Abstract
Despite more than a century of debate, the evolutionary position of turtles (Testudines) relative to other amniotes (reptiles, birds, and mammals) remains uncertain. One of the major impediments to resolving this important evolutionary problem is the highly distinctive and enigmatic morphology of turtles that led to their traditional placement apart from diapsid reptiles as sole descendants of presumably primitive anapsid reptiles. To address this question, the complete (16,787-bp) mitochondrial genome sequence of the African side-necked turtle (Pelomedusa subrufa) was determined. This molecule contains several unusual features: a (TA)n microsatellite in the control region, the absence of an origin of replication for the light strand in the WANCY region of five tRNA genes, an unusually long noncoding region separating the ND5 and ND6 genes, an overlap between ATPase 6 and COIII genes, and the existence of extra nucleotides in ND3 and ND4L putative ORFs. Phylogenetic analyses of the complete mitochondrial genome sequences supported the placement of turtles as the sister group of an alligator and chicken (Archosauria) clade. This result clearly rejects the Haematothermia hypothesis (a sister-group relationship between mammals and birds), as well as rejecting the placement of turtles as the most basal living amniotes. Moreover, evidence from both complete mitochondrial rRNA genes supports a sister-group relationship of turtles to Archosauria to the exclusion of Lepidosauria (tuatara, snakes, and lizards). These results challenge the classic view of turtles as the only survivors of primary anapsid reptiles and imply that turtles might have secondarily lost their skull fenestration.
Keywords: Pelomedusa subrufa/phylogeny/amniota/anapsids
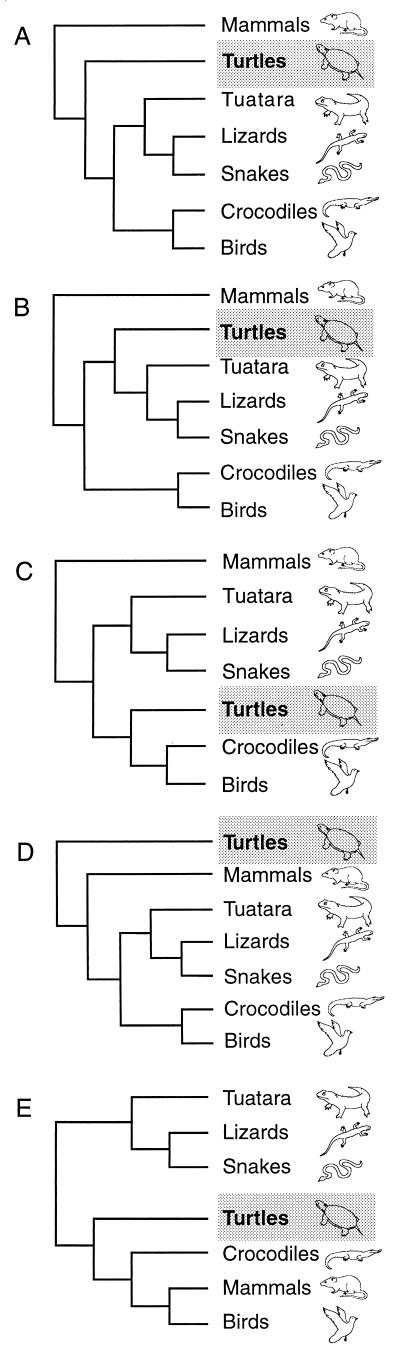
Traditionally, living turtles (Testudines) have been considered to be the only surviving representatives of anapsid reptiles (i.e., those that lack temporal fenestrae in their skulls). The morphology of the anapsid skull is generally regarded as primitive with respect to the more advanced condition of diapsid reptiles (i.e., the presence of two fenestrae in the temporal region of the skull). Therefore, turtles are widely considered basal to all extant reptiles (Lepidosauria and Archosauria; refs. 1 and 2; Fig. 1A). However, after more than a century of debate (3), the phylogenetic position of turtles remains uncertain. Most recently, based on an extensive morphological data set, turtles have been proposed to have diapsid affinities (ref. 4; Fig. 1B). A current molecular study (5) suggested that turtles are the sister group of the Archosauria (crocodiles and birds) to the exclusion of Lepidosauria (tuatara, lizards, and snakes; refs. 6–9; Fig. 1C). Further, it was hypothesized (10) that mammals and diapsid reptiles are sister groups because they share a lower temporal fenestra and that turtles are their sister group (Fig. 1D). A hypothesis in which birds are the closest relatives of mammals, reviving the clade Haematothermia (3), also has been proposed to explain amniote phylogenetic relationships (refs. 11–13; Fig. 1E).
Figure 1.
Proposed hypotheses explaining the phylogenetic relationships among living amniotes. (A) Mammals represent the sister group of all other extant amniotes. Turtles are the only living representatives of the Anapsida and are the sister group of diapsid reptiles (Lepidosauria and Archosauria; refs. 1, 2, 18–20). (B) Turtles have diapsid affinities and are the sister group of Lepidosauria (tuatara, lizards, and snakes; ref 4). (C) Turtles are diapsids as the sister group of Archosauria (birds and crocodiles) to the exclusion of Lepidosauria (5–7). (D) Mammals and diapsids as sister groups to the exclusion of turtles, the Parareptilia hypothesis (10). (E) The Haematothermia hypothesis: birds are the sister group of mammals (3, 11–13).
