Abstract
Background
Considerable interest in the bioconversion of lignocellulosic biomass into ethanol has led to metabolic engineering of Saccharomyces cerevisiae for fermentation of xylose. In the present study, the transcriptome and proteome of recombinant, xylose-utilising S. cerevisiae grown in aerobic batch cultures on xylose were compared with those of glucose-grown cells both in glucose repressed and derepressed states. The aim was to study at the genome-wide level how signalling and carbon catabolite repression differ in cells grown on either glucose or xylose. The more detailed knowledge whether xylose is sensed as a fermentable carbon source, capable of catabolite repression like glucose, or is rather recognised as a non-fermentable carbon source is important for further engineering this yeast for more efficient anaerobic fermentation of xylose.
Results
Genes encoding respiratory proteins, proteins of the tricarboxylic acid and glyoxylate cycles, and gluconeogenesis were only partially repressed by xylose, similar to the genes encoding their transcriptional regulators HAP4, CAT8 and SIP1-2 and 4. Several genes that are repressed via the Snf1p/Mig1p-pathway during growth on glucose had higher expression in the cells grown on xylose than in the glucose repressed cells but lower than in the glucose derepressed cells. The observed expression profiles of the transcription repressor RGT1 and its target genes HXT2-3, encoding hexose transporters suggested that extracellular xylose was sensed by the glucose sensors Rgt2p and Snf3p. Proteome analyses revealed distinct patterns in phosphorylation of hexokinase 2, glucokinase and enolase isoenzymes in the xylose- and glucose-grown cells.
Conclusion
The results indicate that the metabolism of yeast growing on xylose corresponds neither to that of fully glucose repressed cells nor that of derepressed cells. This may be one of the major reasons for the suboptimal fermentation of xylose by recombinant S. cerevisiae strains. Phosphorylation of different isoforms of glycolytic enzymes suggests that regulation of glycolysis also occurred at a post-translational level, supporting prior findings.
Introduction
On hexose sugars, the yeast Saccharomyces cerevisiae has an exceptional capacity for rapid anaerobic growth and fermentation of sugar to carbon dioxide and ethanol. However, S. cerevisiae exhibits only negligible metabolism of xylose even under aerobic conditions [1,2]. The interest in production of fuel ethanol from renewable plant material, often rich in pentose sugars such as xylose, has encouraged extensive metabolic engineering of S. cerevisiae for xylose metabolism [3,4].
Recombinant xylose-utilising S. cerevisiae strains have been constructed by introducing xylose reductase (XR; preferring NADPH over NADH) and xylitol dehydrogenase (XDH; strictly NAD+ specific) encoding genes from the xylose-fermenting yeast Pichia stipitis, and additionally overexpressing the endogenous xylulokinase encoding gene (XKS1) [5-8]. This oxidoreductive xylose pathway has enabled xylose fermentation by S. cerevisiae [7,8] but with significantly lower rates compared with glucose fermentation. The different cofactor specificities of XR and XDH disturb the cellular redox cofactor balance during xylose metabolism, particularly under anaerobic conditions. Recently, an evolutionarily engineered strain with xylose isomerase from the anaerobic fungus Piromyces sp. E2 was shown to have improved anaerobic growth and fermentation on xylose compared with S. cerevisiae strains expressing the oxidoreductive pathway [9-11]. However, even this strain utilised glucose and xylose sequentially, with preference for glucose, and the anaerobic specific growth rate remained lower on xylose than on glucose [11]. It thus seems that the rate of xylose metabolism in engineered S. cerevisiae is not solely restricted by problems in the balance of redox cofactors. Uptake of xylose [12] and the low capacity of the pentose phosphate pathway (PPP) in S. cerevisiae have been identified as plausible limitations of xylose metabolism in this yeast [5,13].
The overall metabolism and changes generated by mutagenesis in xylose-metabolising recombinant S. cerevisiae strains have been analysed by transcriptional, proteome and metabolic flux analyses [14-20]. A moderate increase in transcript levels of some of the genes encoding enzymes of the PPP, as well as in the flux through the pathway was observed in cells metabolising xylose [14,16,18]. Additionally, expression of several genes encoding redox-related enzymes was enhanced [18,21]. Starvation-related but not general stress-related responses were activated during growth on xylose, and furthermore, metabolism on xylose seemed neither fully fermentative nor fully respiratory [21]. Importantly, anaerobic growth on xylose was suggested to be limited by the rate of ATP production [18]. Jin et al. compared the transcription of xylose-metabolising S. cerevisiae growing on xylose alone or on glucose alone in shake flask batch cultures under aerobic and oxygen-limited conditions [17]. They observed up-regulation of genes encoding activities of the tricarboxylic acid (TCA) cycle and gluconeogenesis, and of respiratory genes under oxygen limitation in cells grown on xylose compared with glucose repressed cells.
Most studies of xylose metabolism in the recombinant S. cerevisiae have focused on assessing the reasons behind its inability to grow on or to ferment efficiently this pentose sugar under anaerobic conditions. However, the effect of xylose on the dominant regulatory circuit of carbon metabolism, i.e. glucose repression, has so far been largely overlooked. Unravelling how xylose affects regulation of metabolism, i.e. is xylose sensed as a fermentable carbon source and capable of catabolite repression like glucose, or is it rather sensed as a non-fermentable carbon source, is highly important in achieving understanding for engineering this yeast for efficient anaerobic fermentation of xylose. Thus, our aim was to determine how signalling and carbon catabolite repression differ in cells growing on either glucose or xylose. We carried out batch fermentations on these sugars under fully aerobic conditions to avoid the effect of oxygen limitation on the redox balance in cells grown on xylose. In previous transcriptome and proteome studies the metabolism on xylose was compared only with either glucose repressed [17] or derepressed cells [21], whereas comparison of xylose-grown cells with both glucose repressed and derepressed cells in the present study enabled responses solely due to the absence of glucose repression to be distinguished from those directly linked to the metabolism of xylose. The data reveal novel information on transcript and protein levels of xylose-metabolising cells indicating that they have both fermentative and respirative features. Importantly, although xylose was able to repress many of the glucose-repressible genes, repression was only partial compared with the glucose-grown and fully repressed cells. Additionally, signalling in the Snf1p/Mig1p glucose repression and Snf3p/Rgt2p-Rgt1p glucose induction pathways appeared different from that in the glucose-grown cells.
