 170, 343, 686, 678, 686, 859, … 1030, 684, 344, 344, 855, 15909, 171, 172, 339, 172, 342, 171, 168, 171, 344, 173, 384, 340, 172, 762, 68638, 171, 1023, 848, 1697, 355, 855, 1206, 852, 1206, 851, 1206, 851, 1206, 851, 1206, 851, 1206, 851, 1206, 851, 1206, 851, 75455, 54330, 27340, 27698, 108803, … |
Section Displayed is from Position 4984368 to 6646040) and pJα Key String CCTTTTCCAC and CCTTTTTCAC (S-Strand) were used.
Fragment lengths belonging to alpha monomer islands are underlined.
Table 21.
Fragment Lengths for Chromosome X and CENP-B Box Key String CCCG--T----CGAA (S-Strand)
| 758879, 5573664, 1862, 510, 532, 1529, 529, 1018, 511, 528, 167, 851, 511, 528, 167, 851, 511, 528, 167, 851, 511, 528, 167, 851, 511, 528, 167, 851, 511, 528, 167, 851, 511, 528, 167, 851, 511, 4039689, 28741184, 8106989, 511, 528, 167, 851, 511, 528, 1018, 512, 528, 1018, 511, 528, 167, 851, 510, 528, 167, 851, 510, 528, 167, 851, 510, 528, 1018, 511, 528, 167, 851, 511, 528, 167, 851, 511, 528, 167, 851, 510, 528, 167, 851, 511, 528, 167, 851, 511, … 1533, 1194, 340, 1193, 340, 856, 847, 8078, 5243, 4192878, 4262461, 46482867, 4782757, 13330196, 20005599, 8788730, 2760759 |
Fragment Lengths Belonging to Alpha Monomer Islands are Underlined.
In Table 20 we observe five islands of approximate multiples of 171, i.e., of alpha monomers. The first four islands are without pronounced periodicity of fragment lengths, revealing monomeric alpha satellites. The last island, between the fragment lengths 68638 and 75455, consists of approximate multiples of 171, i.e., of alpha monomers. Furthermore, we easily recognize the (851 bp, 1206 bp) periodic cell (eight copies), representing the 5mer+7mer = 12mer HOR (2057 bp). Two additional islands, one monomeric and one HOR, are not shown in Table 20.
In Table 21 we observe two HOR islands characterized by the (511 bp, 528 bp, 167 bp, 851 bp) periodic cell, representing the same 2057-bp 12mer HOR.
TANDEM OF LONG MONOMERS
KSA is convenient to identify and analyze any type of tandem or dispersed repeats using just PC for computations, from small (of several bp) to very long monomers (thousands or tens of thousands of bp). They are also convenient for identification and studies of palindromes, no matter how large.
As an illustrative case let us present a tandem of 3434-bp monomers in contig NT_006576.15 in chromosome 5 (Build 36.1). For a key string GTTTCG the KSA segmentation provides a local periodicity of the fragment length array from position 17523954 to 17578897, without any periodicity elsewhere. A section of length array encompassing a tandem repeat of 3434-bp monomers is shown in Table 22. Consensus 3434-bp monomer is displaced in Table 23. Sixteen 3434-bp monomers are highly convergent (average divergence from consensus is 0.3%, while the average pairwise divergence between monomers is 0.5%).
Table 22.
A Section of Length Array (from Position 174145793 to 18294206) in Contig NT_006576.15 in Chromosome 5 Showing a Tandem of 16 3434-bp Monomers
| 12284, 3738, 10130, 14427, 2652, 41407, 24523, 3434, 3434, 3434, 3434, 3434, 3434, 3434, 3434, 3434, 3434, 3434, 3434, 3434, 3434, 3434, 3434, 1588, 72632, 39723, 88188, 135119, 12210, 68390, 1976, 33152, 54931, 963, 53944,41916, 8535, 89879 |
Table 23.
Consensus 3434-bp Monomer in Chromosome 5 Determined Using Key String GTTTCG
| 1 | GTTTCGCCGT | AACCGGACAC | GGCTCCCGGC | CGCCCCTTCC | CACACACAAA | CACACACACT |
| 61 | GAATTTTCTC | GCTTCCACAG | TGTGAAGAAA | CTTGTGGAAG | GAGAGTATGT | TAGTTTTAGG |
| 121 | TCAATGCAGA | ACGAATTCTC | ACCAATTTTG | GGTATTTAAA | ACAAACACCA | GCTCACAGGT |
| 181 | CAGAAGTTCT | GCTAGGCCAA | GTGACTGCCT | CCTGCTCAGA | GTCCCACGAG | GGACCTCCAG |
| 241 | GATGGGTCTG | GCTGTGCGGT | CGTTGCCTCC | ACCTGAGAAG | GGTCTGGCTT | CGATCCGATT |
| 301 | CGAGTTGGTG | GCAGAATTCA | ACAATGCCTC | AGGGTTGCGA | GCCCCAGGCC | CACTTGTTTG |
| 361 | TTCTGCCTGC | TGCCGTGAGG | ATGCTCTCAG | CTCCTACCCG | TGCTGCCCAG | GTCTGGGCCG |
| 421 | TGAGGCTCCC | TGGGTGTGCA | CAGCCAGTGC | TGGGGAATCT | CCCACAGGGG | AGCGTAATCA |
| 481 | CAGGGGGGTT | CAGTCCTCCC | TTATAAAGGG | CTCAGATGAC | TGCATTAGAC | CCAGCCCTTA |
| 541 | GCAGCCGTTG | GTTCAGGATA | CCCCCCAATC | TAATGAGGAA | GTCGGGCGGG | CACATCAATT |
| 601 | CGTGCTTCCG | CCCACACCCA | AGGGAGGGGC | AGACACAGGG | CGACTCTCTG | AGGGGCGGGA |
| 661 | AATGCAGGGG | GCATTTCAGA | ATTCAGTCCT | CTTCACAGAA | TCGCAAAGTT | CACATCTCAC |
| 721 | AACAGTAAAG | AAACTATTTA | CAGTAAAAAT | GAGACATTTT | ACGAAGTTGA | GCATTAGAAA |
| 781 | ACTTCGATGT | CTGAGAAAAA | AAACTCTCTA | ACGCACAGGG | AAGAAAGCGG | TTTATCAAAT |
| 841 | ACTCTGAAAA | TAAAATGGGC | TGGGTGAGGG | AAACGTGAAA | ATATTATTTC | AATTTTATTT |
