Abstract
The development of the epidermis, a stratified squamous epithelium, is dependent on the regulated differentiation of keratinocytes. Differentiation begins with the initiation of stratification, a process tightly controlled through proper gene expression. AP-2γ is expressed in skin and previous research suggested a pathway where p63 gene induction results in increased expression of AP-2γ which in turn is responsible for induction of K14. This study uses a conditional gene ablation model to further explore the role of AP-2γ in skin development. Mice deficient for AP-2γ exhibited delayed expression of p63, K14, and K1, key genes required for development and differentiation of the epidermis. In addition, microarray analysis of E16.5 skin revealed delayed expression of additional late epidermal differentiation genes: filaggrin, repetin and secreted Ly6/Plaur domain containing 1, in mutant mice. The genetic delay in skin development was further confirmed by a functional delay in the formation of an epidermal barrier. These results document an important role for AP-2γ in skin development, and reveal the existence of regulatory factors that can compensate for AP-2γ in its absence.
Keywords: AP-2, Tcfap2c, skin, epidermis, keratinocyte, filaggrin, keratin, differentiation, embryo development
INTRODUCTION
The mammalian AP-2 transcription factor family consists of five members AP-2α, β, γ, δ, and ε (Eckert et al., 2005). These transcription factors are known to regulate both proliferation and differentiation (Hilger-Eversheim et al., 2000; Imagawa et al., 1987; Luscher et al., 1989), are critical for embryonic development (Auman et al., 2002; Winger et al., 2006), and are important regulators of epidermal development and differentiation (Leask et al., 1991; Byrne et al., 1994; Wang et al., 1997; Oyama et al., 2002; Koster and Roop, 2004). Furthermore, they display differential expression in developing and mature epidermis. AP-2α is only expressed in the basal layer, whereas, AP-2γ is expressed in the basal, spinous and granular layers and expression of AP-2β is restricted to sweat glands (Byrne et al 1994; Takahashi et al., 2000; Oyama et al., 2002; Panteleyev et al., 2003). Functional AP-2 binding sites have been detected in the promoter regions of the genes encoding the keratin intermediate filament proteins K1, K5, K10 and K14 and the epidermal growth factor receptor (EGFR), all of which play critical roles in epidermal development and differentiation (Leask et al., 1991; Byrne et al., 1994; Wang et al., 1997; Maytin et al., 1999; Koster et al., 2006). Expression of AP-2α represses EGFR gene transcription thereby functioning as a checkpoint for proliferation and differentiation (Wang et al., 2006). Mice lacking AP-2α in the epidermis have an increased expression of EGFR resulting in hyperproliferation. A combination of in vivo and in vitro studies has suggested that AP-2γ is required for the induction of gene expression critical for the commitment of keratinocytes to stratification during embryogenesis (Koster et al., 2006). This study was initiated to further investigate the role of AP-2γ during embryonic development of the epidermis using a conditional AP-2γ mouse model.
Development of the epidermis from a single layer of cells into a fully stratified epithelium is controlled by a tightly regulated pattern of gene expression (Koster and Roop, 2007; Dai and Segre, 2004). The single-layered surface ectoderm surrounding the embryo is initially characterized by the expression of K8 and K18 (Moll et al., 1982). The expression of K8 and K18 is lost at the time the surface ectoderm commits to stratification and expression of K5 and K14 is induced (Byrne et al., 1994). Commitment to stratification is dependent on the expression of the transcription factor p63 (Koster et al., 2004). The p63 gene produces six transcripts that contain (TA) or lack (ΔN) a transactivation domain (Yang et al., 1999). The TAp63α isoform has been shown to be a master molecular switch in initiating epithelial stratification (Koster et al., 2004). TAp63α and AP-2γ seem to be working in coordination, with TAp63α directly inducing AP-2γ expression in the embryonic epidermis. AP-2γ mRNA was not detected in the surface epithelial cells in p63−/− mice (Koster et al., 2006). In turn, increased AP-2γ levels result in the initiation of K5 and K14 gene expression (Byrne et al., 1994; Koster et al., 2006). In vitro studies further demonstrated that in the absence of AP-2γ, TAp63α is unable to induce the expression of K14 (Koster et al., 2006). To further study this genetic regulatory pathway in the embryo we created a conditional AP-2γ mutant mouse model to investigate embryonic skin development in mice lacking AP-2γ.
AP-2γ is necessary for development of the extraembryonic tissues, which produce the placenta, and conventional AP-2γ knock-out mice die around E7.5 due to defects in this tissue (Auman et al., 2002). Therefore, in order to avoid the embryonic lethal phenotype, the conditional ablation of AP-2γ was produced using the Cre/loxP recombination system. Sox2Cre mice were used to induce an epiblast specific ablation of AP-2γ (Hayashi et al., 2002). Sox2Cre is ubiquitously expressed in the epiblast beginning at E6.5 and produces a complete deletion in the embryo while leaving intact AP-2γ expression in the extraembryonic tissues. Therefore, this strategy generates a viable mouse model to explore the role of AP-2γ in skin development
MATERIALS AND METHODS
Production of AP-2γ null mice
Sox2Cre mice were purchased from Jackson Labs (Bar Harbor, ME). This strain expresses an epiblast-specific Cre recombinase gene beginning at E6.5 (Hayashi et al., 2002). Mice containing the AP-2γ null allele, have been previously described (Auman et al., 2002). Mice with the AP-2γ floxed allele have a conditional AP-2γ allele containing loxP sites which flank exon 6, and allow for a gene specific deletion of AP-2γ in the presence of Cre recombinase (Fig 1; TW manuscript in preparation; Winger et al., 2006). The Cre recombinase will recognize the loxP sites in the AP-2γ flox allele creating an AP-2γ allele which is missing exon 6. In order to produce offspring which contained Sox2Cre, AP-2γ null and AP-2γ flox alleles, male mice were generated that contained both the Sox2Cre transgene as well as the AP-2γ null allele. These mice were mated to female mice that were homozygous for AP-2γ flox. Embryos inheriting the AP-2γ null allele and the Sox2Cre transgene needed only one Cre-mediated recombination event of the AP-2γ flox allele per cell to produce a homozygous AP-2γ mutation. Littermates that did not inherit the Sox2Cre or the AP-2γ null allele were used as controls. For timed pregnancies, specific matings were set up in the afternoon, and the mice were checked for vaginal plugs the following mornings. Noon of the day of the vaginal plug was considered 0.5 days of gestation.
Figure 1.