Materials and methods
Strains
Genetically modified S. cerevisiae strain VTT-C-99318 (CEN.PK2-1D ura3::XYL1 XYL2 his3::XKS1 kanMX) [22] was derived from CEN.PK2-1D (MATα, leu2-3/112, ura3-52, trp1-289, his3Δ1, MAL2-8c, SUC2) [23] and contains the genes XYL1 and XYL2 of P. stipitis encoding xylose reductase and xylitol dehydrogenase, respectively, integrated into the URA3 locus. XYL1 is expressed under a PGK1 promoter and XYL2 under a modified ADH1 promoter [24]. In addition, the strain contains an additional copy of the endogenous xylulokinase encoding gene of S. cerevisiae with the modified ADH1 promoter [24] and integrated into the HIS3 locus. The integration of XYL1 and XYL2 was described by Toivari et al., and that of XKS1 by Richard et al. [8,25].
Aerobic batch fermentations
VTT-C-99318 was grown at 30°C in 1.8-litre bioreactors (Chemap CMF Bioreactor, Chemap AG, Volketswil, Switzerland) with a working volume of 1.5 litres. The pH was kept at 5.5 by automatic addition of 2 M KOH, and the stirrer speed at 500 rpm. Mass flow controllers (Bronkhorst High-Tech B.V, Ruurlo, Netherlands) regulated the total gas flow at 0.5 standard litres per minute (SLPM). The composition of the fermentor off-gas was analysed using a QMG 421C quadrupole mass spectrometer (Balzers Pfeiffer Scandinavia AB, Sweden) as described previously [14]. The medium was synthetic complete (SC) (modified from Sherman, 1983) [26]) supplemented with 0.5 ml silicone antifoam l-1 (AnalaR BDH, UK) and 50 g l-1 xylose or 50 g l-1glucose. Triplicate cultures (F0, F1, and F2) were carried out on both carbon sources. Cultures were started with an initial OD600 of 1 to 1.5 from pre-cultures grown for 20 h at 30°C on SC with 30 g/L glucose.
The 25 ml samples for transcriptome and proteome analyses were harvested by centrifugation (2 min, 5000 g) after 5 h and 24 h growth for glucose cultures and after 72 h growth for xylose cultures and frozen in liquid nitrogen. Cell growth was measured as OD600 and cell dry mass as described previously [14]. Extracellular concentrations of glucose, xylose, xylitol, ethanol, acetate and glycerol were analysed by high-performance liquid chromatography as described previously [14].
Samples from the F2 cultivations on each carbon source were hybridised three times on Affymetrix Yeast Genome S98 arrays (hybridisations denoted as H2.1, H2.2 and H2.3; see [Additional files 1 and 2]) and the samples from the cultivations F0 and F1 once (hybridisations denoted as H0 and H1, respectively; see [Additional files 1 and 2]). Proteome analyses were carried out with samples taken at the same time points as the samples for the transcriptome analyses.
Affymetrix hybridisations, data acquisition and analysis
Affymetrix hybridisations and analyses were carried out according to the protocols provided by Affymetrix, Inc. Double-stranded cDNA was synthesised from 5 μg total RNA. An in vitro transcription (IVT) reaction was then carried out to produce biotin-labelled cRNA from the cDNA. The cRNA was fragmented and hybridised to a Test3 array for quality control and then to the Yeast Genome S98 array for 16 h at 45°C. The arrays were washed and stained with streptavidin-phycoerythrin in an Affymetrix Fluidics Station 400 and scanned (Agilent G2500A GeneArray Scanner). The data were extracted with Affymetrix MicroArray Suite 5.0 software.
Array data were pre-processed using the Robust Multichip Average (RMA) [27,28] implemented in the Bioconductor (version 1.5.0) [29] extension to the R software environment for statistical computing and graphics (version 2.0.1). The RMA-method computes log2 scale expression values from cell intensity files using the RMA model of probe-specific correction of perfect match (PM) probes and quantile normalisation [28]. A visual inspection of the scatter plots of the RMA pre-processed expression values, and the Pearson correlations between the replicate arrays did not indicate outlying arrays [Additional files 3, 4, 5].
After the pre-processing, TIGR MultiExperiment Viewer (version 3.1) was used to analyse the expression values [30]. The replicate values were averaged and genes which were differentially expressed between the glucose repressed and derepressed samples and the xylose samples were detected using one-way ANOVA at a p-value 0.01 based on 1000 permutation tests [31]. The expression values of significantly expressed genes (1439) were then mean centred, the biological replicate arrays were averaged and the values clustered using K-means with Euclidean distance [32]. Clustering was performed several times with varying parameters and the most optimal result was obtained with 13 clusters. Since the 13 clusters had 8 distinct expression profiles, and furthermore, as combining the clusters with similar expression profiles lowered the p-values of best gene ontology (GO) classes [33], the clusters were manually reorganised into 8 clusters.
Two-dimensional gel electrophoresis of the proteome
5–10 mg dry mass of cells was suspended in 150 μl of 10% (v/v) trichloroacetic acid (TCA, Merck, USA) in a 1.5 ml microcentrifuge tube. 500 μl glass beads (0.5 mm diameter; Biospec Products, USA) were added and the tubes shaken in a MiniBeadbeater 8 (Biospec Products) at homogenisation speed three times for 30 seconds. The tubes were cooled on ice between each homogenisation step. The supernatants were collected and proteins were precipitated by adding 600 μl of -20°C acetone and incubating 30 min on ice. Precipitated proteins were collected by centrifugation for 30 min, 13 000 rpm, at 4°C, rinsed once with 600 μl of -20°C acetone, and re-suspended in 450 μl of 7 M urea (Promega, USA), 2 M thiourea (Fluka, USA), 4% (w/v) CHAPS (Fluka), 1% (w/v) Pharmalytes 3–10 (Pharmacia, Sweden) and 1% (w/v) DTT (Sigma) by gently shaking for 20 min at room temperature. Supernatants were collected by centrifugation for 5 min, 13 000 rpm. The protein concentration of the supernatants was determined by the Non-Interfering Protein Assay (Geno Technology, Inc., USA) and the samples were stored at -70°C prior to isoelectric focusing.