| 901 | TACGTCACTT | TATTTTAGTT | TATTTTATTT | TATTTGTTTA | TTTCTGAGAC | AGTGCCTCGC |
| 961 | TCTGTCCCCC | AGGCTGGATT | ACAGCGGCCT | CATCTCAGCC | CACTGCAGCC | TCGGCATCCT |
| 1021 | AGGCTCAACG | GATTCTCCTG | CCTCAGCCTC | CAGAGTGGCT | GGGACTAAAT | GTGCGCGCTA |
| 1081 | CCACGCCGGG | CAAATTTTTG | TATTTGCTCA | AGTAGAGACG | AGGTCTCGCC | ATTTTGGCCA |
| 1141 | GGCTGGTCTT | GAACTGCTGA | CTTCAGGTGA | TCTGCCCCAC | CTTGGCTTCC | CAAAGTGAAG |
| 1201 | GGACTATAGG | CGTGAGCCAC | CGCGCCCAGA | CTATGATAGT | TTCACACTGA | AGCCTGACGC |
| 1261 | TGCTCTGCCT | TAGGATTTTT | CCTGAGTTTT | ACTTCCTTGT | CAGGATGAGT | TGCTAGTTCA |
| 1321 | TATTTTCTGT | TGGATCTTTT | AGAAAGGCGT | TACTGATGAG | ATTATGGCTT | TCTCACAAGA |
| 1381 | AATACTACTC | TGGTGAAACT | CTGTTGAAAT | TATCAGTACC | TTAAGTTTCC | AATCCTTATC |
| 1441 | AAGTACAATA | GTTGAACATG | GCGTGGTAGC | TGAAAGTGTA | AGAGGCAGAA | TTTGGCAGAC |
| 1501 | TCCACTTCTT | CCCATTTCGA | TGGTTCCAGG | TTTTTTGGCT | TCAGCCGAAC | TAAAGAATGT |
| 1561 | CCTCACGAGC | TGTGAATTCA | CAGGTCACTA | CAGACAATTT | TTGAAACTGA | ATCACACTGT |
| 1621 | AATTTTTGGC | GTATGCTCTG | TGAGCTGTGC | TGGGAAGGTT | CACGCTGATT | CCGTAATAAA |
| 1681 | TCTCGGGTTT | TTACTCTATA | GCGAAAAATT | ACTCTTTGCC | ATCATGAAGG | CAAAGCAGAG |
| 1741 | TATGTACAAG | TAGAGTGTGG | AATAACTTTG | TCACTCGTGA | CGAACCGACT | TGGTCCAATA |
| 1801 | CTTTAACGAC | TTCTCCAATG | TCTCCGTACT | CAGGTTTGAT | TTTCTGAGTG | GATCATCGGT |
| 1861 | AGAATGAATA | AAATGAAGAA | TCCTCTAAGG | CAATGTTTGG | AACTAAATTT | CAGTGTCTCC |
| 1921 | GGAAGCACTG | GAAAAATCAC | CACGTGTAGC | GAAAGTGAAG | TGTCAATAGG | CTCTCTCTGT |
| 1981 | GTCCTTCAAA | CCGCCCATAT | GGTCGTTACA | AACGGCGGCT | TGAGGAAAGG | TGGTTTTGGA |
| 2041 | ATCGGTTTCT | CTCTGGTCTT | ACATGATGCA | TCTATACTAT | ACTGCATTAT | AATACAGGAA |
| 2101 | AGGGTCACTT | GCTGACATAA | AGCACAGCAG | GCAGGAATAG | AAGAGTCAAC | TTAGGGGAAA |
| 2161 | AAAAGAAAGT | GCTTTGTGAT | TTCAATTTGG | TGTCTGCAGT | TTGGAAAACG | GTTGATCAGT |
| 2221 | TTAACTGTTT | TCGTGGTGAC | TCACAAAAAT | ACATATGAGC | GTTGAAATTC | TACAGAAGAA |
| 2281 | CAACAATCGG | GGAAACATTT | CTGCAAGCTC | CAATTACTGG | AACCCAGACA | TAAGCCTACA |
| 2341 | AGCTAAGACA | GAGCTACACC | AGGCTTCAGC | AGGAAACCAT | ACAGATCTCC | TGGGAAGGGC |
| 2401 | TTCCCTCTCT | GAATGCAGCT | GCCTGTCCAC | AGGATGCTCT | AGGCCCAGGC | ACCTTGATTC |
| 2461 | CTCCAGCTGG | AAAGACATAG | AGAAACGCCT | CCACATCCCA | TTAAAATGCC | CAAAGATTTA |
| 2521 | GCCAAGGCTC | CTATGAAGCG | ATCTGCTGTC | TTCATCCAGG | TAAGGGCAAC | TTCGCATTTT |
| 2581 | AAGACACGAA | GATCGTGGGT | AAATCCAGGT | GGGACTGAGA | TGCGGGAGCT | CCGGCGCACA |
| 2641 | CACTCCTGTC | ATTGGAAGAT | GAACGCGGTA | CTTATTCCTG | CACAAACAGA | CCCTGCCCTC |
| 2701 | TGGCCCTGGG | CCTAGAACAT | GATTCTTTTG | CAGTTGCTGT | TGGGGAAGAG | GCCCTTGGGC |
| 2761 | TTTAACCTGC | GAACGGCCTC | CCTTAAATGC | TTGGGCTGCA | GCGGGGGCGT | CTCTCCCCAC |
| 2821 | ATCTCACACA | CGTCCAGGGC | CTCTTCCACC | ACCTCTCCAA | CAAAGAGCTT | GGCTATTCCA |
| 2881 | GCCATGGCAA | TGGCCGCGTT | CTCCGACACC | GAACTGCCAG | TGATAGCCCG | CATCAGACCC |
| 2941 | GCAACGCGTG | CTCTCGGGAA | CGCTGACCGG | CGACACACTT | CGTAGCGGGA | CAGCTGCTCC |
| 3001 | TCAGACATGG | CAGACAGCAG | GGTTGTCATC | CTCTGAGCCT | CCTCCGCATC | CACGGTGGGC |
| 3061 | TTGCTCTCCT | TCTTGCCTTT | CGTATGTGTT | TTCCGTCTTT | TGGCTGCAGG | AGGAGCTGAG |
| 3121 | GCTGAGGCCT | CACTGTCACC | TTCTGTGAGG | TCCATGACAT | CCTCACTCCT | GAGCTCACCT |
| 3181 | TCCTGATCCC | TGGGTGCTTC | CAAGTTCCCG | TCTAGGTCCT | CAGGGATTCC | ATCCTTCTTG |
| 3241 | CTGCCCTTCA | GACCTCGGGG | CATGGCGAGC | ATCTCAGCAG | ACACACCTGT | TTGCCTGCCG |
| 3301 | GTCTCCATGG | GTGAGATTCA | AGTCTGCTCC | GTGACAGCAG | CTGTACAGGC | AGAAGTTCCG |
| 3361 | GCTGGGGTGG | TTTGATTGTG | GATCTGCGAT | GAGAACCTTT | CAAAGATTTT | AGCTGCTGTG |
| 3421 | TTTCTGCTGA | GCCA |
Using the key string CGAAAC (reverse complement of GTTTCG) we identified three highly convergent 3434-bp monomers (average divergence 0.3% with respect to consensus) in tandem (positions 17508344 to 17518645). These three 3434-bp monomers are reverse complement to sixteen 3434-bp monomers in the neighboring region. Reverse complement of these three monomers are almost identical to 16 monomers which follow (divergence of only 0.09%).
REFERENCES
- 1.Maio JJ. DNA strand reassociation and polyribonucleotide binding in the African green monkey. Cercopithecus aethiops. J MolBiol. 1971;56:579–595. doi: 10.1016/0022-2836(71)90403-7. [DOI] [PubMed] [Google Scholar]
- 2.Manuelidis L. Repeating restriction fragments of human DNA. Nucleic Acids Res. 1976;3:3063–3076. doi: 10.1093/nar/3.11.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manuelidis L. Complex and simple sequences in human repeated DNAs. Chromosoma. 1978;66:1–21. doi: 10.1007/BF00285812. [DOI] [PubMed] [Google Scholar]