AP-2γ mutant and conditional alleles. A) AP-2γ flox has loxP sites flanking exon 6, AP-2γ flox deleted represents the floxed allele after the Cre recombination event and AP-2γ null represents the allele that contains a deletion including part of exon 7. P1 and P2 shows the primer locations used to verify deletion of AP-2γ flox allele by RT-PCR. B) Confirmation of AP-2γ flox deletion in skin of E16.5 control (+/+) and AP-2γ mutant embryos (−/−). Product size 200 bp. C) AP-2γ mutant embryos (−/−) are visibly smaller than control embryos (+/+) at E16.5. Weights were significantly different at E16.5 and E18.5 (mean ± SD) significance was determined by Student’s T-test, p<0.05a and p<0.001b. D) Skeletal preparations show the curly tail phenotype in the mutant (−/−) but did not show other skeletal defects compared to the control (+/+). Bar in (C) = 5mm.
Genotyping of embryos was performed using PCR analysis of either placental yolk sac samples or tail DNA. PCR reactions were carried out for 35 cycles (94°C, 40 sec; 67°C, 40 sec; 72°C, 40 sec) using Taq DNA polymerase (Fermentas, Hanover, MD) in the supplied Taq buffer at 1X and 2mM MgCl2. χ2-tests were carried out using the resulting genotypes to determine if the AP-2γ mutant embryos were found in expected Mendelian ratios at E10.5, E16.5 and E18.5. Primers that detect the AP-2γ flox deleted allele were used to assay for Cre mediated deletion in RNA collected from yolk sac and placental samples. Skeletal preparations were prepared by staining with alcian blue and alizarin red as described previously (Martin et al., 1995).
Detection of AP-2γ mRNA transcripts
Total RNA was collected from E16.5 mouse skin using the RNeasy isolation kit (Qiagen, Santa Clarita, CA) according to manufacturer’s specifications. cDNA was then prepared from 1μg of total RNA and was analyzed for AP-2γ transcripts by PCR. Primer sequences were designed to anneal in exons 6 and 7 (P1-5′-AAG CGG TGG CTG ACT ATT TAA C- 3′; P2-5′-CAG GCT GAA ATG AGA CAA ACA G -3′) of the AP-2γ gene producing a 206bp product from the WT and AP-2γ flox, and no amplification of the AP-2γ null or AP-2γ flox-deleted alleles (Figure 1). This set of primers was used to verify that AP-2γ conditional knock-out animals were not expressing any functional AP-2γ in the skin.
Histology and immunofluorescence
Embryos or tissues were washed in PBS and then fixed in 10% buffered formalin overnight at 4°C. Samples were then dehydrated, embedded and sectioned at 5μm and stained with H&E or used for immunofluorescence. Primary antibodies used for immunofluorescence were guinea pig anti-K14 (Yuspa et al., 1989), rabbit anti-K1 (Yuspa et al., 1989) and mouse-anti-p63 (mAb4A4; Yang et al., 1999). Secondary antibody conjugates used were Alexa-conjugated fluorochromes 488 goat-anti-rabbit, 594 goat anti-guinea pig and 488 goat anti-mouse (Molecular Probes). Images were acquired using a Nikon Eclipse E600 microscope (Nikon Corporation). Objectives used were Nikon Plan Apo 20x/0.75 and Nikon Plan Fluor 40x/1.30 Oil. Pictures were taken using a Photometrics Coolsnap fx camera (Roper Scientific) and MetaVue imaging software (Universal Imaging Corporation).
Affymetrix microarray analysis
Three biological replicates each of AP-2γ mutant and wild type skin RNA were extracted from E16.5 embryos. Total RNA was extracted using an RNeasy kit (Qiagen) according to manufacturer’s instructions, and RNA was submitted to the Center for Integrated Biosystems at Utah State University (Logan, UT) for preparation of cRNA and hybridization to the GeneChip® Mouse Genome 430_2.0 Array from Affymetrix. The expression data were analyzed using tools from the Bioconductor project (Gentleman et al., 2004). After preprocessing by Affymetrix’s PLIER correction (including quantile normalization), a nonspecific filter (Chiaretti et al., 2004; Scholtens and von Heydebreck, 2005) was used, restricting attention to probesets satisfying both of the following two conditions: expression level above 100 on at least 20 percent of the arrays, and coefficient of variability exceeding 0.3. About 42 percent of all probe sets satisfied the first condition, and about 27 percent satisfied the second. The purpose of this filter was to restrict attention to a more interesting set of genes (that were both expressed and exhibited some appreciable change in expression levels), and also to reduce the severity of the multiple comparisons adjustment. The filter settings were selected based on graphical checks of the expression data (not shown), and reduced the number of probesets tested from 45,101 to 1,414. Significant differences in gene expression from wild type to mutant were determined based on the moderated t-statistic of the limma/eBayes test for differential expression (Smyth, 2004), with P-values adjusted to control the false discovery rate (Benjamini and Hochberg, 1995) at 0.05.
Quantitative real time RT-PCR
Skin samples were taken from E16.5 and E18.5 mice. RNA was extracted using the RNeasy isolation kit according to manufacturer’s specifications (Qiagen) and cDNA was generated using reverse transcription. Real-time RT-PCR reactions were prepared to compare filaggrin mRNA levels using Taqman probes (Applied Biosystems, Foster City, CA). Reactions were set up according to manufacturer’s specifications and run on a BioRad iCycler (2 min 50°C, 10 min 95°C, Cycle 40 times at 15 sec 95°C and 1 min 60°C). The cycle threshold (Ct) values were obtained from each of three replicates. Relative expression was calculated using standard curves of ten fold cDNA dilutions for both filaggrin and the β-actin internal control. The relative expression of filaggrin was normalized to endogenous β-actin expression. AP-2γ mutant levels were calculated as a percentage of expression compared to control samples which were determined to be 100% of expression. Results were analyzed using Student’s t-test, values of p<0.05 were considered significant.
Western Blots
Protein was extracted from E18.5 mouse embryonic skin using T-Per tissue protein extraction reagent (Pierce, Rockford, IL) and samples were quantitated on a Versa Max tunable microplate reader (Molecular Devices, Sunnyvale, CA) using the BCA™ Protein Assay kit (Pierce) according to manufacturer’s specifications. Western blots were performed to compare filaggrin protein levels between AP-2γ mutant and control mice. 25 μg of protein was loaded per well for SDS-PAGE using 12% Tris-HCl Ready Gels (BioRad, Hercules, CA). The samples were transferred to a PVDF membrane (BioRad) for blotting.
Membranes were blocked in Super Block (Pierce) for 1 h at room temperature, the membrane was then incubated in primary antibody against filaggrin (Provided by Dr. Roop; Waikel et al., 1999) at a dilution of 1:5000 followed by HRP-anti-rabbit secondary antibody (Rockland, Gilbertsville, PA) and visualized using SuperSignal (Pierce) on a digital gel documentation system (UVP). Quantification of filaggrin protein was performed by standardization with primary antibodies against β-actin (Rockland Immunochemicals, Gilbertsville, PA).