During the last decade, various forms of molecular data have been collected with the explicit goal of resolving the controversy of amniote relationships and thereby the phylogenetic position of turtles. However, phylogenetic analyses of amino acid sequences from αA-crystallin, α- and β-hemoglobin, myoglobin, histone H2B, cytochrome c, and insulin turned out to be rather inconclusive, leaving the relationships of birds, turtles, and crocodiles unresolved (ref. 14 and references therein). A mammal–bird clade (Haematothermia) seemed plausible, based on maximum-parsimony (MP) and neighbor-joining (NJ) analyses of the 18S rRNA gene sequences of 21 tetrapods (14). However, all reanalyses of these nuclear data (14) with weighted parsimony (15), an approach that combines molecular and morphological data (16), and a tree reconstruction method that takes into account the variability of the rRNA molecule (17) rejected the idea of a Haematothermia clade and supported a phylogeny with mammals as a sister group to all reptiles (including the turtle) and birds (Fig. 1A). Recently, the phylogenetic NJ analysis of the complete αA- and αB-crystallin amino acid sequences supported the traditional amniote phylogeny (18). The Archosauria clade (the bird and crocodile relationship) was confirmed further by the analyses of the complete mitochondrial 12S and 16S rRNA sequences (19) and an analysis that included the complete mitochondrial genome of the alligator (20) but lacked information from turtles.
To resolve the phylogenetic position of turtles and to test the traditional amniote phylogeny, we have determined the complete mitochondrial genome sequence of the African side-necked turtle, Pelomedusa subrufa (suborder Pleurodira). The need for more molecular data on turtles was stressed repeatedly by those who study phenotypic traits (4, 5, 8). Phylogenetic analyses of complete mitochondrial genomes have been used successfully to infer phylogenetic relationships among major groups of vertebrates. Therefore, complete mitochondrial genomes were expected to be appropriate markers for resolving this evolutionary question.
MATERIALS AND METHODS
DNA Extraction, Cloning, PCR, and Sequencing.
Mitochondrial DNA was purified from the liver of a single specimen of the African side-necked turtle (P. subrufa) as described previously (21). After homogenization, intact nuclei and cellular debris were removed by low-speed centrifugation. Mitochondria were pelleted and subjected to a standard alkaline lysis procedure followed by a phenol/chloroform extraction. The isolated mtDNA was cleaved with ApaI and HindIII restriction enzymes. One ApaI fragment of 5 kb and three HindIII fragments of 3.3, 0.6, and 0.23 kb were cloned into pUC18. To complete the cloning of the mitochondrial molecule, PCR primers were designed to amplify two fragments of 6 and 1 kb by using the mtDNA extraction as template source. The amplification of the 6-kb fragment was achieved by using the Expand long template PCR system of Boehringer Mannheim (with PCR buffer 1) and the following program: 2 min at 94°C, 10 cycles of 10 s at 94°C, 30 s at 58°C, and 4 min at 68°C, 20 cycles of 10 s at 94°C, 30 s at 58°C, and 4 min at 68°C with an elongation that increases 20 s for each cycle, and finally, 1 cycle of 5 min at 68°C. PCR fragments were cloned into the pGEM-T vector (Promega).
Recombinant plasmids were used as a template for Taq Dye Deoxy Terminator cycle-sequencing reactions (Applied Biosystems) according to manufacturer’s instructions. Sequencing was performed with an automated DNA sequencer (Applied Biosystems 373A Stretch). Sequences were obtained by using both M13 universal sequencing primers and several specific oligonucleotide primers. The sequences obtained from each clone averaged 450 bp in length, and each sequence overlapped the next contig by about 100 bp. In no case were differences in sequence observed between the overlapping regions. The location and sequence of these primers will be provided by the authors on request.
Sequence Analysis, Phylogenetic Reconstruction, and Statistical Methods.
The complete nucleotide sequence of the side-necked-turtle mitochondrial genome was aligned with the clustal w program (22) and refined by eye based on the corresponding deduced amino acid sequences and rRNA and tRNA secondary structures. Gaps resulting from the alignment were treated as missing data. Ambiguous alignments were excluded from the phylogenetic analyses. (Aligned sequences and exclusion sets are available from the authors on request.)