The isoelectric focusing and the second dimension on 11% (w/v) SDS-PAGE were carried out as described earlier [15]. After electrophoresis the gels were fixed for 1.5 h in 30% (v/v) ethanol and 0.5% (v/v) acetic acid, and stained either only with Sypro Ruby (Molecular Probes, USA) or with phosphoprotein-specific Pro-Q Diamond (Molecular Probes) followed by Sypro Ruby, according to instructions of the manufacturer. The stained gels were scanned with a resolution of 100 microns on a Typhoon instrument (GE Healthcare, USA). The gel images were analysed using the PDQuest software (Bio-Rad, USA). The gel patterns from different gels were automatically matched with some additional manual editing, and the quantities of matching spots in different gels were compared. For each condition, average spot quantities were calculated from two to four gels of each sample taken from each of the triplicate glucose and xylose cultivations. The quantification of the resolved proteins was normalised to the total optical density in each gel image. The data from 2-DE (two-dimensional electrophoresis) gels were analysed using the TIGR MultiExperiment Viewer (version 3.1) [30]. Proteins which had different abundance in glucose compared with xylose cultures were identified using the one-way ANOVA at a p-value of 0.01 [31]. The 220 protein-spot intensity values obtained were mean centred, the spots from replicate gels averaged, and the signal intensity values for identified protein spots (in total 70) were clustered using hierarchical clustering with Euclidean distance and average linkage [34].
The degree of phosphorylation was calculated by dividing the Pro-Q Diamond signal of a protein spot with the total protein amount of the spot determined by the Sypro Ruby signal. The averages and standard errors of the mean (SEM) were calculated from six replicate gels (2 replicate gels per sample from each of the three fermentations).
Protein identification by mass spectrometry and proteome data analysis
Excised gel spots were destained in a freshly prepared solution of 30 mM potassium ferricyanide and 100 mM sodium thiosulfate (1:1) and then dehydrated in acetonitrile (ACN). For protein alkylation, the dried gel pieces were incubated in 5 mM tris (2-carboxyethyl) phosphine and 55 mM iodoacetamide (Sigma, USA) in dark at room temperature for 1 h, followed by washing with 100 mM ammonium hydrogen carbonate (NH4HCO3) and dehydration with ACN. The gel pieces were rehydrated in 10 to 15 μl 10 mM NH4HCO3, 10% (v/v) ACN containing 0.01 μg/μl sequencing grade trypsin (Promega, USA), and digested overnight at 37°C. To elute the tryptic peptides from the gel, the pieces were incubated twice in 150 μl 66% ACN (v/v) and 0.1% (v/v) trifluoroacetic acid (TFA; Fluka) for 30 min at 37°C. The peptide eluents were dried to a minimal amount of liquid in a SpeedVac, and suspended in 5 μl of 0.1% (v/v) TFA.
For co-crystallisation of peptides with the matrix, an aliquot of the peptide solution was mixed with a saturated matrix solution in a ratio of 1:10, and 1 to 2 μl was dispensed on a MALDI target and let to dry at room temperature. The matrix solution used was prepared by dissolving 17 mg α-cyano-4-hydroxycinnamic acid (Sigma) in 1 ml of 33% (v/v) ACN and 0.1% (v/v) TFA.
MALDI-TOF analyses were carried out with Autoflex (Bruker Daltonics, Bremen, Germany), equipped with a nitrogen pulsed laser (337 nm), operated in positive mode. Typically, mass spectra were acquired by accumulating spectra of 400 laser shots. External calibration was performed for molecular assignments using a peptide calibration standard (Bruker Daltonik GmbH, Leipzig, Germany).
Protein identifications were performed by searching the peptide masses against the National Center for Biotechnology Information (NCBI) non-redundant database using Prowl's ProFound – Peptide Mapping (Rockefeller University) [35], Protein Prospector ms-fit (University of California, San Francisco) [36] and Matrix Science's Mascot – Peptide Mass Fingerprint (Matrix Science Ltd, UK) [37]. Protein identifications by peptide mass fingerprinting were further evaluated by comparing the calculated and observed molecular masses and pI-values, as well as the number of peptides matched and the percentage of sequence coverage.
Results
Experimental design and data analysis of the transcriptome and proteome
Three aerobic batch fermentations were carried out both on 50 g l-1 glucose and on 50 g l-1 xylose to compare the yeast transcriptome and proteome of cells growing on xylose with that of glucose repressed and glucose derepressed cells. Samples of the xylose-grown cells were harvested at 72 h from the start of the xylose cultures with 32 ± 1 g l-1 of residual xylose present. Samples of the glucose repressed cells were harvested at 5 h from the start of the glucose cultures with 37 ± 2 g l-1 of residual glucose present. Samples of the glucose derepressed cells were harvested at 24 h from the start of the glucose cultures containing no glucose but 13 ± 1 g l-1 of accumulated ethanol.
The volumetric consumption and production profiles of xylose, glucose, biomass, xylitol and ethanol (g l-1) of the cultures are shown in Figure 1. In addition, some acetate and glycerol (less than 2 g l-1 each) were produced in the glucose cultures, and less than 4 g l-1acetate, 3 g l-1 glycerol and 2 g l-1 ethanol were measured in the xylose cultures. The doubling time of the cells in the xylose cultures was 28 ± 2 h (during 5 to 72 h), and in the glucose cultures 1.8 ± 0.1 h (during 2 to 5 h) and 73 ± 3 h (during 24 to 53 h).
Figure 1.
Recombinant xylose-metabolising S. cerevisiae in aerobic batch cultures on xylose (A) and glucose (B) showing concentrations (g l-1) of A) xylose (■), cellular dry weight; DW (▲), ethanol (□) and xylitol (○) and B) glucose (■), cellular dry weight; DW (▲), and ethanol (□). Cultures were maintained at pH 5.5, 30°C, 500 rpm, 0.33 volume air [volume culture]-1 min-1. Data represent the average and standard deviation of three separate batch cultures on each carbon source. The arrows indicate times at which samples were taken for transcriptome and proteome analyses.