Barrier staining
Embryos were collected at E16.5, E17.5 and E18.5 and washed in PBS. Embryos were then submerged overnight at 37°C in X-gal (5-bromo-4-chloro-e-indolyl-b,D-galactopyranoside) solution (100 mM NaPO4, 1.3 mM MgCl2, 3 mM K3Fe(CN)6, 3 mM K4Fe(CN)6 and 1 mg/ml X-Gal) at pH4.5. Embryos were then rinsed in PBS and fixed in formalin for 12 h and photographed. This assay relies on the ability of X-gal to penetrate embryonic skin prior to barrier formation where X-gal is cleaved by endogenous β-galactosidase activity in the skin producing a blue precipitate (Hardman 1998). If the impermeable skin barrier has already formed, the X-gal is unable to penetrate and no precipitate is produced.
RESULTS
Characterization of the AP-2γ mutant mice
In order to produce AP-2γ mutant mice males containing an AP-2γ null allele and the Sox2Cre transgene were bred to females homozygous for the AP-2γ flox allele. Offspring inheriting the AP-2γ null allele, the AP-2γ flox allele and the Sox2Cre transgene then underwent a single Cre recombination event of the AP-2γ flox allele in each cell of the epiblast to produce an AP-2γ homozygous mutant embryo (Figure 1). After genotyping 3 week old offspring from 10 litters, no AP-2γ mutant pups were found, suggesting that homozygous deletion of AP-2γ in the epiblast beginning at E6.5 was embryonic lethal. Embryos were collected to determine the stage of embryonic lethality of AP-2γ mutant embryos. Genotyping embryos obtained at E10.5, E16.5 and E18.5 showed that AP-2γ mutant embryos were present at each developmental stage (Table 1). Embryos collected at E10.5 showed fewer mutants but numbers were not significantly different from expected Mendelian ratios. At E16.5 and E18.5 AP-2γ mutant embryos were found alive but were significantly decreased in number compared to expected ratios (Table 1). Genotyping of newborn litters showed that AP-2γ mutant pups were born, but these mutants were not alive upon collection, suggesting that mutants can survive until birth but not after.
Table 1.
Detection of AP-2γ mutant embryos at E10.5, E16.5 and E18.5. Genotypes of embryos from crosses between AP-2γ flox/flox females and Sox2Cre gene and AP-2γ null heterozygous males.
| Genotype | AP-2γ null/AP-2γ flox No Cre | AP-2γ flox-deleted/+ Sox2Cre | AP-2γ flox/+ No Cre | AP-2γ null/AP-2γflox-deleted Sox2Cre | ||||
|---|---|---|---|---|---|---|---|---|
| Exp | Ob | Exp | Ob | Exp | Ob | Exp | Ob | |
| E 10.5c | 17 | 17 | 17 | 19 | 17 | 20 | 17 | 10 |
| E16.5d | 57 | 67 | 57 | 64 | 57 | 75 | 57 | 22a |
| E18.5e | 32 | 35 | 32 | 36 | 32 | 39 | 32 | 18b |
χ2 test p< 0.01
66 embryos
228 embryos
128 embryos
“+” indicates the presence of a wild type allele
Comparison of the Sox2Cre AP-2γ mutant pups to littermate controls revealed some phenotypic developmental differences. The weight of AP-2γ mutant embryos was significantly less than control littermates at E16.5 and E18.5 (Figure 1). At E16.5 the body weight of control embryos averaged 0.71g ± 0.20 (n=15), while AP-2γ mutant embryos weighed 0.35g ± 0.20 (n=15). Embryos collected at E18.5 showed similar results with control embryos weighing 1.4g ± 0.05 (n=20) and AP-2γ mutant embryos weighing 0.73g ± 0.13 (n=20). Curly tails were detected in 31% (10/32) of mutant embryos collected suggesting a potential caudal neural tube closure and/or skeletal defect; however, no other major skeletal defects were detected in skeletal preparations (Figure 1).
Low levels of Sox2Cre expression have been previously detected in the yolk sac and we were able to detect the AP-2γ mutant allele in 100% of DNA samples collected from Sox2Cre AP-2γ mutant yolk sacs. The AP-2γ deletion in the yolk sac is not likely to contribute to the developmental delayed phenotype because AP-2γ is not expressed in the yolk sac (Shi and Kellems, 1998). Sox2Cre expression has not previously been reported in the placenta; however, PCR analysis of placental DNA from embryos that contained the Sox2Cre and the AP-2γ flox allele detected the presence of the AP-2γ flox deleted allele in 56% (9/16) of the samples. Deletion of the allele was therefore not present in all mutant embryos and was presumably restricted to the embryonic/fetal portion of the placenta. This level of placental deletion in mutant embryos at E16.5 and E18.5 did not result in any morphology or weight differences in the placenta compared to littermate controls (Data not shown).
Abnormal skin development in AP-2γ mutant mice
The AP-2γ mutant mice display abnormal epidermal stratification as shown histologically by H&E staining of skin sections at E11.5 and E16.5 (Figure 2). The induction of skin differentiation markers p63 at E7.5 and K14 at E9.5 indicates keratinocyte commitment to stratification in control skin (Koster et al., 2004; Byrne et al., 1994). Immunofluorescence analysis on E11.5 embryos demonstrated that, unlike in control skin, p63 and K14 protein expression was absent from the skin of AP-2γ mutant embryos (Figure 2). However, p63 and K14 were detected in E16.5 AP-2γ mutant skin indicating that p63 and K14 were not entirely lost in the mutant but that the expression was delayed.
Figure 2.