Three data sets were analyzed: (i) all protein coding genes combined at the amino acid level (excluding ND6, because it is encoded by the L strand, and ND3 and ND4L, because their putative ORFs are interrupted by extra nucleotides), (ii) 12S and 16S rRNA genes combined, and (iii) all 22 tRNA gene sequences combined. Each of these DNA data sets was subjected to the MP method (paup*, version d64; ref. 23) by using heuristic searches (tree bisection and reconnection TBR branch swapping; mulpars option in effect) with 10 random stepwise additions of taxa to find the most parsimonious tree. Transitions and transversions were given equal weight. NJ (24) analyses (based on HKY85 distance matrices; ref. 25) of the sequences were also performed with paup* (23). Maximum-likelihood (ML) analyses (based on HKY85 distance matrices; ref. 25) were performed with paup* (23), molphy, version 2.3 (26), and puzzle, version 4.0 (27). In the NJ and ML analyses, transition/tranversion ratios were optimized to maximize the likelihood, and empirical base frequencies were used. In the protein ML analyses, a NJ tree was inferred as the starting tree for a local rearrangement search for the ML tree with the mtREV model (28) by using molphy (26), and puzzle (27).
The robustness of the phylogenetic results was tested by (i) bootstrap analyses (29), implemented in paup* (23) with 100 pseudoreplications each, (ii) the RELL resampling of the estimated log likelihood method (30), implemented in molphy (26), with 10,000 pseudo-replications, and (iii) quartet puzzling, implemented in puzzle (27) with 10,000 pseudoreplications.
Statistical confidence of the resulting best trees of each ML analysis was evaluated by calculating the standard error of the difference in log likelihood between the resulting best tree and the competing hypotheses by using the formula of Kishino and Hasegawa (31) implemented in molphy (26) and paup* (23). Similarly, for MP analyses, statistical confidence was assessed by calculating the standard deviation of the difference in the number of steps between the resulting most parsimonious tree and the alternative trees by using a two-tailed Wilcoxon signed-rank test (32) implemented in paup* (23). If the difference in log likelihoods or the difference in the number of steps between two competing phylogenetic hypotheses were more than 1.96 times the standard deviation, then the two phylogenies were declared significantly different.
RESULTS AND DISCUSSION
The African Side-Necked-Turtle Mitochondrial Genome Shows Several Unusual Features.
The complete mitochondrial genome of P. subrufa is 16,787 bp long. The overall base composition of the L strand is 34% A, 27% T, 27% C, and 12% G. The organization of the turtle mitochondrial genome conforms to the consensus vertebrate mitochondrial gene order (Table 1). However, this mitochondrial genome is characterized by a set of unusual features: the absence of an origin of replication for the light strand, a long noncoding region separating the ND5 and ND6 genes, an overlap between ATPase 6 and COIII genes, and the existence of extra nucleotides in ND3 and ND4L putative ORFs.
Table 1.
Features of the African side-necked turtle mitochondrial genome
| Feature | Nucleotide no.
|
Codon
|
|||
|---|---|---|---|---|---|
| From | To | Size, bp | Start | Stop | |
| tRNAPhe | 1 | 71 | 71 | ||
| 12S rRNA | 72 | 1039 | 968 | ||
| tRNAVal | 1040 | 1104 | 65 | ||
| 16S rRNA | 1105 | 2708 | 1604 | ||
| tRNALeu (UUR) | 2709 | 2784 | 76 | ||
| NADH 1 | 2785 | 3752 | 968 | ATA | TA− |
| tRNAIle | 3753 | 3823 | 71 | ||
| tRNAGln | 3893 | 3824 | 70 (L) | ||
| tRNAMet | 3894 | 3962 | 69 | ||
| NADH 2 | 3963 | 5002 | 1040 | ATC | TA− |
| tRNATrp | 5003 | 5077 | 75 | ||
| tRNAAla | 5147 | 5079 | 69 (L) | ||
| tRNAAsn | 5221 | 5149 | 73 (L) | ||
| tRNACys | 5294 | 5225 | 70 (L) | ||
| tRNATyr | 5366 | 5296 | 71 (L) | ||
| COI | 5368 | 6912 | 1545 | GTG | AGA |
| tRNASer (UCN) | 6978 | 6908 | 71 (L) | ||
| tRNAAsp | 6980 | 7047 | 68 | ||
| COII | 7049 | 7736 | 688 | ATG | T-- |
| tRNALys | 7737 | 7806 | 70 | ||
| ATPase 8 | 7808 | 7975 | 168 | ATA | TAA |
| ATPase 6 | 7966 | 8652 | 687 | ATG | TAA |
| COIII | 8645 | 9428 | 784 | ATA | T-- |
| tRNAGly | 9429 | 9495 | 67 | ||
| NADH 3 | 9575 | 9845 | 271 | ATA* | T-- |
| tRNAArg | 9846 | 9914 | 69 | ||
| NADH 4L | ? | ? | ? | ? | ? |
| NADH 4 | 10209 | 11586 | 1378 | ATG | T-- |
| tRNAHis | 11587 | 11656 | 70 | ||
| tRNASer (AGY) | 11658 | 11714 | 57 | ||
| tRNALeu (CUN) | 11715 | 11786 | 72 | ||
| NADH 5 | 11789 | 13606 | 1818 | ATG | TAA |
| NADH 6 | 14252 | 13731 | 522 (L) | ATA | AGG |
| tRNAGlu | 14323 | 14256 | 68 (L) | ||
| Cyt b | 14316 | 15458 | 1143 | ATA | TAG |
| tRNAThr | 15460 | 15526 | 67 | ||
| tRNAPro | 15593 | 15527 | 67 (L) | ||
| Control region | 15594 | 16787 | 1194 | ||
The control region in the turtle mitochondrial genome is 1194 bp long, and it is localized between the tRNAPro and tRNAPhe genes (Table 1; ref. 33). Analysis of this control-region sequence permitted the identification of one conserved-sequence block, a putative conserved-sequence block, and three termination-associated sequences (34). Moreover, the presence of six direct repeats localized in tandem at the 3′ end of the control region was also detected (34). Each repeat was composed of a 45-bp sequence followed by a (TA)n microsatellite with a variable number of repeat units (n = 10–11) (34).