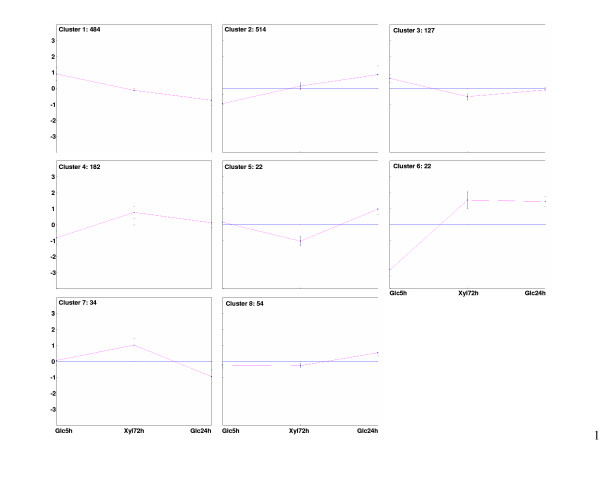
The 1439 genes, which were expressed differently (based on ANOVA at a p-value 0.01) between the glucose repressed (Glc5h), glucose derepressed (Glc24h) and xylose (Xyl72h) samples were organised into eight separate clusters with distinct expression profiles shown in Figure 2. The top three gene ontology classes of each cluster and the number of genes found in each class are listed in Table 1. All genes in each cluster of Figure 2 are shown in additional files [Additional files 6, 7, 8, 9, 10, 11, 12, 13].
Figure 2.
The eight distinct clusters of the 1439 differentially expressed genes determined by K-means with Euclidean distance. The y-axis corresponds to the difference of a gene relative to the mean expression of the gene in all samples on a log2-scale (values above zero-level represent up-regulation and below it down-regulation). The red lines represent the average expression pattern of each cluster. The x-axes are the 5 h and 24 h glucose and 72 h xylose samples (Glc5h, Glc24h and Xyl72h, respectively). The total number of genes in each cluster was: 484, 514, 127, 182, 22, 22, 34 and 54 for clusters 1 to 8, respectively.
Table 1.
The top three gene ontology (GO) classes in each of the eight clusters in Figure 1. n is the number of the genes with the specified function, % is the percentage of the genes in the cluster with the specified function, and p-value is the binomial distribution probability [33]. (Total number of genes in clusters 1 to 8: 484, 514, 127, 182, 22, 22, 34 and 54, respectively)
| Cluster | GO-class | Annotation | n | % | p-value |
| 1 | GO:0009058 | Biosynthesis | 159 | 32.9 | 1.96E-23 |
| GO:0044249 | Cellular biosynthesis | 148 | 30.6 | 1.95E-21 | |
| GO:0044238 | Primary metabolism | 286 | 59.1 | 5.51E-19 | |
| 2 | GO:0006091 | Generation of precursor metabolites and energy | 67 | 13.0 | 5.89E-25 |
| GO:0015980 | Energy derivation by oxidation of organic compounds | 57 | 11.1 | 6.24E-21 | |
| GO:0006119 | Oxidative phosphorylation | 20 | 3.9 | 5.25E-11 | |
| 3 | GO:0006896 | Golgi to vacuole transport | 5 | 3.9 | 1.39E-05 |
| GO:0006892 | Post-Golgi transport | 6 | 4.7 | 2.10E-04 | |
| GO:0042147 | Retrograde transport, endosome to Golgi | 3 | 2.4 | 6.40E-04 | |
| 4 | GO:0044242 | Cellular lipid catabolism | 2 | 1.1 | 4.99E-03 |
| GO:0016042 | Lipid catabolism | 2 | 1.1 | 4.99E-03 | |
| GO:0006000 | Fructose metabolism | 2 | 1.1 | 4.99E-03 | |
| 5 | GO:0006790 | Sulphur metabolism | 6 | 27.3 | 8.16E-09 |
| GO:0000096 | Sulphur amino acid metabolism | 4 | 18.2 | 1.71E-06 | |
| GO:0015837 | Amine transport | 4 | 18.2 | 8.44E-06 | |
| 6 | GO:0000017 | Alpha-glucoside transport | 2 | 9.1 | 1.00E-04 |
| GO:0042946 | Glucoside transport | 2 | 9.1 | 1.00E-04 | |
| GO:0008643 | Carbohydrate transport | 2 | 9.1 | 4.21E-03 | |
| 7 | GO:0009082 | Branched chain family amino acid biosynthesis | 5 | 14.7 | 2.50E-09 |
| GO:0009081 | Branched chain family amino acid metabolism | 5 | 14.7 | 7.00E-09 | |
| GO:0009098 | Leucine biosynthesis | 3 | 8.8 | 1.30E-06 | |
| 8 | GO:0000002 | Mitochondrial genome maintenance | 4 | 7.4 | 3.24E-05 |
| GO:0051294 | Establishment of spindle orientation | 2 | 3.7 | 1.66E-03 | |
| GO:0051293 | Establishment of spindle localization | 2 | 3.7 | 1.66E-03 |
70 out of 547 protein spots separated on the 2-DE gels had different abundances between the glucose repressed (Glc5h), glucose derepressed (Glc24h) and xylose (Xyl72h) samples and were identified by MALDI-TOF peptide mass mapping. They represented 55 different proteins, with 12 proteins present as multiple protein spots on the gels (Table 2) [Additional files 14 and 15]. 73% of the responses at the protein level correlated qualitatively with the microarray data (Table 2).
Table 2.