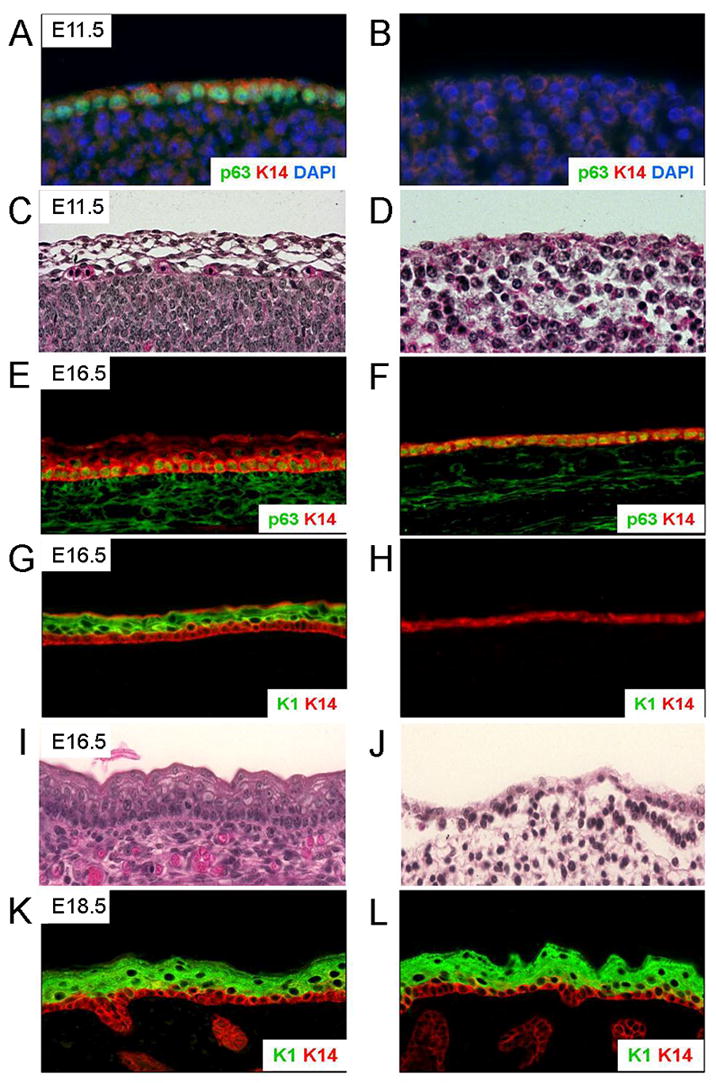
AP-2γ mutant skin shows delayed expression of proteins involved in epidermal differentiation. Skin samples from control (A, C, E, G, I and K) and AP-2γ mutant (B, D, F, H, J and L) embryos were sectioned and stained with antibodies specific for epidermal development markers. A, B, E and F) p63 and K14 were detected in control but not in mutant skin at E11.5 but were detected in mutant skin at E16.5. G, H, K and L) K1 was detected in control skin at E16.5 but not in AP-2γ mutant skin. K1 expression was detected in the AP-2γ mutant skin at E18.5. C, D, I, and J) H&E staining shows a lack of stratification in mutant samples compared to controls at both E11.5 and E16.5.
In wild type mice, K1 expression is induced around E15.5 indicating the initiation of spinous layer formation during keratinocyte differentiation (Bickenbach et al., 1995). As expected, K1 protein was detected in the developing spinous layer of control E16.5 embryos (Figure 2). However, at this stage AP-2γ mutant skin did not express K1, indicating that the spinous layer had not yet formed. We did, however, detect K1 protein in E18.5 AP-2γ mutant skin, a delay of about three days compared to control skin (Figure 2).
Differential gene expression in E16.5 control and AP-2γ mutant skin
AP-2γ regulation of gene expression in skin development was further investigated by microarray analysis to detect differentially expressed genes in skin samples from mice lacking AP-2γ. RNA was isolated from E16.5 control and AP-2γ mutant skin and hybridized to Mouse 430_2.0 Affymetrix chips. The data set for the microarrays has been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo) as accession number GSE10175. The expression data were analyzed using tools available from the Bioconductor project to determine gene expression differences. Nineteen probesets representing 18 genes were found to have statistically significant differences in expression and are summarized in Figure 3. Of the 18 differentially expressed genes 9 were down-regulated and 9 were up-regulated due to the AP-2γ mutation. Interestingly, AP-2γ mutant skin showed decreased expression of late-epidermal differentiation markers filaggrin (Flg), repetin (Rptn) and secreted Ly6/Plaur domain containing 1 (Slurp1). In addition to these known epidermal genes the expression of keratin 12 (K12), keratin 72 (K72) and serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 (Serpina12) were significantly decreased in the mutant skin. All of these identified down-regulated genes, with the exception of K12 which is cornea specific, are potentially involved in skin development. Ten probesets representing 9 genes were determined to be up-regulated in AP-2γ mutant skin. Surfactant associated protein D (Sftpd), lectin, galactose binding, soluble 6 (Lgals6), hydroxysteroid 11-beta dehydrogenase1 (Hsd11b1), glutathione peroxidase 3 (Gpx3) and proline-rich acidic protein 1 (Prap1) were all determined to have increased expression in the mutant; however, to date none of these genes have well defined roles in the epidermis.
Figure 3.

Heatmap of nineteen significant differentially expressed probesets. Of the 1,414 probesets tested for differential expression from control (1–3) and AP-2γ mutant skin (4–6), 19 showed statistical significance. Colors in this heatmap correspond to expression levels on a scale of dark red for lowest values to dark blue for highest values, with lighter colors for moderate values. The dendrograms on top and at left are based on divisive analysis clustering with average linkage.
Analysis of filaggrin expression in AP-2γ mutant skin
To verify the differential gene expression of filaggrin, mRNA levels were compared by real-time RT-PCR (TaqMan) for AP-2γ mutant and control skin. Filaggrin is an important factor for skin differentiation and showed the largest differential fold change and level of significance of the down-regulated genes. AP-2γ mutant skin samples showed an 80% decrease in filaggrin transcript expression at E16.5 compared to control samples (Figure 4). The difference in expression was no longer present at E18.5.
Figure 4.