The origin of light-strand replication, which in vertebrates is located normally in a cluster of five tRNA genes (WANCY region), was not found in the turtle mitochondrial genome. The same condition has been reported in other reptiles and in the chicken (35–37). It has been suggested that the tRNAs in the WANCY region, which have the potential to fold into a stem–loop secondary structure, might replace the function of the light strand in these species (37).
There are only 14 noncoding intergenic spacer nucleotides that show a slight A + C bias (57%). Several unusual overlaps in the same strand (ATPase 8/ATPase 6/COIII) and in opposite strands (COI/tRNASer(UCN); tRNAGlu/cyt b) account for this small number of intergenic spacer nucleotides. Interestingly, this extreme economy of intergenic spacers in most of the mitochondrial genome contrasts with the existence of a long 124-bp noncoding stretch separating ND5 and ND6 genes. No significant matches for this sequence were found in a GenBank search.
The 12S and 16S rRNA genes in turtle mitochondria are 937 and 1591 nucleotides long, respectively. Our sequence shows only a single difference (a transition) with respect to a partial 12S rRNA gene sequence of P. subrufa that has been reported (38). All of the 22 tRNA gene sequences can be folded into a canonical cloverleaf secondary structure with the exception of tRNASer(AGY). These tRNAs range in size from 65 to 76 nucleotides.
The turtle mitochondrial genome contains at least 12 of the 13 large ORFs found in other vertebrate mitochondrial genomes (Table 1). The only ORF that cannot be inferred directly from the sequence is that of the ND4L gene, because detection is prevented by the existence of at least two extra nucleotides in the sequence. Posttranscriptional editing is likely involved in the translation of this ORF. It is unlikely that ND4L is not functional in P. subrufa, because the deletion of the extra bases leads to a putative amino acid sequence which is conserved with respect to other ND4L protein sequences. Additionally, the ND3 ORF is truncated in its 3′ end with respect to other vertebrate ND3 genes because of an extra base. This gene likely starts with an internal initiation codon (as shown in Table 1). Alternatively, posttranscriptional editing of the extra nucleotide may occur, and then the first initiation codon after the tRNAGly may be used to start the translation. There are two cases of reading-frame overlap in two genes encoded by the same strand (ATPases 8 and 6 overlap by 10 nucleotides; ATPase 6 and COIII share 8 nucleotides). The ND4L/ND4 overlap found in other vertebrate mitochondrial genomes could not be determined in the turtle (see above). Turtle mtDNA protein-encoding genes use ATG, ATA, ATC, and GTG as initiation codons (Table 1). Interestingly, a wide variety of stop codons is also found (AGA, AGG, TAG, TAA, TA-, and T--).
Phylogenetic Analyses of the Complete Mitochondrial Genome Data Set Placed Turtle Basal to Archosauria and Ruled out the Haematothermia and the Parareptilia Hypotheses.
The complete nucleotide sequence of the side-necked-turtle mitochondrial genome was aligned with 12 complete vertebrate mitochondrial DNA sequences: rainbow trout, Oncorhynchus mykiss (L29771; ref. 21); carp, Cyprinus carpio (X61010; ref. 39); loach, Crossostoma lacustre (M91245; ref. 40); African lungfish, Protopterus dolloi (L42813; ref. 41); coelacanth, Latimeria chalumnae (U82228; ref. 42); clawed frog, Xenopus laevis (M10217; ref. 43); alligator, Alligator mississippiensis (Y13113; ref. 20); chicken, Gallus gallus (X52392; ref. 35); platypus, Ornithorhynchus anatinus (X83427; ref. 44); opossum, Didelphis virginiana (Z29573; ref. 45); blue whale, Balaenoptera musculus (X72204; ref. 46); and human, Homo sapiens (D38112; ref. 47).
The new molecular data were analyzed in three data sets. (i) The deduced amino acids of all the mitochondrial protein-coding genes (with the exception of ND3, ND4L, and ND6) were aligned, producing 3,484 positions, of which 489 were excluded because of ambiguity. Of the remaining sites, 1,755 were variable, and 1,150 were parsimony-informative. Amino acid sequence divergence varied between 11% and 35%. (ii) The 12S and 16S rRNA genes produced 2,903 positions, of which 1,120 were excluded. Of the remaining sites, 1,087 were variable, and 783 were phylogenetically informative based on the parsimony criterion. Nucleotide sequence divergence varied between 11% and 35%. (iii) All tRNA genes combined produced an alignment of 1,621 positions. Of them, 1,182 were variable, and 847 were parsimony-informative. Also here nucleotide sequence divergence varied between 11% and 35%.