Proteins which were differentially translated in the glucose repressed, glucose derepressed and xylose-grown cells, identified from the 2-DE gels [Additional files 14 and 15].
| log2 X72/G24 | log2 X72/G5 | log2 G24/G5 | |||||||
| Swiss Prot Id | Proteina | ORF | Function | 2D | array | 2D | array | 2D | array |
| P32316 | Ach1p | YBL015W | acetyl-CoA hydrolase | 0.0 | -1.0 | 3.4 | 2.7 | 3.4 | 3.7 |
| P00330 | Adh1p_a | YOL086C | alcohol dehydrogenase | 1.4 | 0.9 | 0.2 | 0.3 | -1.2 | -0.6 |
| P00330 | Adh1p_b | YOL086C | alcohol dehydrogenase | 2.1 | 0.9 | 0.5 | 0.3 | -1.6 | -0.6 |
| P00331 | Adh2p | YMR303C | alcohol dehydrogenase | 4.8 | -2.6 | 5.7 | 1.4 | 0.9 | 4.0 |
| P47143 | Ado1p_a | YJR105W | adenosine kinase | -1.1 | 0.4 | -1.4 | -1.3 | -0.2 | -1.7 |
| P47143 | Ado1p_b | YJR105W | adenosine kinase | -1.4 | 0.4 | -1.9 | -1.3 | -0.5 | -1.7 |
| P38013 | Ahp1p | YLR109W | thioredoxin peroxidase | -0.4 | 0.0 | 0.5 | -0.2 | 1.0 | -0.2 |
| P54114 | Ald3p | YMR169C | aldehyde dehydrogenase | 0.4 | 0.8 | 1.8 | 1.8 | 1.4 | 1.0 |
| P46367 | Ald4p | YOR374W | aldehyde dehydrogenase (NAD+) | 1.7 | -0.2 | 3.6 | 1.0 | 1.9 | 1.1 |
| P54115 | Ald6p_a | YPL061W | aldehyde dehydrogenase (NADP+) | -0.9 | -2.6 | 0.2 | -1.9 | 1.0 | 0.7 |
| P54115 | Ald6p_b | YPL061W | aldehyde dehydrogenase (NADP+) | -2.0 | -2.6 | -2.8 | -1.9 | -0.8 | 0.7 |
| P28777 | Aro2p | YGL148W | chorismate synthase | -0.7 | 0.4 | -1.4 | -1.4 | -0.7 | -1.8 |
| P53090 | Aro8p | YGL202W | aromatic-amino-acid transaminase | -1.7 | 1.5 | -0.6 | -1.0 | 1.1 | -2.5 |
| P38011 | Asc1p | YMR116C | molecular function unknown | -0.6 | 1.8 | -2.4 | -1.1 | -1.8 | -3.0 |
| P07251 | Atp1p | YBL099W | hydrogen-transporting ATP synthase | 0.1 | -0.2 | 1.8 | 0.7 | 1.7 | 0.9 |
| P00830 | Atp2p | YJR121W | hydrogen-transporting ATP synthase | 0.2 | -0.4 | 1.6 | 0.9 | 1.4 | 1.2 |
| P38891 | Bat1p | YHR208W | branched-chain-amino-acid transaminase | 1.9 | 3.9 | 0.4 | 0.7 | -1.5 | -3.2 |
| P07256 | Cor1p_a | YBL045C | ubiquinol-cytochrome-c reductase | 0.6 | -0.2 | 1.3 | 1.0 | 0.8 | 1.2 |
| P07256 | Cor1p_b | YBL045C | ubiquinol-cytochrome-c reductase | 0.3 | -0.2 | 1.3 | 1.0 | 1.0 | 1.2 |
| P07256 | Cor1p_c | YBL045C | ubiquinol-cytochrome-c reductase | -0.1 | -0.2 | 1.3 | 1.0 | 1.5 | 1.2 |
| P06115 | Ctt1p | YGR088W | catalase | 1.4 | 1.8 | 3.4 | 2.4 | 2.0 | 0.6 |
| P31373 | Cys3p | YAL012W | cystathionine gamma-lyase | -2.3 | -0.6 | -2.9 | -3.1 | -0.6 | -2.4 |
| P32582 | Cys4p_a | YGR155W | cystathionine beta-synthase | -2.5 | -0.8 | -0.2 | -1.1 | 2.3 | -0.3 |
| P32582 | Cys4p_b | YGR155W | cystathionine beta-synthase | -0.9 | -0.8 | -1.7 | -1.1 | -0.9 | -0.3 |
| P06634 | Ded1p | YOR204W | RNA helicase | 0.5 | 0.3 | -2.2 | 0.5 | -2.7 | 0.2 |
| P14306 | Dka1p | YLR178C | carboxypeptidase Y inhibitor | -1.2 | -0.6 | 1.7 | 0.8 | 2.9 | 1.4 |
| P39976 | Dld3p | YEL071W | D-lactate dehydrogenase (cytochrome) | -2.0 | -0.2 | -2.4 | -0.6 | -0.3 | -0.4 |
| P00924 | Eno1p_a | YGR254W | phosphopyruvate hydratase | 1.5 | 0.6 | 5.2 | 0.5 | 3.7 | -0.1 |
| P00924 | Eno1p_b | YGR254W | phosphopyruvate hydratase | 3.5 | 0.6 | 5.7 | 0.5 | 2.2 | -0.1 |
| P00924 | Eno1p_c | YGR254W | phosphopyruvate hydratase | 1.2 | 0.6 | 2.6 | 0.5 | 1.3 | -0.1 |
| P14065 | Gcy1p | YOR120W | aldo-keto reductase | 2.0 | 0.3 | 3.7 | 3.1 | 1.7 | 2.8 |
| Q00055 | Gpd1p | YDL022W | glycerol-3-phosphate dehydrogenase | 0.7 | 0.0 | 1.9 | 0.2 | 1.2 | 0.2 |
| P00950 | Gpm1p | YKL152C | phosphoglycerate mutase | 1.7 | 0.5 | 1.6 | -0.1 | 0.0 | -0.7 |
| P38625 | Gua1p | YMR217W | GMP synthase | -0.8 | 1.3 | -1.8 | -1.0 | -1.0 | -2.3 |
| P15454 | Guk1p | YDR454C | guanylate kinase | -1.7 | 0.3 | -1.4 | -1.3 | 0.3 | -1.6 |
| P15992 | Hsp26p_a | YBR072W | heat shock protein | 0.1 | -0.2 | 3.0 | 2.0 | 2.9 | 2.2 |
| P15992 | Hsp26p_b | YBR072W | heat shock protein | -0.6 | -0.2 | 2.2 | 2.0 | 2.8 | 2.2 |