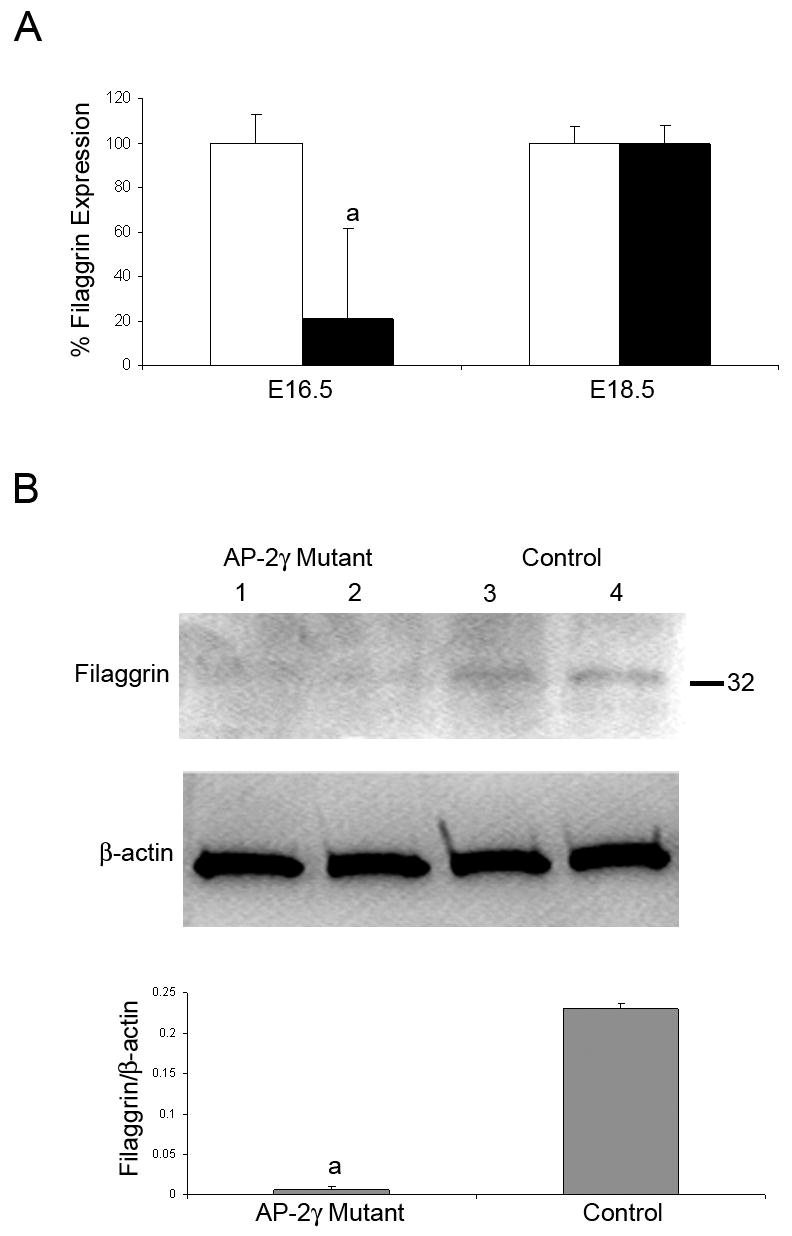
Quantitative real-time RT-PCR analysis on AP-2γ mutant skin. A) Relative filaggrin mRNA levels were significantly lower in AP-2γ mutant skin when compared to control skin at E16.5, but were not different at E18.5. Bars represent mean ± SD, significance determined by Student t-test, ap<0.05. B) Filaggrin protein monomer levels were determined in E18.5 AP-2γ mutant (lanes 1,2) and control (lanes 3,4) skin by Western blot. Filaggrin levels are reduced in AP-2γ mutant skin. Bars represent mean ± SD, significance determined by Student t-test, ap<0.05.
Expression of filaggrin protein, a late epidermal marker of skin differentiation, is normally induced around E18.0. Quantification of filaggrin protein by Western blot at E18.5 confirms that filaggrin protein level is lower in the mutant compared to the control (Figure 4).
AP-2γ mutant mice show a delay in skin barrier formation
To investigate the time of skin barrier formation, AP-2γ mutant embryos from E16.5–E18.5 were subjected to x-gal staining. At E16.5 both control and AP-2γ mutant embryos stained blue indicating the barrier had not yet formed in the skin of either genotype (Figure 5). At E17.5, control embryos showed complete barrier formation as indicated by the lack of any blue staining, while AP-2γ mutant embryos absorbed stain on the ventral side of the embryos. At E18.5, AP-2γ mutant embryos blocked x-gal penetration indicating complete formation of the barrier (Figure 5).
Figure 5.
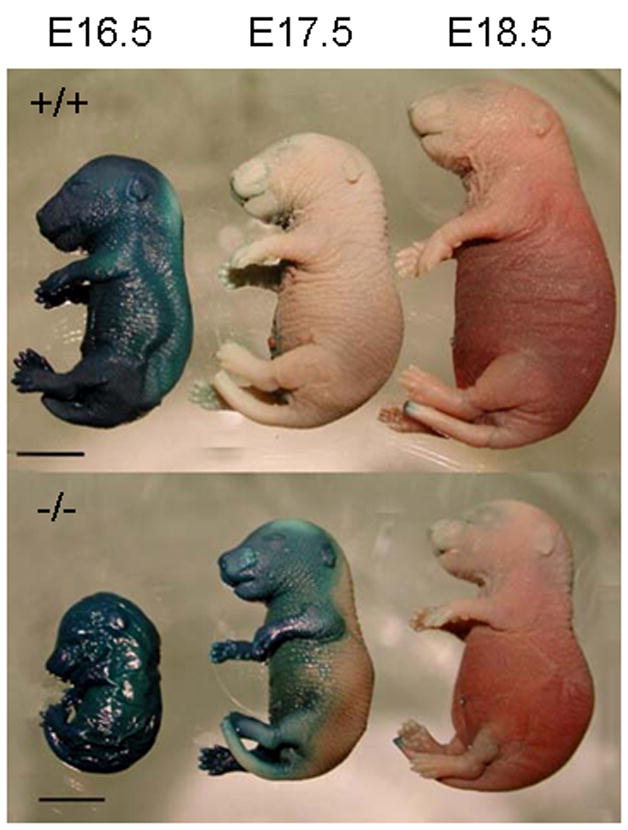
AP-2γ mutant embryos have delayed skin barrier formation. Control embryos (+/+) have established an epidermal barrier by E17.5 as shown by the lack of blue x-gal staining. AP-2γ mutant embryos (−/−) have an incomplete barrier at E17.5 as shown by the uptake of x-gal on the ventral half of the embryo. Bar = 5mm.
DISCUSSION
The commitment of keratinocytes to stratification is dependent on the transcription factor p63 (Koster et al., 2004, 2006; Mills et al., 1999; Yang et al., 1999). A previous study reported that in the absence of AP-2γ, TAp63α is unable to induce K14 expression (Koster et al., 2006). The current study investigates epidermal development in mice that are deficient for AP-2γ. Our results show that the AP-2γ mutation delays the expression of several proteins involved in skin development including p63, K14 and K1. In wild type embryos the developing epidermis begins to express p63 at E7.5 and K14 at E9.5, which indicates the commitment of the keratinocytes to stratification (Byrne et al., 1994; Koster et al., 2004). However, our data show that p63 and K14 proteins were still not detected at E11.5 in AP-2γ mutant skin, indicating a severe delay in expression. These proteins were both detected in mutant skin by E16.5. We also found a similar delay for the differentiation-specific gene K1, which is normally induced at E15.5 in the future spinous layer of the epidermis (Bickenbach et al., 1995). In AP-2γ mutant skin however, K1 protein was not detected at E16.5 indicating a delay in the development of the spinous layer. Similar to p63 and K14, K1 protein expression was not delayed indefinitely in AP-2γ mutant mice and was detected at E18.5 in the spinous layer.