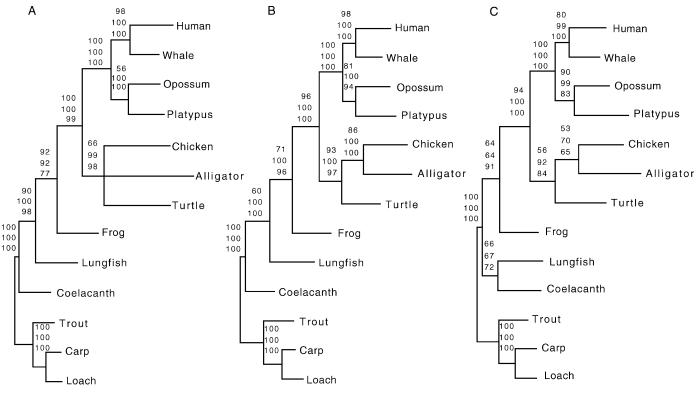
The phylogenetic analysis of the combined protein-coding genes at the amino acid level produced a single most-parsimonious tree of 5,919 steps with a consistency index for informative characters of 0.72. According to this tree, the alligator and the chicken are sister groups, and the turtle clusters with the Archosauria group. NJ (with mean character distances) and ML analyses (with the mtREV model) arrived at a topology identical to that of MP. Bootstrap analyses with MP, NJ, and ML showed support (66%, 99%, and 98%, respectively) for the placement of the turtle with the alligator and the chicken (Fig. 2A). However, phylogenetic relationships among the chicken, the alligator, and the turtle could not be resolved confidently, because the bootstrap values that clustered alligator and chicken were below 50% (Fig. 2A). This result might be caused by long-branch attraction (48) between the turtle and alligator, which have somewhat longer branches than the chicken (Fig. 2A).
Figure 2.
Bootstrap consensus trees (which used the 50% majority rule) of amniotes based on 100 pseudoreplications. The three data sets were subjected to MP, NJ, and ML analyses (bootstrap values are listed as upper, middle, and lower numbers above branches, respectively). (A) The first data set included mitochondrial protein-coding genes combined at the amino acid level. (B) The second data set combined all mitochondrial rRNA genes (C). The third data set combined all 22 tRNA genes. Teleosts (carp, loach, and rainbow trout) were used as outgroup taxa.
The phylogenetic analyses of the rRNA data set with MP (a single most-parsimonious tree of 3,234 steps with a consistency index of 0.55), NJ (HKY85 distances; A = 0.33%, C = 0.24%, G = 0.21%, T = 0.22%; transition/transversion ratio = 1.37), and ML (−ln L = 15943.68) resulted in topologies identical to that produced by the protein-coding-gene data set (Fig. 2A). The Archosauria (alligator and chicken), as well as the sister-group relationship between turtle and Archosauria, achieved convincing bootstrap support (Fig. 2B).
The phylogenetic analyses of the combined tRNA genes produced a single most-parsimonious tree of 3,299 steps (consistency index of 0.54) with a slightly different topology. In this tRNA tree, the lungfish is not identified as a sister group of tetrapods as in the other two data sets. This tree groups the lungfish with the coelacanth (66% bootstrap value; Fig. 2C). NJ (HKY85 distances; A = 0.31, C = 0.20, G = 0.21, T = 0.28; transition/transversion ratio = 2.23) and ML (−ln L = 14859.62) analyses of the tRNA data set resulted in identical topologies (Fig. 2C).
All phylogenetic analyses of the mitochondrial sequences, regardless of the weighting scheme (data not shown), agreed on a grouping of turtles, crocodiles, and birds with the exclusion of mammals (Fig. 2), ruling out the two hypotheses shown in Fig. 1 D and E. All mitochondrial DNA data, therefore, reject the Haematothermia hypothesis (refs. 3 and 11–14; Fig. 1E and Table 2) and provide evidence that crocodiles are the living sister group of birds (5, 10, 18–20, 49). The hypothesis, based only on living taxa, that turtles branched off from the amniotes before mammals and diapsid reptiles (the Parareptilia hypothesis, the standard textbook hypothesis, shown in Fig. 1D; refs. 10 and 49) is therefore invalidated by the analyses of our mitochondrial data (Fig. 2). Importantly, the statistical support for four alternative placements of turtles (Fig. 1) was determined further by tests developed by Templeton (32) and Kishino-Hasegawa (ref. 31; Table 2).
Table 2.