| P04807 | Hxk2p_a | YGL253W | hexokinase | 2.6 | 2.9 | 10.7 | -0.5 | 8.1 | -3.4 |
| P04807 | Hxk2p_b | YGL253W | hexokinase | 0.7 | 2.9 | -1.1 | -0.5 | -1.8 | -3.4 |
| P28241 | Idh2p | YOR136W | isocitrate dehydrogenase (NAD+) | -0.9 | -0.5 | 0.7 | 0.2 | 1.7 | 0.7 |
| P00817 | Ipp1p | YBR011C | inorganic diphosphatase | -1.7 | -0.5 | -1.5 | -0.9 | 0.3 | -0.5 |
| P53598 | Lsc1p | YOR142W | succinate-CoA ligase (ADP-forming) | -0.5 | -0.4 | 0.6 | 0.1 | 1.1 | 0.4 |
| P36060 | Mcr1p | YKL150W | cytochrome-b5 reductase | 1.8 | 0.1 | 3.3 | 1.5 | 1.4 | 1.4 |
| P05694 | Met6p | YER091C | methionine synthase | -2.5 | -1.3 | -2.9 | -1.0 | -0.4 | 0.3 |
| P04147 | Pab1p | YER165W | poly(A) binding protein | 0.4 | 0.2 | -1.1 | -0.3 | -1.6 | -0.5 |
| P06169 | Pdc1p_ab | YLR044C | pyruvate decarboxylase | -1.7 | 2.0 | -1.8 | -0.2 | 0.0 | -2.2 |
| P06169 | Pdc1p_bb | YLR044C | pyruvate decarboxylase | -2.1 | 2.0 | -1.2 | -0.2 | 0.9 | -2.2 |
| P17967 | Pdi1p | YCL043C | protein disulfide isomerase | -0.7 | -0.1 | -2.6 | -0.5 | -1.9 | -0.4 |
| P00560 | Pgk1p_a | YCR012W | phosphoglycerate kinase | 1.1 | 0.6 | 1.8 | 0.1 | 0.7 | -0.5 |
| P00560 | Pgk1p_b | YCR012W | phosphoglycerate kinase | 1.9 | 0.6 | 2.6 | 0.1 | 0.7 | -0.5 |
| P34227 | Prx1p | YBL064C | thioredoxin peroxidase | -0.2 | -0.9 | 1.7 | 0.7 | 1.9 | 1.6 |
| Q12335 | Pst2p | YDR032C | molecular function unknown | -0.6 | -0.1 | 0.8 | -0.4 | 1.5 | -0.3 |
| P07703 | Rpc5p | YPR110C | DNA-directed RNA polymerase | -0.3 | 0.5 | -1.5 | -1.3 | -1.2 | -1.8 |
| P26783 | Rps5p | YJR123W | structural constituent of ribosome | -0.6 | 0.7 | -3.0 | -0.6 | -2.4 | -1.2 |
| P19358 | Sam2p | YDR502C | methionine adenosyltransferase | -2.2 | -2.0 | -2.5 | -2.6 | -0.3 | -0.6 |
| P07283 | Sec53p_a | YFL045C | phosphomannomutase | -0.1 | 0.2 | -2.5 | -1.6 | -2.4 | -1.8 |
| P07283 | Sec53p_b | YFL045C | phosphomannomutase | -1.1 | 0.2 | -2.0 | -1.6 | -0.8 | -1.8 |
| P33330 | Ser1p | YOR184W | phosphoserine transaminase | -0.8 | -0.3 | -0.9 | -1.0 | -0.1 | -0.8 |
| P37291 | Shm2p | YLR058C | glycine hydroxymethyltransferase | 0.1 | -0.1 | -1.5 | 0.6 | -1.6 | 0.7 |
| Q03144 | Sno1p | YMR095C | molecular function unknown | 0.3 | 0.4 | -0.8 | 2.0 | -1.1 | 1.6 |
| P00447 | Sod2p | YHR008C | manganese superoxide dismutase | -0.5 | -0.2 | 1.9 | 1.1 | 2.4 | 1.2 |
| P15705 | Sti1p | YOR027W | Hsp90 cochaperone | 0.6 | 0.2 | 1.7 | 0.3 | 1.1 | 0.1 |
| P23254 | Tkl1p | YPR074C | transketolase | -0.6 | 1.1 | -1.0 | -0.6 | -0.5 | -1.7 |
| P00942 | Tpi1p | YDR050C | triose-phosphate isomerase | -1.1 | 0.2 | 0.5 | -0.1 | 1.5 | -0.3 |
| P17649 | Uga1p | YGR019W | 4-aminobutyrate transaminase | -0.8 | -0.1 | 1.4 | 0.2 | 2.2 | 0.3 |
| Q12363 | Wtm1p_a | YOR230W | transcriptional modulator | 0.1 | -0.1 | 1.7 | 0.2 | 1.6 | 0.3 |
| Q12363 | Wtm1p_b | YOR230W | transcriptional modulator | -0.1 | -0.1 | 1.3 | 0.2 | 1.4 | 0.3 |
| P23180 | Yhl021p | YHL021C | molecular function unknown | -0.5 | -0.8 | 1.6 | 0.6 | 2.1 | 1.3 |
| P35691 | Ykl056cp | YKL056C | molecular function unknown | -1.0 | 0.5 | -1.8 | -1.4 | -0.9 | -1.9 |
| Q04869 | Ymr315wp | YMR315W | molecular function unknown | 2.5 | 2.3 | 3.1 | 1.8 | 0.6 | -0.4 |
Log2 values of the protein abundance ratios, and the expression ratios for the corresponding gene of cells grown on xylose for 72 h (X72) to cells grown on glucose for 24 h (G24), or 5 h (G5), and of cells grown on glucose for 24 h to cells grown on glucose for 5 h are given. Proteins whose encoding genes showed a statistically significant expression difference in the samples studied are marked with bold text.
aDifferent pI forms of a protein indicated by a, b and c
bThese spots of Pdc1p had substantially lower molecular weight compared with the major Pdc1p protein spot identified earlier [15] and are likely either degraded or post-translationally processed forms of the enzyme.