- 4.Manuelidis L. Chromosomal localization of complex and simple repeated human DNAs. Chromosoma. 1978;66:23–32. doi: 10.1007/BF00285813. [DOI] [PubMed] [Google Scholar]
- 5.Rosenberg H, Singer M, Rosenberg M. Highly iterated sequences of SIMIANSIMIANSIMIANSIMIANSIMIAN. Science. 1978;200:394–402. doi: 10.1126/science.205944. [DOI] [PubMed] [Google Scholar]
- 6.Wu JC, Manuelidis L. Sequence definition and organization of a human repeated DNA. J Mol Biol. 1980;142:363–386. doi: 10.1016/0022-2836(80)90277-6. [DOI] [PubMed] [Google Scholar]
- 7.Warburton PE, Willard HF. Evolution of centromeric alpha satellite DNA: molecular organization within and between human and primate chromosomes. In: Jackson M, Strachan T, Dover G, editors. Human Genome Evolution. Oxford: BIOS Scientific Pub-lishers; 1996. pp. 121–145. [Google Scholar]
- 8.Cho KHA. The Centromere. Oxford: Oxford University Press; 1997. [Google Scholar]
- 9.Willard HF. Chromosome-specific organization of human alpha satellite DNA. Am J Hum Genet. 1985;37:524–532. [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell AR, Gosden JR, Miller DA. A cloned sequence, p82H, of the alphoid repeated DNA family found at the centro-meres of all human chromosomes. Chromosoma. 1985;92:369–377. doi: 10.1007/BF00327469. [DOI] [PubMed] [Google Scholar]
- 11.Willard HF, Waye JS. Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol. 1987;25:207–214. doi: 10.1007/BF02100014. [DOI] [PubMed] [Google Scholar]
- 12.Waye JS, Willard HF. Chromosome-specific alpha satellite DNA: nucleotide sequence analysis of the 2.0 kilobasepair repeat from the human X chromosome. Nucleic Acids Res. 1985;13:2731–2743. doi: 10.1093/nar/13.8.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wevrick R, Willard VP, Willard HF. Structure of DNA near long tandem arrays of alpha satellite DNA at the centromeres of human chromosome 7. Genomics. 1992;14:912–923. doi: 10.1016/s0888-7543(05)80112-0. [DOI] [PubMed] [Google Scholar]
- 14.Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC. Human centromeric DNAs. Hum Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- 15.Vogt P. Potential genetic functions of tandem repeated DNA sequence blocks in the human genome are based on a highly conserved “chromatin folding code“. Hum Genet. 1990;84:301–336. doi: 10.1007/BF00196228. [DOI] [PubMed] [Google Scholar]
- 16.Willard HF. Evolution of alpha satellite. Curr Opin Genet Dev. 1991;1:509–514. doi: 10.1016/s0959-437x(05)80200-x. [DOI] [PubMed] [Google Scholar]
- 17.Choo KH, Vissel B, Nagy A, Earle E, Kalitsis P. A survey of the genomic distribution of alpha satellite DNA on all human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 1991;19:1179–1182. doi: 10.1093/nar/19.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandrov IA, Kazakov A, Tumeneva I, Shepelev V, Yurov Y. Alpha-satellite DNA of primates: old and new families. Chromosoma. 2001;110:253–266. doi: 10.1007/s004120100146. [DOI] [PubMed] [Google Scholar]
- 19.Rudd MK, Wray GA, Willard HF. The evolutionary dynamics of alpha-satellite. Genome Res. 2006;16:88–96. doi: 10.1101/gr.3810906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexandrov IA, Mashkova TD, Romanova LY, Yurov YB, Kisselev LL. Segment substitutions in alpha satellite DNA. Unusual structure of human chromosome 3-specific alpha satellite repeat unit. J MolBiol. 1993;20:516–520. doi: 10.1006/jmbi.1993.1302. [DOI] [PubMed] [Google Scholar]
- 21.Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- 22.Yang TP, Hansen SK, Oishi KK, Ryder OA, Hamkalo BA. Characterization of a cloned repetitive DNA sequence concentrated on the human X chromosome. Proc Natl Acad Sci USA. 1982;79:6593–6597. doi: 10.1073/pnas.79.21.6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willard HF, Smith KD, Sutherland J. Isolation and characterization of a major tandem repeat family from the human X chromosome. Nucleic Acids Res. 1983;11:2017–2033. doi: 10.1093/nar/11.7.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jabs EW, Wolf SF, Migeon BR. Characterization of a cloned DNA sequence that is present at centromeres of all human autosomes and the X chromosome and shows polymorphic variation. Proc Natl Acad Sci USA. 1984;81:4884–4888. doi: 10.1073/pnas.81.15.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolfe J, Darling SM, Erickson RP, Craig IW, Buckle VJ, Rigby PWJ, Willard HF, Goodfellow PN. Isolation and characterization of an alphoid centromeric repeat family from the human Y chromosome. J Mol Biol. 1985;182:477–485. doi: 10.1016/0022-2836(85)90234-7. [DOI] [PubMed] [Google Scholar]