Statistical support for alternative hypotheses on amniote phylogenetic relationships
| Hypothesis | Templeton test, Δ ± SE
|
Kishino-Hasegawa test, Δ ± SE
|
||||
|---|---|---|---|---|---|---|
| Protein-coding | rRNA | tRNA | Protein-coding | rRNA | tRNA | |
| out, (mammals, (turtle, (alligator, chicken))) | 5,919 | 3,234 | 3,299 | −4.53 ± 19 | −15,943.7 | −14,859.6 |
| out, (mammals, (chicken, (turtle, alligator))) | 2 ± 9.8 | 20 ± 8.5 | 3 ± 9.2 | −40102.1 | −15.9 ± 9.4 | −6.5 ± 9.4 |
| out, ((mammals, chicken), (turtle, alligator)) | 25 ± 13.2 | 39 ± 11.5 | 11 ± 11.3 | −75.3 ± 20.3 | −50.1 ± 17.4 | −24.4 ± 12.7 |
| out, (turtle, (mammals, (alligator, chicken))) | 23 ± 9 | 20 ± 8.1 | 7 ± 7 | −77.8 ± 28.3 | −26.2 ± 13 | −10.2 ± 6.6 |
| out, ((tuatara, iguana), (turtles, (alligator, chicken))) | 4,080 | −19,435.4 | ||||
| out, (turtles, ((tuatara, iguana), (alligator, chicken))) | 2 ± 5.8 | −5.7 ± 8.9 | ||||
| out, ((turtles, (tuatara, iguana)), (alligator, chicken)) | 7 ± 5.4 | −7.9 ± 8.5 | ||||
The differences in number of steps (Δ) from that of the MP tree are shown ± their SE, which were estimated by the Wilcoxon signed-rank test32. The differences in log likelihood of alternative trees (Δ) from that of the ML tree are shown ± their SE, which were estimated by the formula of Kishino and Hasegawa31. Two phylogenies were declared significantly different when the difference in log likelihoods or number of steps was more than 1.96 times the SE.
Phylogenetic Analyses of the Complete 12S and 16S rRNA Gene Sequences Supported the Diapsid Affinities of Turtles.
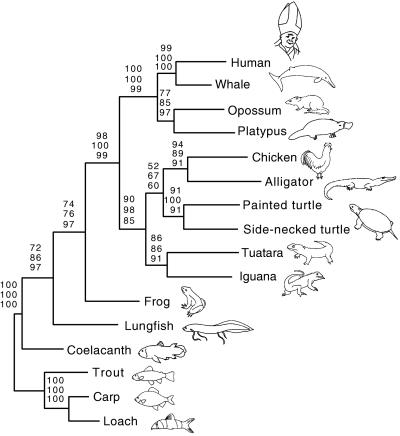
To test the anapsid or diapsid affinities of the turtle, the complete nucleotide sequences of the 12S and 16S rRNA genes of the painted turtle, the iguana, and the tuatara (19) were added to the rRNA data set. In the MP analysis a single most-parsimonious tree of 4,441 steps and a consistency index of 0.46 were found. According to this tree, the turtles are the sister group of the alligator and chicken clade (Fig. 1C). The tuatara and the iguana form a monophyletic group (Lepidosauria), which is a sister group to turtles, alligators, and chickens (Fig. 3). NJ (HKY85 distances; A = 0.34%, C = 0.24%, G = 0.20%, T = 0.22%; transition/transversion ratio = 1.30) and ML (−ln L = 20811.56) produced trees with topology identical to the MP tree (Fig. 3). The sister-group relationship of turtles to the alligator and chicken clade is supported by moderate bootstrap values (52%, 67%, and 60% for MP, NJ, and ML, respectively), which are higher in a data set that excludes the monotreme sequence (67%, 97%, and 79% for MP, NJ, and ML, respectively; data not shown). The placement of the tuatara and iguana with the turtle and Archosauria clade (the monophyly of Reptilia argues against the Parareptilia hypothesis) is supported by high bootstrap values for MP, NJ, and ML (90%, 98%, and 85%, respectively; Fig. 3). According to tests designed by Templeton (32) and Kishino-Hasegawa (31), alternative hypotheses (Fig. 1) could not be rejected statistically based on this rRNA data set (Table 2). However, in the data set that excludes the platypus sequence, the hypothesis depicted in Fig. 1C was favored, and only a sister-group relationship between turtles and Lepidosauria (Fig. 1B) could not be rejected statistically from all other phylogenetic hypotheses for the phylogenetic position of turtles (data not shown).
Figure 3.
Phylogenetic position of the turtle. A data set combining the two rRNA mitochondrial genes (12S and 16S) was analyzed with MP, NJ, and ML phylogenetic methods (upper, middle, and lower numbers above branches, respectively). Numbers shown above branches represent bootstrap values from 100 pseudoreplicates. Rainbow trout, carp, and loach were used as outgroup taxa.