To identify genes that are potentially regulated by AP-2γ during skin development, we compared gene expression levels in the mutant to control skin at E16.5. At this age, expression of genes involved in the formation of the granular layer and the stratum corneum is expected (Bickenbach et al., 1995; Mehrel et al., 1990). A comparison of differential transcript expression by microarray detected nine probesets that were down-regulated in AP-2γ mutant skin compared to control. Three of these probsets represent filaggrin, repetin and secreted Ly6/Plaur domain containing 1 (Slurp1), which are all known late-epidermal differentiation markers. Filaggrin and repetin are both members of the “fused” gene family that are associated with the keratin intermediate filaments and are necessary for formation of the cornified envelope (Huber et al., 2005; Presland et al., 2004; List et al., 2006). Slurp1 is a late epidermal differentiation marker and is predominately expressed in the granular layer. Mutations in the Slurp1 gene result in Mal de Meleda (MDM) a disease characterized by inflammatory palmoplantar keratoderma (Favre et al., 2007). The Slurp1 gene has an AP-2 binding site just upstream of the first exon indicating a potential for direct regulation by AP-2γ (Mastrangeli et al., 2003). These three genes represent key epidermal differentiation factors that are necessary for proper formation of skin. Three additional genes, K12, K72 and Serpina12, were found to be significantly decreased in the mutant skin. K12 and K72 are keratin family member genes that may be involved in skin differentiation; however, K12 has been shown to be a corneal specific keratin and K72 to be an inner-root-sheath-specific type II epithelial keratin in humans and mice (Tanifuji-Terai et al., 2006; Porter et al., 2001). Serpina12 has previously been detected in the epidermis where expression levels were increased by treatment with the corticosteroid dexamethasone, and by the over-expression of the transcription factor Klf4, which is a known factor required for barrier formation (Segre et al., 1999; Patel et al., 2006).
Nine genes were identified as increased in the mutant skin including surfactant associated protein D (Sftpd), lectin, galactose binding, soluble 6 (Lgals6), hydroxysteroid 11-beta dehydrogenase1 (Hsd11b1), glutathione peroxidase 3 (Gpx3) and proline-rich acidic protein 1 (Prap1). None of these genes have well defined roles in the epidermis. However, the role of these genes in inflammatory response and response to oxidative stress might suggest that they are induced due to the abnormal skin phenotype produced in the mutant and may not be the direct result of the lack of AP-2γ gene regulation.
Transcripts for filaggrin, a major component of the epidermal barrier, were dramatically down-regulated in E16.5 AP-2γ mutant skin. At E18.5, we found that transcript levels were comparable to control; however, filaggrin protein levels were lower in AP-2γ mutant skin than in control skin. The developmental delay was further verified by a delay in detection of the epidermal barrier. In mice, the barrier is completely formed by E17.0 (Hardman et al., 1998); however, the barrier was incomplete in mutant embryos at E17.5 but was complete by E18.5 even though the filaggrin protein levels were decreased.
The microarray experiments detected relatively few differentially expressed genes considering the obvious phenotype at E16.5. At this stage, the epidermis in the mutant is essentially one layer thick and K1 protein was not detected. However, the microarray analysis did not detect a significant difference in K1 mRNA expression. It is possible that K1 transcripts are induced at E16.5 in the mutant to levels similar to the control but K1 protein is not yet at detectable levels. We have shown that the mutant embryos undergo rapid epidermal differentiation between E16.5 when the barrier is absent to E17.5 when a partial barrier is detected. Therefore, a second possible explanation for not detecting more differentially expressed genes is that the embryos used in the microarray were slightly more advanced in their development than the mutant embryos used for histology.
Together, our data demonstrate that Sox2Cre AP-2γ mutant embryos have a developmental delay, one aspect of which is the delayed initiation of epidermal morphogenesis. The absence of p63 protein at E11.5 suggests that in the absence of AP-2γ the early processes necessary for epidermal initiation cannot occur normally. One possible explanation is that AP-2γ is necessary in the mesenchyme for the development of the dermis and for the initial induction of p63 expression in the overlying epidermis. During later stages of epidermal development, AP-2γ mutant mice appear to overcome this delay and induce K14 protein. Previous research indicated that AP-2γ was necessary for the induction of K14 (Koster et al., 2006). Our Sox2Cre conditional model now demonstrates that without AP-2γ expression the embryo can compensate for the absence of AP-2γ and induce K14 expression at later developmental stages. Redundant gene regulatory mechanisms, which may or may not include other AP-2 family members, must compensate for the absence of AP-2γ and allow for the initiation of skin development. In an earlier study AP-2β expression was found to be increased in p63−/− surface epithelial cells, which lack AP-2γ expression (Koster et al., 2006). In AP-2γ mutant skin we did not detect differences in the expression of the other AP-2 family members based on an examination of the microarray data. However, functional redundancy could occur as a result of the endogenous expression of a family member without an induction of expression. Previous research has highlighted redundancy between AP-2α and AP-2γ in various processes including pre-implantation embryonic development and cranial neural tube closure (Winger et al., 2006; Kohlbecker et al., 2002). Therefore, the presence of AP-2α in the skin could rescue the initiation of skin development in the AP-2γ mutants.
Mice deficient for some of the genes which displayed delayed expression in AP-2γ mutants result in either lethal phenotypes or skin that fails to function properly. For example, K14 mutations have been implicated in epidermolysis bullosa simplex and K14 null mice die a few days after birth due to cracking and drying of the skin (Paladini and Coulombe, 1999; El Ghalbzouri et al., 2003;) and K1 mutant mice develop epidermolytic hyperkeratosis (Arin et al., 2000). Mutations in p63 are implicated in ectodermal dysplasias (Rinne et al., 2007). In addition, abnormal processing of filaggrin is observed in a mouse model of Netherton syndrome (Hewett et al., 2005) and mice that contain the Matriptase/MT-SP1−/− mutation are unable to proteolytically process filaggrin resulting in death within 48 hours of birth (List et al., 2002, 2006). In humans, mutations in the filaggrin gene cause moderate or severe ichthyosis vulgaris (IV), the most common inherited disorder of keratinization (Smith et al., 2006).
The Sox2Cre AP-2γ mutant mice did not display skin abnormalities such as blistering or hyperkeratosis, and formed a functional barrier at E18.5. Therefore, the data is inconclusive for a role of the epidermal defect in the lethal phenotype. Mice that contain a homozygous AP-2γ gene mutation from conception fail to produce proper extraembryonic tissues preventing placental development resulting in death around E7.5 (Auman et al., 2002). The developmental delay and lethal phenotype seen in this Sox2Cre model indicate that AP-2γ is necessary for additional aspects of embryonic development after E7.5. The detection of floxed gene deletion in the placenta is a novel observation with the Sox2Cre model and should be taken into account when using this transgene in the future. Nevertheless, since this phenomenon was observed in only a fraction of the mutant mice it is unlikely to be the cause of the embryonic developmental delay seen in all mutants. In conclusion, the Sox2Cre mediated loss of AP-2γ in the epiblast results in developmental delay which is particularly evident in the epidermis. At present it remains unclear whether the post-natal lethality in this model is due to a subtle defect inherent to the epidermis and its derivatives, or is caused by the loss of AP-2γ in another location. Therefore, additional research is necessary to determine the full effects of the AP-2γ mutation during embryonic development.