Carbon source sensing and signalling in cells metabolising xylose
One of the first responses of S. cerevisiae to glucose is the induction of genes encoding hexose transporters (Hxt) that are also used for xylose uptake [38,39]. The signal for the presence of glucose is mediated via the glucose sensor proteins Snf3p and Rgt2p, responding to low and high glucose concentrations, respectively [40,41]. The expression of SNF3 is repressed at high concentrations of glucose via Mig1p ([42] and references therein). In the xylose-grown cells, the transcript levels of SNF3 were higher than in the glucose repressed cells, but lower than in the glucose derepressed cells (Fig. 2, cluster 2), while RGT2 had its highest expression in the xylose-grown cells (Fig. 2, cluster 4). The expression of RGT2 is reported to be independent of glucose concentration [43], but in the present study derepression was observed (Fig. 2, cluster 4).
RGT1, encoding a transcription factor of HXT genes had, like SNF3, higher transcript levels in the xylose-grown cells than in the glucose repressed cells, but lower than in the glucose derepressed cells (Fig. 2, cluster 2). The expression of RGT1 is reported not to be regulated in response to glucose ([42] and references therein), but the expression profile in cluster 2 suggests that its expression was derepressed in the absence of glucose, and to a lesser extent also on xylose (Fig. 2). Consistently, HXT2 and HXT3, which are repressed by RGT1, had highest expression in the presence of glucose and lowest in the glucose derepressed cells (Fig. 2, cluster 1). In addition to its function in repressing HXT genes in the absence of glucose [44], Rgt1p is required for repression of HXK2 at low levels of glucose together with the transcription cofactor Med8p [45]. The expression of RGT1 and HXK2 was as expected: while RGT1 was expressed at its highest level in the glucose derepressed cells, HXK2 had its lowest expression in these cells (Fig. 2, cluster 1). MTH1 encoding a corepressor of RGT1 had its highest expression in the xylose-grown cells (Fig. 2, cluster 4). Mth1p is involved in maintaining the repression of HXT genes when glucose is not present [46]. Nevertheless, in the cells grown on xylose, HXT2 and HXT3 had higher transcript levels compared with the glucose derepressed cells (Fig. 2, cluster 1), HXT6 compared with the glucose repressed cells (Fig. 2, cluster 2), and HXT4 and HXT16 compared with both the glucose repressed and derepressed cells (Fig. 2, clusters 7 and 4, respectively).
The metabolism of the recombinant S. cerevisiae on xylose appears neither as fully glucose repressed nor as fully derepressed
In xylose-grown cells, the genes encoding activities of the TCA and glyoxylate cycles, respiration and gluconeogenesis were expressed more strongly than in glucose repressed cells but less strongly than in derepressed cells (Fig. 2, cluster 2). On glucose these genes are repressed via the pathway involving Snf1p, Mig1p and Hxk2p [47]. In more detail, cluster 2 contained genes encoding enzymes of respiration (e.g. COX4, COX5a, COX7, COX14-15, COR1, QCR2, QCR7, QCR9, ATP3-5, ATP16), gluconeogenesis (e.g. FBP1, PCK1), the TCA and glyoxylate cycles (e.g. KGD1-2, SDH1-4, FUM1, MDH1, CIT1-3, MLS1, ICL1), alcohol catabolism (e.g. ADH2), and trehalose and glycogen synthesis (e.g. TSL1, GLC1-3, GSY2) (Table 1). In addition, genes encoding transcriptional regulators of the aforementioned genes, such as ADR1, CAT8, HAP4, SIP1-2 and 4, and REG2 [48] were present in cluster 2 (Fig. 2). In the proteome analysis of the corresponding samples, a similar trend was seen in the abundance of proteins with respiratory functions, such as ATP synthases Atp1p and Atp2p, ubiquinol-cytochrome-c reductase (Cor1p; two isoforms detected), isocitrate dehydrogenase Idh2p (expression of IDH2 was comparable in the glucose repressed and xylose-grown cells; Fig. 2, cluster 8), and the α-subunit of succinyl-CoA ligase Lsc1p (Table 2) [Additional file 15].
Distinct profiles in the abundance of transcripts and enzyme levels of the glycolytic and ethanol pathways were observed. HXK2 and PGI1 had lower expression in the xylose-grown cells compared with the glucose repressed cells (Fig. 2, cluster 1). On the other hand, the xylose-grown cells had higher amounts of Pgk1p, Gpm1p and Adh1p, compared with the glucose repressed cells (Table 2) [Additional file 15]. In addition, Eno1p and Adh2p (whose encoding gene had higher transcript abundance on xylose compared with the glucose repressed cells; Fig. 2, cluster 2) were most abundant in the xylose-grown cells (Table 2) [Additional file 15]. The latter two proteins are typically synthesised on non-fermentable carbon sources [49,50].
Overlaying the comparison of gene expression in the glucose repressed, derepressed and xylose-grown cells on the main metabolic network (Fig. 3) further demonstrates how the genes encoding activities of the TCA and glyoxylate cycles had higher expression profiles on xylose compared to the glucose repressed cells, whereas the opposite was observed with many genes of glycolysis and the PPP. The differences indicated in the expression of all individual genes in Figure 3 were not statistically significant based on ANOVA, however, the analysis clearly illustrates the distinct trends in the expression of genes of individual pathways in the central carbon metabolism on xylose compared to the glucose repressed and derepressed cells.
Figure 3.
Expression profiles of genes involved in the main metabolic networks of S. cerevisiae. Transcription factors are presented with boldfacing and connected to the respective target genes with gray lines. The connections shown between transcription factors and their target genes are based on compilations in Yeast Proteome Database [70][80] and on the transcription factor binding network by Young and co-workers [82]. Expression of the genes presented in white boxes with black text was highest in the glucose derepressed cells (Derep.) and lowest in the glucose repressed cells (Rep.). Expression of the genes presented in black boxes with white text was highest in the glucose repressed cells and lowest in the glucose derepressed cells. Expression of the genes presented in dark gray boxes with black text was highest in the xylose-grown cells and lowest in the glucose derepressed cells. Expression of the genes presented in light gray boxes with black text was highest in the xylose-grown cells and lowest in the glucose repressed cells. Expression of the genes presented in gray boxes with white text was lowest in the xylose-grown cells. In addition to the genes shown in the figure, 89% of the genes (31 out of 35) annotated to GO category "Oxidative phosphorylation" and its daughter categories [33] had highest expression in the glucose derepressed cells, lowest expression in the glucose repressed cells and intermediate expression in the cells grown on xylose (data not shown).