The recently proposed diapsid affinities of turtles (4, 5) were supported by the analysis of the rRNA gene data set that included the nucleotide sequences of the iguana and the tuatara (19). Mitochondrial rRNA evidence particularly supports the results obtained from the phylogenetic analyses of the pancreatic polypeptide, a nuclear-encoded protein (5). Both data sets suggest the placement of the turtle as the sister group of Archosauria (Fig. 1C; ref. 5) within the diapsid reptiles. The hypothesis of a sister-group relationship between turtles and Archosauria also has been proposed based on morphological evidence (6, 7, 50) and has important implications for several key features of vertebrate evolution, such as the patterns of evolution of fenestration in the amniote skull. Our mitochondrial phylogeny (Fig. 3) suggests that living turtles are relatives of archosaurs and that they might be secondary anapsids (i.e., they lost their skull fenestration secondarily rather than never having evolved it). The new mitochondrial phylogenies (Figs. 2 and 3) also show that the monotremes and marsupials are sister groups, as was recently shown (reviewed in ref. 51), rather than supporting the classical view that monotremes predate the divergence of marsupials and placental mammals.
A recent phylogenetic analysis of mostly paleontological data (4), also placed turtles with diapsid reptiles, although as a sister group of Lepidosauria and not of the Archosauria as suggested here. A phylogenetic analysis of αA-crystallin (18), an analysis of ND2 (52), and a study on α- and β-hemoglobin, myoglobin, and cytochrome c (16) also supported a turtle and Archosauria clade. However, in the latter analysis, Lepidosauria was unexpectedly grouped with mammals (16).
To decide between the two remaining alternative placements of turtles within the diapsid reptiles as close relatives of the Lepidosauria or the Archosauria (Figs. 1 B and C), it would be desirable to determine the complete mitochondrial genome sequences of representatives of the Lepidosauria (tuatara, lizards, and snakes).
Acknowledgments
We thank two anonymous reviewers for providing helpful suggestions, Nicole Valenzuela (State University of New York, Stony Brook) for the gift of the side-necked turtle, David Mindell for sharing advance results on the painted turtle mitochondrial genome and for comments on the manuscript, Axel Janke for providing the alligator sequence before publication, Dave Swofford for permission to publish results based on the test version of his excellent paup* program, and Heike Haunstetter for drawing the figures. R.Z. was sponsored by a postdoctoral grant of the Ministerio de Educación y Ciencia of Spain. This work received partial financial support from U.S. National Science Foundation Grants BSR-9119867 and DEB-9615178, the Max Planck Society, and the University of Konstanz to A.M.
ABBREVIATIONS
- ML
maximum likelihood
- MP
maximum parsimony
- NJ
neighbor joining
Footnotes
Data deposition: The sequences reported in this paper have been deposited in GenBank database (accession no. AF039066).
References
- 1.Carroll R L. Vertebrate Paleontology and Evolution. New York: Freeman; 1988. [Google Scholar]
- 2.Benton M J. J Mol Evol. 1990;30:409–424. doi: 10.1007/BF02101113. [DOI] [PubMed] [Google Scholar]
- 3.Owen R. On the Anatomy of Vertebrates. London: Longmans and Green; 1866. [Google Scholar]
- 4.Rieppel O, deBraga M. Nature (London) 1996;384:453–455. [Google Scholar]
- 5.Platz J E, Conlon J M. Nature (London) 1997;389:246. [Google Scholar]
- 6.Hennig W. In: Testudines. Hennig W, editor. Hamburg, Germany: Parey; 1983. pp. 132–139. [Google Scholar]
- 7.Ax P. Das Phylogenetische System. Stuttgart: G. Fischer; 1984. [Google Scholar]
- 8.Wilkinson M, Thorley J, Benton M J. Nature (London) 1997;387:466. [Google Scholar]
- 9.Lee M. Nature (London) 1997;389:245–246. [Google Scholar]
- 10.Gaffney E S. In: Phylogenetic Relationships of the Major Groups of Amniotes. Panchen A L, editor. London: Academic; 1980. pp. 593–610. [Google Scholar]
- 11.Løvtrup S. Syst Zool. 1985;34:463–470. [Google Scholar]
- 12.Gardiner B G. Zool J Linn Soc London. 1982;74:207–232. [Google Scholar]
- 13.Gardiner B G. Cladistics. 1993;9:369–395. doi: 10.1111/j.1096-0031.1993.tb00232.x. [DOI] [PubMed] [Google Scholar]
- 14.Hedges S B, Moberg K D, Maxson L R. Mol Biol Evol. 1990;7:607–633. doi: 10.1093/oxfordjournals.molbev.a040628. [DOI] [PubMed] [Google Scholar]
- 15.Marshall C R. Mol Biol Evol. 1992;9:370–373. doi: 10.1093/oxfordjournals.molbev.a040726. [DOI] [PubMed] [Google Scholar]