- 26.Devilee P, Cremer T, Slagboom P, Bakker E, Scholl HP, Hager HD, Stevenson AFG, Cornelisse CJ, Pearson PL. Two subsets of human alphoid repetitive DNA show distinct preferential localization in the pericentric regions of chromosomes 13, 18, and 21. Cytogenet Cell Genet. 1986;41:193–201. doi: 10.1159/000132229. [DOI] [PubMed] [Google Scholar]
- 27.Waye JS, Willard HF. Structure, organization, and sequence of alpha satellite DNA from human chromosome 17: evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Moll Cell Biol. 1986;6:3156–3165. doi: 10.1128/mcb.6.9.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDermid HE, Duncan AM, Higgins MJ, Hamerton JL, Rector E, brasch KR, et al. Isolation and characterization of an alpha satellite repeated sequence from human chromosome 22. Chromosoma. 1986;94:228–234. doi: 10.1007/BF00288497. [DOI] [PubMed] [Google Scholar]
- 29.Devilee P, Slagboom P, Cornelisse CJ, Pearson PL. Sequence heterogeneity within the human alphoid repetitive DNA family. Nucleic Acids Res. 1986;14:2059–2073. doi: 10.1093/nar/14.5.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jorgensen AL, Bostock CJ, Bak AL. Chromosome specific subfamilies within human alphoid repetitive DNA. J Mol Biol. 1986;187:185–196. doi: 10.1016/0022-2836(86)90227-5. [DOI] [PubMed] [Google Scholar]
- 31.Choo KH, Brown R, Webb G, Craig IW, Filby RG. Genomic organization of human centromeric alpha satellite DNA: characterization of a chromosome 17 alpha satellite sequence. DNA. 1987;6:297–305. doi: 10.1089/dna.1987.6.297. [DOI] [PubMed] [Google Scholar]
- 32.Waye JS, Creeper LA, Willard HF. Organization and evolution of alpha satellite DNA from human chromosome 11. Chromosoma. 1987;95:182–188. doi: 10.1007/BF00330349. [DOI] [PubMed] [Google Scholar]
- 33.Yurov YB, Mitkevich SP, Alexandrov IA. Application of cloned satellite DNA sequences to molecular-cytogenetic analysis of constitutive heterochromatin heteromorphisms in man. Hum Genet. 1987;76:157–164. doi: 10.1007/BF00284914. [DOI] [PubMed] [Google Scholar]
- 34.Waye JS, Durfy SJ, Pinkel D, Kenwrick S, Patterson M, Davies KE, Willard HF. Chromosome-specific alpha satellite DNA from human chromosome 1: hierarchical structure and genomic organization of a polymorphic domain spanning several hundred kilobase pairs of centromeric DNA. Genomics. 1987;1:43–51. doi: 10.1016/0888-7543(87)90103-0. [DOI] [PubMed] [Google Scholar]
- 35.Waye JS, England SB, Willard HF. Genomic organization of alpha satellite DNA on human chromosome 7: evidence for two distinct alphoid domains on a single chromosome. Mol Cell Biol. 1987;7:349–356. doi: 10.1128/mcb.7.1.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donlon TA, Bruns GA, Latt SA, Mulholland J, Wyman AR. A chromosome 8-enriched alphoid repeat. Cytogenet Cell Genet. 1987;46:607. [Google Scholar]
- 37.Jabs EW, Persico MG. Characterization of human centro-meric regions of specific chromosomes by means of alphoid DNA sequences. Am J Hum Genet. 1987;41:374–390. [PMC free article] [PubMed] [Google Scholar]
- 38.Jorgensen AL, Bostock CJ, Bak AL. Homologous sub-families of human alphoid repetitive DNA on different nucleolus organizing chromosomes. Proc Natl Acad Sci USA. 1987;84:1075–1079. doi: 10.1073/pnas.84.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waye JS, Willard HF. Nucleotide sequence heterogeneity of alpha satellite repetitive DNA: a survey of alphoid sequences from different human chromosomes. Nucleic Acids Res. 1987;15:7549–7569. doi: 10.1093/nar/15.18.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyler-Smith C, Brown WRA. Structure of the major block of alphoid satellite DNA on the human Y chromosome. J Mol Biol. 1987;195:457–470. doi: 10.1016/0022-2836(87)90175-6. [DOI] [PubMed] [Google Scholar]
- 41.Delattre O, Bernard A, Malfoy B, Marlhens F, Viegas-Pequinot E, Brossard C, Haguenauer O, Creau-Goldberg N, Van Cong N, Dutrillaux B, Thomas G. Studies on the human chromosome 3 centromere with a newly cloned alphoid DNA probe. Hum Hered. 1988;38:156–167. doi: 10.1159/000153777. [DOI] [PubMed] [Google Scholar]
- 42.Hulsebos T, Schonk D, Dalen Iv, Coerwinkel-Driessen M, Schepens J, Ropers HH, Wieringa B. Isolation and characterization of alphoid DNA sequences for the pericentric regions of chromosomes 4, 5, 9, and 19. Cytogenet Cell Genet. 1988;47:144–148. doi: 10.1159/000132533. [DOI] [PubMed] [Google Scholar]
- 43.Devilee P, Kievits T, Waye JS, Pearson PL, Willard HF. Chromosome-specific alpha satellite DNA: isolation and mapping of a polymorphic alphoid repeat from human chromosome 10. Genomics. 1988;3:1–7. doi: 10.1016/0888-7543(88)90151-6. [DOI] [PubMed] [Google Scholar]