Acknowledgments
The authors would like to thank Frances Bhushan for her work maintaining the animals and for assistance with the research.
QAW was supported by NIH grant HD049624, The Center for Integrated Biosystems, Functional Genomics Seed Grant, Utah State University, and the Utah Agriculture Experiment Station project UTA0043. This paper is published as UAES number 7908. MIK and DRR were supported by NIH grant (AR47898 and AR052263), TW was supported by NIH grant (DE12728).
Footnotes
Summary Sentence: Conditional ablation of AP-2γ results in a delay in skin development and abnormal expression of p63, K14, K1, filaggrin, repetin and secreted Ly6/Plaur domain containing 1, key genes required for epidermal development and differentiation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arin MJ, Longley MA, Epstein EH, Jr, Rothnagel JA, Roop DR. Identification of a novel mutation in keratin 1 in a family with epidermolytic hyperkeratosis. Exp Dermatol. 2000;9:16–19. doi: 10.1034/j.1600-0625.2000.009001016.x. [DOI] [PubMed] [Google Scholar]
- Auman HJ, Nottoli T, Lakiza O, Winger Q, Donaldson S, Williams T. Transcription factor AP-2gamma is essential in the extra-embryonic lineages for early postimplantation development. Development. 2002;129:2733–2747. doi: 10.1242/dev.129.11.2733. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- Bickenbach JR, Greer JM, Bundman DS, Rothnagel JA, Roop DR. Loricrin expression is coordinated with other epidermal proteins and the appearance of lipid lamellar granules in development. J Invest Dermatol. 1995;104:405–410. doi: 10.1111/1523-1747.ep12665896. [DOI] [PubMed] [Google Scholar]
- Byrne C, Tainsky M, Fuchs E. Programming gene expression in developing epidermis. Development. 1994;120:2369–2383. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- Chiaretti S, Li X, Gentleman R, Vitale A, Vignetti M, Mandelli F, Ritz J, Foa R. Gene expression profile of adult T-cell acute lymphocytic leukemia identifies distinct subsets of patients with different response to therapy and survival. Blood. 2004;103:2771–2778. doi: 10.1182/blood-2003-09-3243. [DOI] [PubMed] [Google Scholar]
- Dai X, Segre JA. Transcriptional control of epidermal specification and differentiation. Curr Opin Genet Dev. 2004;14:485–491. doi: 10.1016/j.gde.2004.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D, Buhl S, Weber S, Jager R, Schorle H. The AP-2 family of transcription factors. Genome Biol. 2005;6:246. doi: 10.1186/gb-2005-6-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ghalbzouri A, Jonkman M, Kempenaar J, Ponec M. Recessive epidermolysis bullosa simplex phenotype reproduced in vitro: ablation of keratin 14 is partially compensated by keratin 17. Am J Pathol. 2003;163:1771–1779. doi: 10.1016/S0002-9440(10)63537-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre B, Plantard L, Aeschbach L, Brakch N, Christen-Zaech S, de Viragh PA, Sergeant A, Huber M, Hohl D. SLURP1 is a late marker of epidermal differentiation and is absent in Mal de Meleda. J Invest Dermatol. 2007;127:301–308. doi: 10.1038/sj.jid.5700551. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman MJ, Sisi P, Banbury DN, Byrne C. Patterned acquisition of skin barrier function during development. Development. 1998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Gene Expr Patterns. 2002;2:93–97. doi: 10.1016/s0925-4773(02)00292-7. [DOI] [PubMed] [Google Scholar]
- Hewett DR, Simons AL, Mangan NE, Jolin HE, Green SM, Fallon PG, McKenzie AN. Lethal, neonatal ichthyosis with increased proteolytic processing of filaggrin in a mouse model of Netherton syndrome. Hum Mol Genet. 2005;14:335–346. doi: 10.1093/hmg/ddi030. [DOI] [PubMed] [Google Scholar]
- Hilger-Eversheim K, Moser M, Schorle H, Buettner R. Regulatory roles of AP-2 transcription factors in vertebrate development, apoptosis and cell-cycle control. Gene. 2000;260:1–12. doi: 10.1016/s0378-1119(00)00454-6. [DOI] [PubMed] [Google Scholar]
- Huber M, Siegenthaler G, Mirancea N, Marenholz I, Nizetic D, Breitkreutz D, Mischke D, Hohl D. Isolation and characterization of human repetin, a member of the fused gene family of the epidermal differentiation complex. J Invest Dermatol. 2005;124:998–1007. doi: 10.1111/j.0022-202X.2005.23675.x. [DOI] [PubMed] [Google Scholar]
- Imagawa M, Chiu R, Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987;51:251–260. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- Kohlbecker A, Lee AE, Schorle H. Exencephaly in a subset of animals heterozygous for AP-2alpha mutation. Teratology. 2002;65:213–218. doi: 10.1002/tera.10037. [DOI] [PubMed] [Google Scholar]
- Koster MI, Kim S, Huang J, Williams T, Roop DR. TAp63alpha induces AP-2gamma as an early event in epidermal morphogenesis. Dev Biol. 2006;289:253–261. doi: 10.1016/j.ydbio.2005.10.041. [DOI] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Roop DR. Genetic pathways required for epidermal morphogenesis. Eur J Cell Biol. 2004;83:625–629. doi: 10.1078/0171-9335-00387. [DOI] [PubMed] [Google Scholar]
- Koster MI, Roop DR. Mechanisms regulating epithelial stratification. Annu Rev Cell Dev Biol. 2007;23:93–113. doi: 10.1146/annurev.cellbio.23.090506.123357. [DOI] [PubMed] [Google Scholar]
- Leask A, Byrne C, Fuchs E. Transcription factor AP2 and its role in epidermal-specific gene expression. Proc Natl Acad Sci U S A. 1991;88:7948–7952. doi: 10.1073/pnas.88.18.7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List K, Haudenschild CC, Szabo R, Chen W, Wahl SM, Swaim W, Engelholm LH, Behrendt N, Bugge TH. Matriptase/MT-SP1 is required for postnatal survival, epidermal barrier function, hair follicle development, and thymic homeostasis. Oncogene. 2002;21:3765–3779. doi: 10.1038/sj.onc.1205502. [DOI] [PubMed] [Google Scholar]