- 16.Eernisse D J, Kluge A G. Mol Biol Evol. 1993;10:1170–1195. doi: 10.1093/oxfordjournals.molbev.a040071. [DOI] [PubMed] [Google Scholar]
- 17.Van de Peer Y, Neefs J M, de Rijk P, de Wachter R. J Mol Evol. 1993;37:221–232. doi: 10.1007/BF02407359. [DOI] [PubMed] [Google Scholar]
- 18.Caspers G J, Reinders G J, Leunissen J A, Watte J, de Jong W W. J Mol Evol. 1996;42:580–586. doi: 10.1007/BF02352288. [DOI] [PubMed] [Google Scholar]
- 19.Hedges S B. Proc Natl Acad Sci USA. 1994;91:2621–2624. doi: 10.1073/pnas.91.7.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janke A, Arnason U. Mol Biol Evol. 1997;14:1266–1272. doi: 10.1093/oxfordjournals.molbev.a025736. [DOI] [PubMed] [Google Scholar]
- 21.Zardoya R, Garrido-Pertierra A, Bautista J M. J Mol Evol. 1995;41:942–951. doi: 10.1007/BF00173174. [DOI] [PubMed] [Google Scholar]
- 22.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland, MA: Sinauer Associates; 1997. , Version d64. [Google Scholar]
- 24.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 25.Hasegawa M, Kishino H, Yano T. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 26.Adachi J, Hasegawa M. molphy, Programs for Molecular Phylogenetics Based on Maximum Likelihood. Tokyo: Institute of Statistical Mathematics; 1996. , Version 2.3. [Google Scholar]
- 27.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 28.Adachi J, Hasegawa M. J Mol Evol. 1996;42:459–468. doi: 10.1007/BF02498640. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. Evolution (Lawrence, KS) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 30.Kishino H, Miyata T, Hasegawa M. J Mol Evol. 1990;30:151–160. doi: 10.1007/BF02109497. [DOI] [PubMed] [Google Scholar]
- 31.Kishino H, Hasegawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 32.Templeton A R. Evolution (Lawrence, KS) 1983;37:221–244. doi: 10.1111/j.1558-5646.1983.tb05533.x. [DOI] [PubMed] [Google Scholar]
- 33.Quinn T W, Mindell D P. Mol Phylogenet Evol. 1996;5:344–351. doi: 10.1006/mpev.1996.0029. [DOI] [PubMed] [Google Scholar]
- 34.Zardoya R, Meyer A. Gene. 1998;216:149–153. doi: 10.1016/s0378-1119(98)00332-1. [DOI] [PubMed] [Google Scholar]
- 35.Desjardins P, Morais R. J Mol Biol. 1990;212:599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- 36.Kumazawa Y, Nishida M. Mol Biol Evol. 1995;12:759–772. doi: 10.1093/oxfordjournals.molbev.a040254. [DOI] [PubMed] [Google Scholar]
- 37.Macey J R, Larson A, Ananjeva N B, Fang Z, Papenfuss T J. Mol Biol Evol. 1997;14:91–104. doi: 10.1093/oxfordjournals.molbev.a025706. [DOI] [PubMed] [Google Scholar]
- 38.Seddon J M, Georges A, Baverstock P R, McCord W. Mol Phylogenet Evol. 1997;7:55–61. doi: 10.1006/mpev.1996.0372. [DOI] [PubMed] [Google Scholar]
- 39.Chang Y S, Huang F L, Lo T B. J Mol Evol. 1994;38:138–155. doi: 10.1007/BF00166161. [DOI] [PubMed] [Google Scholar]
- 40.Tzeng C S, Hui C F, Shen S C, Huang P C. Nucleic Acids Res. 1992;20:4853–4858. doi: 10.1093/nar/20.18.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zardoya R, Meyer A. Genetics. 1996;142:1249–1263. doi: 10.1093/genetics/142.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zardoya R, Meyer A. Genetics. 1997;146:995–1010. doi: 10.1093/genetics/146.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roe B A, Din-Pow M, Wilson R K, Wong J F. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- 44.Janke A, Gemmell N J, Feldmaier-Fuchs G, von Haeseler A, Paabo S. J Mol Evol. 1996;42:153–159. doi: 10.1007/BF02198841. [DOI] [PubMed] [Google Scholar]
- 45.Janke A, Feldmaier-Fuchs G, Thomas K, von Haeseler A, Paabo S. Genetics. 1994;137:243–256. doi: 10.1093/genetics/137.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arnason U, Gullberg A. J Mol Evol. 1993;37:312–322. doi: 10.1007/BF00178861. [DOI] [PubMed] [Google Scholar]
- 47.Horai S, Hayasaka K, Kondo R, Tsugane K, Takahata N. Proc Natl Acad Sci USA. 1995;92:532–536. doi: 10.1073/pnas.92.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Felsenstein J. Syst Zool. 1978;27:27–33. [Google Scholar]
- 49.Gauthier J, Kluge A G, Rowe T. Cladistics. 1988;4:105–209. doi: 10.1111/j.1096-0031.1988.tb00514.x. [DOI] [PubMed] [Google Scholar]
- 50.Løvtrup S. The Phylogeny of Vertebrata. New York: Wiley; 1977. [Google Scholar]
- 51.Penny D, Hasegawa M. Nature (London) 1997;387:549–550. doi: 10.1038/42352. [DOI] [PubMed] [Google Scholar]
- 52.Seutin G, Lang B F, Mindell D P, Morais R. Mol Biol Evol. 1994;11:329–340. doi: 10.1093/oxfordjournals.molbev.a040116. [DOI] [PubMed] [Google Scholar]