- 44.Choo KH, Vissel B, Brown R, Filby RG, Earle E. Homologous alpha satellite sequences on human acrocentric chromosomes with selectivity for chromosomes 13, 14 and 21: implications for recombination between nonhomologues and Robertsonian translocations. Nucleic Acids Res. 1988;16:1273–1284. doi: 10.1093/nar/16.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jorgensen AL, Kolvraa S, Jones C, Bak AL. A subfamily of alphoid repetitive DNA shared by the NOR-bearing human chromosomes 14 and 22. Genomics. 1988;3:1000–1009. doi: 10.1016/0888-7543(88)90139-5. [DOI] [PubMed] [Google Scholar]
- 46.Waye JS, Mitchell AR, Willard HF. Organization and genomic distribution of «82H» alpha satellite DNA. Evidence for a low copy or single-copy alphoid domain located on human chromosome 14. Hum Genet. 1988;78:27–32. doi: 10.1007/BF00291229. [DOI] [PubMed] [Google Scholar]
- 47.Jabs EW, Carpenter N. Molecular cytogenetic evidence for amplification of chromosome-specific alphoid sequences at enlarged C-bands on chromosome 6. Am J Hum Genet. 1988;43:69–74. [PMC free article] [PubMed] [Google Scholar]
- 48.Baldini A, Smith DI, Rocchi M, Miller OJ, Miller DA. A human alphoid DNA clone from the EcoRI dimeric family: genomic and internal organization and chromosomal assignment. Genomics. 1989;5:822–828. doi: 10.1016/0888-7543(89)90124-9. [DOI] [PubMed] [Google Scholar]
- 49.Waye JS, Willard HF. Chromosome specificity of satellite DNAs: short- and long-range organization of a diverged dimeric subset of human alpha satellite from chromosome 3. Chromosoma. 1989;97:475–480. doi: 10.1007/BF00295032. [DOI] [PubMed] [Google Scholar]
- 50.Alexandrov IA, Akopian TA, Vinnik EA, Mitkevich SP, Kisselev LL, Yurov YB. Cloned alpha satellite fragment - the molecular marker of human chromosome 4: sequence, genomic organization, polymorphism. Cytogenet Cell Genet. 1989;1989:949. [Google Scholar]
- 51.Greig GM, England SB, Bedford HM, Willard HF. Chromosome-specific alpha satellite DNA from the centromere of human chromosome 16. Am J Hum Genet. 1989;45:862–872. [PMC free article] [PubMed] [Google Scholar]
- 52.Wevrick R, Willard HF. Long-range organization of tandem arrays of alpha satellite DNA at the centromeres of human chromosomes: High frequency array-length polymorphism and meiotic stability. Proc Natl Acad Sci USA. 1989;86:9394–9398. doi: 10.1073/pnas.86.23.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexandrov IA, Akopian TA, Vinnik EA, Mitkevich SP, Kisselev LL, Yurov YB. Two alpha satellite domains on human chromosome 18: A novel 18-specific repeated unit. Cytogenet Cell Genet. 1989;51:949. [Google Scholar]
- 54.Jabs EW, Goble CA, Cutting GR. Macromolecular organization of human centromeric regions reveals high-frequency, polymorphic macro DNA repeats. Proc Natl Acad Sci USA. 1989;86:202–206. doi: 10.1073/pnas.86.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carine K, Jacquemin-Sablon A, Waltzer E, Mascarello J, Scheffler IE. Molecular characterization of human minichromosomes with centromere from chromosome 1 in human-hamster hybrid cells. Somat Cell Mol Genet. 1989;15:445–460. doi: 10.1007/BF01534895. [DOI] [PubMed] [Google Scholar]
- 56.Rocchi M, Baldini A, Archidiacono N, Lainwala S, Miller OJ, Miller DA. Chromosome-specific subsets of human alphoid DNA identified by a chromosome 2-derived clone. Genomics. 1990;8:705–709. doi: 10.1016/0888-7543(90)90258-v. [DOI] [PubMed] [Google Scholar]
- 57.Carson NL, Simpson NE. Two HinfI RFLPs detected by p-alpha-10RP8 at D10Z1. Nucleic Acids Res. 1990;18:1932. doi: 10.1093/nar/18.7.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baldini A, Rocchi M, Archidiacono N, Miller OJ, Miller DA. A human alpha satellite DNA subset specific for chromosome 12. Am J Hum Genet. 1990;46:784–788. [PMC free article] [PubMed] [Google Scholar]
- 59.Looijenga LH, Smit VT, Wessels JW, Mollewanger P, Oosterhuis JW, Cornelisse CJ, et al. Localization and polymorphism of a chromosome 12-specific alpha satellite DNA sequence. Cytogenet Cell Genet. 1990;53:216–218. doi: 10.1159/000132934. [DOI] [PubMed] [Google Scholar]
- 60.Wu JS, Kidd KK. Extensive sequence polymorphisms associated with chromosome 10 alpha satellite DNA and its close linkage to markers from the pericentromeric region. Hum Genet. 1990;84:279–282. doi: 10.1007/BF00200575. [DOI] [PubMed] [Google Scholar]