- List K, Szabo R, Molinolo A, Nielsen BS, Bugge TH. Delineation of matriptase protein expression by enzymatic gene trapping suggests diverging roles in barrier function, hair formation, and squamous cell carcinogenesis. Am J Pathol. 2006;168:1513–1525. doi: 10.2353/ajpath.2006.051071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B, Mitchell PJ, Williams T, Tjian R. Regulation of transcription factor AP-2 by the morphogen retinoic acid and by second messengers. Genes Dev. 1989;3:1507–1517. doi: 10.1101/gad.3.10.1507. [DOI] [PubMed] [Google Scholar]
- Martin JF, Bradley A, Olson EN. The paired-like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 1995;9:1237–49. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- Mastrangeli R, Donini S, Kelton CA, He C, Bressan A, Milazzo F, Ciolli V, Borrelli F, Martelli F, Biffoni M, Serlupi-Crescenzi O, Serani S, Micangeli E, El Tayar N, Vaccaro R, Renda T, Lisciani R, Rossi M, Papoian R. ARS Component B: structural characterization, tissue expression and regulation of the gene and protein (SLURP-1) associated with Mal de Meleda. Eur J Dermatol. 2003;13:560–570. [PubMed] [Google Scholar]
- Maytin EV, Lin JC, Krishnamurthy R, Batchvarova N, Ron D, Mitchell PJ, Habener JF. Keratin 10 gene expression during differentiation of mouse epidermis requires transcription factors C/EBP and AP-2. Dev Biol. 1999;216:164–181. doi: 10.1006/dbio.1999.9460. [DOI] [PubMed] [Google Scholar]
- Mehrel T, Hohl D, Rothnagel JA, Longley MA, Bundman D, Cheng C, Lichti U, Bisher ME, Steven AC, Steinert PM, Yuspa SH, Roop DR. Identification of a major keratinocyte cell envelope protein, loricrin. Cell. 1990;61:1103–1112. doi: 10.1016/0092-8674(90)90073-n. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- Oyama N, Takahashi H, Tojo M, Iwatsuki K, Iizuka H, Nakamura K, Homma Y, Kaneko F. Different properties of three isoforms (alpha, beta, and gamma) of transcription factor AP-2 in the expression of human keratinocyte genes. Arch Dermatol Res. 2002;294:273–280. doi: 10.1007/s00403-002-0327-x. [DOI] [PubMed] [Google Scholar]
- Paladini RD, Coulombe PA. The functional diversity of epidermal keratins revealed by the partial rescue of the keratin 14 null phenotype by keratin 16. J Cell Biol. 1999;146:1185–1201. doi: 10.1083/jcb.146.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panteleyev AA, Mitchell PJ, Paus R, Christiano AM. Expression patterns of the transcription factor AP-2alpha during hair follicle morphogenesis and cycling. J Invest Dermatol. 2003;121:13–19. doi: 10.1046/j.1523-1747.2003.12319.x. [DOI] [PubMed] [Google Scholar]
- Patel GK, Wilson CH, Harding KG, Finlay AY, Bowden PE. Numerous keratinocyte subtypes involved in wound re-epithelialization. J Invest Dermatol. 2006;126:497–502. doi: 10.1038/sj.jid.5700101. [DOI] [PubMed] [Google Scholar]
- Porter RM, Corden LD, Lunny DP, Smith FJ, Lane EB, McLean WH. Keratin K6irs is specific to the inner root sheath of hair follicles in mice and humans. Br J Dermatol. 2001;145:558–568. doi: 10.1046/j.1365-2133.2001.04463.x. [DOI] [PubMed] [Google Scholar]
- Presland RB, Coulombe PA, Eckert RL, Mao-Qiang M, Feingold KR, Elias PM. Barrier function in transgenic mice overexpressing K16, involucrin, and filaggrin in the suprabasal epidermis. J Invest Dermatol. 2004;123:603–606. doi: 10.1111/j.0022-202X.2004.23226.x. [DOI] [PubMed] [Google Scholar]
- Rinne T, Brunner HG, van Bokhoven H. p63-associated disorders. Cell Cycle. 2007;6:262–268. doi: 10.4161/cc.6.3.3796. [DOI] [PubMed] [Google Scholar]
- Scholtens D, von Heydebreck A. Analysis of Differential Gene Expression Studies. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; 2005. pp. 229–248. [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Shi D, Kellems RE. Transcription factor AP-2gamma regulates murine adenosine deaminase gene expression during placental development. J Biol Chem. 1998;273:27331–27338. doi: 10.1074/jbc.273.42.27331. [DOI] [PubMed] [Google Scholar]
- Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, Liao H, Evans AT, Goudie DR, Lewis-Jones S, Arseculeratne G, Munro CS, Sergeant A, O’Regan G, Bale SJ, Compton JG, DiGiovanna JJ, Presland RB, Fleckman P, McLean WH. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–342. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Oyama N, Itoh Y, Ishida-Yamamoto A, Kaneko F, Iizuka H. Transcriptional factor AP-2gamma increases human cystatin A gene transcription of keratinocytes. Biochem Biophys Res Commun. 2000;278:719–723. doi: 10.1006/bbrc.2000.3850. [DOI] [PubMed] [Google Scholar]
- Tanifuji-Terai N, Terai K, Hayashi Y, Chikama T, Kao WW. Expression of keratin 12 and maturation of corneal epithelium during development and postnatal growth. Invest Ophthalmol Vis Sci. 2006;47:545–551. doi: 10.1167/iovs.05-1182. [DOI] [PubMed] [Google Scholar]
- Waikel RL, Wang XJ, Roop DR. Targeted expression of c-Myc in the epidermis alters normal proliferation, differentiation and UV-B induced apoptosis. Oncogene. 1999;18:4870–4878. doi: 10.1038/sj.onc.1203040. [DOI] [PubMed] [Google Scholar]
- Wang D, Shin TH, Kudlow JE. Transcription factor AP-2 controls transcription of the human transforming growth factor-alpha gene. J Biol Chem. 1997;272:14244–14250. doi: 10.1074/jbc.272.22.14244. [DOI] [PubMed] [Google Scholar]
- Wang X, Bolotin D, Chu DH, Polak L, Williams T, Fuchs E. AP-2alpha: a regulator of EGF receptor signaling and proliferation in skin epidermis. J Cell Biol. 2006;172:409–421. doi: 10.1083/jcb.200510002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger Q, Huang J, Auman HJ, Lewandoski M, Williams T. Analysis of transcription factor ap-2 expression and function during mouse preimplantation development. Biol Reprod. 2006;75:324–333. doi: 10.1095/biolreprod.106.052407. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yuspa SH, Kilkenny AE, Steinert PM, Roop DR. Expression of murine epidermal differentiation markers is tightly regulated by restricted extracellular calcium concentrations in vitro. J Cell Biol. 1989;109:1207–1217. doi: 10.1083/jcb.109.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]