- 61.Choo KH, Earle E, Vissel B, Filby RG. Identification of two distant subfamilies of alpha satellite DNA that are highly specific for human chromosome 15. Genomics. 1990;7:143–151. doi: 10.1016/0888-7543(90)90534-2. [DOI] [PubMed] [Google Scholar]
- 62.Alexandrov IA, Mashkova TD, Akopian TA, Medvedev LI, Kisselev LL, Mitkevich SP, Yurov YB. Chromosome specific alpha satellites: two distinct families on human chromosome 18. Genomics. 1991;11:15–23. doi: 10.1016/0888-7543(91)90097-x. [DOI] [PubMed] [Google Scholar]
- 63.Greig GM, Parikh S, George J, Powers VE, Willard HF. Molecular cytogenetics of alpha satellite DNA from chromosome 12: fluorescence in situ hybridization and description of DNA and array length polymorphisms. Cytogenet Cell Genet. 1991;56:144–148. doi: 10.1159/000133071. [DOI] [PubMed] [Google Scholar]
- 64.Vissel B, Choo KH. Four distinct satellite subfamilies shared by human chromosomes 13, 14, and 21. Nucleic Acids Res. 1991;19:271–277. doi: 10.1093/nar/19.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rocchi M, Archidiacono N, Ward DC, Baldini A. A human chromosome 9-specific alphoid DNA repeat spatially resolvable from satellite 3 DNA by fluorescent in situ hybridization. Genomics. 1991;9:517–523. doi: 10.1016/0888-7543(91)90419-f. [DOI] [PubMed] [Google Scholar]
- 66.Wevrick R, Willard HF. Physical map of the centromeric region of human chromosome 7: relationship between two distinct alpha satellite arrays. Nucleic Acids Res. 1991;19:2295–2301. doi: 10.1093/nar/19.9.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Willard HF, Waye JS. Hierarchical order in chromosome-specific human alpha satellite DNA. Trends Genet. 1991;3:192–198. [Google Scholar]
- 68.Marcais B, Bellis M, Gerard A, Pages M, Boublik Y, Roizes G. Structural organization and polymorphism of the alpha satellite DNA sequences of chromosomes 13 and 21 as revealed by pulsed field gel electrophoresis. Hum Genet. 1991;86:311–316. doi: 10.1007/BF00202418. [DOI] [PubMed] [Google Scholar]
- 69.Haaf T, Willard HF. Organization, polymorphism, and molecular cytogenetics of chromosome-specific alpha-satellite DNA from the centromere of chromosome 2. Genomics. 1992;13:122–128. doi: 10.1016/0888-7543(92)90211-a. [DOI] [PubMed] [Google Scholar]
- 70.Ge Y, Wagner MJ, Siciliano M, Wells DE. Sequence, higher order repeat structure, and long-range organization of alpha satellite DNA specific to human chromosome 8. Genomics. 1992;13:585–593. doi: 10.1016/0888-7543(92)90128-f. [DOI] [PubMed] [Google Scholar]
- 71.Looijenga LHJ, Oosterhuis JW, Smit VTH, Wessels JW, Mollevanger P, Devilee P. Alpha satellite DNAs on chromosome 10 and 12 are both members of the dimeric suprachromosomal subfamily, but display little identity at the nucleotide sequence level. Genomics. 1992;13:1125–1132. doi: 10.1016/0888-7543(92)90027-p. [DOI] [PubMed] [Google Scholar]
- 72.Baldini A, Archidiacono N, Carbone R, Bolino A, Shridhar V, Miller OJ, et al. Isolation and comparative mapping of a human chromosome 20-specific alpha satellite DNA clone. Cytogenet Cell Genet. 1992;59:12–16. doi: 10.1159/000133188. [DOI] [PubMed] [Google Scholar]
- 73.Jackson MS, Mole SE, Ponder BAJ. Characterisation of a boundary between satellite III and alphoid sequences on human chromosome 10. Nucleic Acids Res. 1992;20:4781–4787. doi: 10.1093/nar/20.18.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.D’Aiuto L, Antonacci R, Marzella R, Archidiacono N, Rocchi M. Cloning and comparative mapping of a human chromosome 4-specific alpha satellite DNA sequence. Genomics. 1993;18:230–235. doi: 10.1006/geno.1993.1460. [DOI] [PubMed] [Google Scholar]
- 75.Cooper KF, Fisher RB, Tyler-Smith C. The major centromeric array of alphoid satellite DNA on the human Y chromosome is non-palindromic. Hum Mol Genet. 1993;2:1267–1270. doi: 10.1093/hmg/2.8.1267. [DOI] [PubMed] [Google Scholar]
- 76.Greig GM, Warburton PE, Willard HF. The organization and evolution of an alpha satellite subset shared by chromosomes 13 and 21. J Mol Evol. 1993;37:464–475. doi: 10.1007/BF00160427. [DOI] [PubMed] [Google Scholar]
- 77.Jackson MS, Slijepcevic P, Ponder BAJ. The organization of repetitive sequences in the pericentromeric region of human chromosome 10. Nucleic Acids Res. 1993;21:5865–5874. doi: 10.1093/nar/21.25.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trowell HE, Nagy A, Vissel B, Choo KH. Long-range analyses of the centromeric regions of human chromosomes 13, 14 and 21: identification of a narrow domain containing two key centromeric DNA elements. Hum Mol Genet. 1993;2:1639–1649. doi: 10.1093/hmg/2.10.1639. [DOI] [PubMed] [Google Scholar]
- 79.Mashkova TD, Akopian TA, Romanova LY, Mitkevich SP, Yurov YB, Kisselev LL, Alexandrov IA. Genomic organization, sequence and polymorphism of the human chromosome 4 specific alpha satellite DNA. Gene. 1994;140:211–217. doi: 10.1016/0378-1119(94)90546-0. [DOI] [PubMed] [Google Scholar]
- 80.Larin Z, Fricker MD, Tyler-Smith C. De novo formation of several features of a centromere following introduction of a Y alphoid YAC into mammalian cells. Hum Mol Genet. 1994;3:689–695. doi: 10.1093/hmg/3.5.689. [DOI] [PubMed] [Google Scholar]
- 81.Ikeno M, Masumoto H, Okazaki T. Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range alpha-satellite DNA arrays of human chromosome 21. Hum Mol Genet. 1994;3:1245–1257. doi: 10.1093/hmg/3.8.1245. [DOI] [PubMed] [Google Scholar]
- 82.Finelli P, Antonacci R, Marzella R, Lonoce A, Archidiacono N, Rocchi M. Structural organization of multiple alphoid sub-sets coexisting on human chromosomes 1, 4, 5, 7, 9, 15, 18, and 19. Genomics. 1996;38:325–330. doi: 10.1006/geno.1996.0635. [DOI] [PubMed] [Google Scholar]
- 83.Sugimoto K, Furukawa K, Kusumi K, Himeno M. The distribution of binding sites for centromere protein B (CENP-B) is partly conserved among diverged higher order repeating units of human chromosome 6-specific alphoid DNA. Chromosome Res. 1997;5:395–405. doi: 10.1023/a:1018448425994. [DOI] [PubMed] [Google Scholar]
- 84.Mahtani MM, Willard HF. Physical and genetic mapping of the human X chromosome centromere - repression of recombination. PCR Meth Appl. 1998;8:100–110. doi: 10.1101/gr.8.2.100. [DOI] [PubMed] [Google Scholar]
- 85.De la Puente A, Velasco E, Perez Jurado LA, Hernandez-Chico C, van de Rijke FM, Scherer SW, Raap AK, Cruces J. Analysis of the monomeric alphoid sequences in the pericentromeric region of human chromosome 7. Cytogenet Cell Genet. 1998;83:176–181. doi: 10.1159/000015175. [DOI] [PubMed] [Google Scholar]
- 86.Puechberty J, Laurent AM, Gimenez S, Billault A, Brun-Laurent ME, Calenda A, Marcais B, Prades C, Ioannou P, Yurov Y, Roizes G. Genetic and physical analyses of the centromeric and pericentromeric regions of human chromosome 5: recombination across 5cen. Genomics. 1999;56:274–287. doi: 10.1006/geno.1999.5742. [DOI] [PubMed] [Google Scholar]
- 87.Lo AWI, Liao GCC, Rocchi M, Choo KHA. Extreme reduction of chromosome-specific alpha satellite array is unusually common in human chromosome 21. Genome Res. 1999;9:895–908. doi: 10.1101/gr.9.10.895. [DOI] [PubMed] [Google Scholar]
- 88.O’Keefe CL, Matera AG. Alpha satellite DNA variant-specific oligoprobes differing by a single base can distinguish chromosome 15 homologs. Genome Res. 2000;10:1342–1350. doi: 10.1101/gr.10.9.1342. [DOI] [PubMed] [Google Scholar]
- 89.Mashkova TD, Oparina NY, Lacroix ML, Fedorova LI, Tumeneva IG, Zinovieva OL, Kisselev LL. Structural rearrangements and insertions of dispersed elements in pericentromeric alpha satellites occur preferably at kinkable DNA sites. J Mol Biol. 2001;305:33–48. doi: 10.1006/jmbi.2000.4270. [DOI] [PubMed] [Google Scholar]
- 90.Rudd MK, Willard HF. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 91.Spence JM, Critcher R, Ebersole TA, Valdivia MM, Earnshaw WC, Fukagawa T, Farr CJ. Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array. EMBO J. 2002;21:5269–5280. doi: 10.1093/emboj/cdf511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vafa O, Sullivan KF. Chromatin containing CENP-A and alpha satellite DNA is a major component of the inner kinetochore plate. Curr Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- 93.Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Harrington JJ, Van Bokkelen G, Mays RW, Gustasshaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 95.Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NJ, Cooke H, Masumoto H. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 96.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 97.Thompson JD, Higgins DG, Gibson TJ. CUSTAL-W-improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sonnhammer EL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- 99.Jurka J, Klonowski P, Dagman V, Pelton P. CENSOR - a program for identification and elimination of repetitive elements from DNA sequences. Comput Chem. 1996;20:119–121. doi: 10.1016/s0097-8485(96)80013-1. [DOI] [PubMed] [Google Scholar]
- 100.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Benson G, Su X. On the distribution of the k-tuple matches for sequence homology: a constant time exact calculation of the variance. J Comput Biol. 1998;5:86–100. doi: 10.1089/cmb.1998.5.87. [DOI] [PubMed] [Google Scholar]
- 102.Sagot MF, Myers EW. Identifying satellites and repetitions in biological sequences. J Comput Biol. 1998;5:539–553. doi: 10.1089/cmb.1998.5.539. [DOI] [PubMed] [Google Scholar]
- 103.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Delgrange O, Dauchet M, Rivals E. Location of repetitive regions in sequences by optimizing a compression method. In: Altman RB, Dunker AK, Hunter L, Klein TE, editors. Pacific Symposium on Biocomputing. The Orchid at Mauna Lani; 1999. pp. 245–265. Co-Chairs. [DOI] [PubMed] [Google Scholar]
- 105.Smit AFA, Hubley R, Green P. RepeatMasker at http://repeatmasker.org [Google Scholar]
- 106.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 107.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Landau GM, Schmidt JP, Sokol D. An algorithm for approximate tandem repeats. J Comput Biol. 2001;8:1–18. doi: 10.1089/106652701300099038. [DOI] [PubMed] [Google Scholar]
- 109.Stoye J, Gusfield D. Simple and flexible detection of contiguous repeats using a suffix tree. Theor Computer Sci. 2001;270:843–856. [Google Scholar]
- 110.Volfovsky N, Haas BJ, Salberg SL. Clustering method for repeat analysis in DNA sequences. Genome Biol. 2001;2:1–11. doi: 10.1186/gb-2001-2-8-research0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Castello AT, Martins W, Gao GR. TROLL-tandem repeat occurrence locator. Bioinformatics. 2002;18:634–636. doi: 10.1093/bioinformatics/18.4.634. [DOI] [PubMed] [Google Scholar]
- 112.Hauth AM, Joseph DA. Beyond tandem repeats: complex pattern structures and distant regions of similarity. Bioinformatics. 2002;S18:31–37. doi: 10.1093/bioinformatics/18.suppl_1.s31. [DOI] [PubMed] [Google Scholar]
- 113.Rosandić M, Paar V, Basar I. Key-string segmentation algorithm and higher-order repeat 16mer (54 copies) in human alpha satellite DNA in chromosome 7. J Theor Biol. 2003;221:29–37. doi: 10.1006/jtbi.2003.3165. [DOI] [PubMed] [Google Scholar]
- 114.Rosandić M, Paar V, Glunčić M, Basar I, Pavin N. Key-string algorithm- Novel approach to computational analysis of repetitive sequences in human centromeric DNA. Croat Med J. 2003;44:386–406. [PubMed] [Google Scholar]
- 115.Paar V, Pavin N, Rosandić M, Glunčić M, Basar I, Pezer R, Durajlija Žinić S. ColorHOR - novel graphical algorithm for fast scan of alpha satellite higher-order repeats and HOR annotation for GenBank sequence of human genome. Bioinformatics. 2005;21:846–852. doi: 10.1093/bioinformatics/bti072. [DOI] [PubMed] [Google Scholar]
- 116.Rosandić M, Paar V, Basar I, Glunčić M, Pavin N, Pilaš I. CENP-B box and pJ sequence distribution in human alpha satellite higher-order repeats (HOR) Chromosome Res. 2006;14:735–753. doi: 10.1007/s10577-006-1078-x. [DOI] [PubMed] [Google Scholar]
- 117.Romanova LY, Deriagin GV, Mashkova TD, Tumeneva IG, Mushegian AR, Kisselev LL, Alexandrov IA. Evidence for selection of alpha satellite DNA: The central role of CENP-B/pJαbinding region. J Mol Biol. 1996;261:334–340. doi: 10.1006/jmbi.1996.0466. [DOI] [PubMed] [Google Scholar]
- 118.Yoda K, Okazaki T. Site-specific base deletions in human alpha-satellite monomer DNAs are associated with regularly distributed CENP-B boxes. Chromosome Res. 1997;5:207–211. doi: 10.1023/a:1018407316908. [DOI] [PubMed] [Google Scholar]
- 119.Ohzeki J, Nakano M, Okada T, Matsumoto H. CENP-B box is required for the novo centromere chromatin assembly on human alphoid DNA. J Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T. Centromere protein B assembles human centromeric alpha satellite DNA at 17-bp sequence, CENP-B box. J Cell Biol. 1992;116:585–596. doi: 10.1083/jcb.116.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yoda K, Nakamura T, Masumoto H, et al. Centromere protein B of African green monkey cells: gene structure, cellular expression and centromeric localization. Mol Cell Biol. 1996;16:5169–5177. doi: 10.1128/mcb.16.9.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yoda K, Ando S, Okuda A, Kikuchi A, Okazaki T. In vitro assembly of the CENP-B/ alpha satellite DNA/core histone complex: CENP-B causes nucleosome positioning. Genes Cells. 1998;3:533–548. doi: 10.1046/j.1365-2443.1998.00210.x. [DOI] [PubMed] [Google Scholar]
- 124.Masumoto H, Nakano M, Ohzeki J. The role of CENP-B and alpha-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres. Chromosome Res. 2004;12:543–556. doi: 10.1023/B:CHRO.0000036593.72788.99. [DOI] [PubMed] [Google Scholar]
- 125.Basu J, Stromberg G, Compitello G, Willard HF, Van Bokkelen G. Rapid creation of BAC-based human artificial chromosome vectors by transposition with synthetic alpha-satellite arrays. Nucleic Acids Res. 2005;33:587–596. doi: 10.1093/nar/gki207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]