In addition to Snf3p and Rgt2p, extracellular glucose is sensed via Gpr1p, which signals the presence of glucose via Gpa2p to the cyclic AMP-protein kinase A (cAMP-PKA) signalling pathway [51]. The glucose-induced activation of cAMP synthesis is subsequently repressed by glucose and therefore not considered to be operative during growth on glucose [52]. Most genes encoding components of the cAMP-PKA pathway had intermediate expression in the xylose-grown cells with highest expression in the glucose derepressed cells and lowest in the glucose repressed cells [Additional file 16]. Consistently, 52% of the genes (28/54 genes) repressed via this pathway in the presence of glucose had the same intermediate expression profile in the xylose-grown cells (data not shown).
Some glucose repressible genes had their highest expression on xylose
HXK1, HXT16 and SUC2, which are all repressed by glucose via the Snf1p/Mig1p-pathway [42,53-55], had their highest expression levels in the xylose-grown cells (Fig. 2, cluster 4). MAL11, MAL31, and MAL32, encoding activities for the utilisation of maltose and representing classical prototype genes of glucose repression by Mig1p, were also relieved from glucose repression (Fig. 2, cluster 6). This was observed despite the fact that GAL83 encoding one of the β-subunits of the Snf1 kinase complex and enabling nuclear localisation of the kinase in the presence of non-fermentable carbon sources [56] had its lowest expression in the cells grown on xylose (Fig. 2, cluster 3), as if xylose was perceived as a fermentable, repressive carbon source.
Growth on xylose affected expression of genes in the pathways of acetyl-CoA synthesis and consumption
The metabolic pathway responsible for the cytoplasmic synthesis of acetyl-CoA from ethanol involves alcohol dehydrogenase 2, cytoplasmic aldehyde dehydrogenase 6, and acetyl-CoA synthase encoded by ADH2, ALD6 and ACS1, respectively. ADH2 and ACS1, and the genes encoding the two transcription factors, Adr1p and Cat8p, which regulate their expression [57] had higher transcript levels on xylose compared to the glucose repressed cells (Fig. 2, cluster 2). However, expression of ALD6 was lower on xylose compared with the glucose repressed and derepressed cells (Fig. 2, cluster 5), as was the abundance of two isoforms of Ald6p in the proteome analysis (Table 2) [Additional file 15]. ALD5 encoding a mitochondrial, NADPH-dependent isoform of acetaldehyde dehydrogenase [58], was up-regulated on xylose (Fig. 2, cluster 7). In the proteome analysis, Ald4p, another mitochondrial, NADPH-dependent isoform, had its highest abundance on xylose (Table 2) [Additional file 15]. The expression of glucose repressed ACH1, encoding a mitochondrial acetyl-CoA hydrolase [59], had higher transcription on xylose compared to the glucose repressed cells (Fig. 2, cluster 2), which was also reflected in the abundance of Ach1p (Table 2) [Additional file 15].
Distinct phosphorylation patterns of some major glycolytic enzymes on xylose and glucose
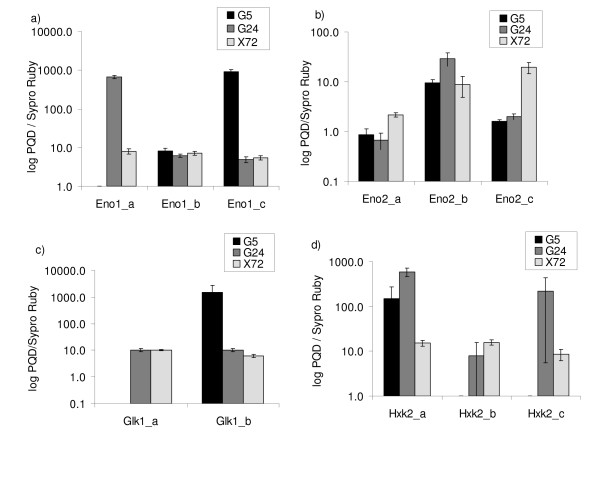
Interestingly, several glycolytic enzymes (i.e. Hxk2p, Pgk1p, Eno1p, Eno2p) had post-translationally modified forms with different abundances in the xylose-grown and glucose repressed and derepressed cells (Table 2) [Additional files 14, 15 and 17]. The fluorescent phosphoprotein-specific Pro-Q Diamond stain revealed reproducible, distinct patterns of phosphorylated forms of Hxk2p, Glk1p, Eno1p and Eno2p that were present in different relative quantities in the three different conditions studied (normalised to the total amount of each protein; Fig. 4) [Additional file 17]. Both Eno1p and Eno2p had three phosphorylated pI forms with different abundances in the xylose- and glucose grown cells (Fig. 4). The expression of ENO1 was expected to be repressed by glucose, but both ENO1 and ENO2 should be expressed during growth on non-fermentable carbon sources [49]. Two phosphorylated isoforms were identified for Glk1p and three for Hxk2p (Fig. 4). GLK1 should be expressed during growth on non-fermentable carbon sources and Hxk2p should be dominant during growth on glucose [55]. The expression of ENO1 and GLK1 did not significantly change between the three samples studied.
Figure 4.
Log-scale ratio of intensity of Pro-Q Diamond phosphoprotein stain to Sypro Ruby signal (total protein amount) of phosphorylated a) enolase 1, b) enolase 2, c) glucokinase, and d) hexokinase 2 protein isoforms identified from 2-DE gel analyses (a to c refer to different isoforms on gels). G5 = cells after 5 h on glucose; G24 = cells after 24 h on glucose; X72 = cells after 72 h on xylose.
Discussion
Transcription of a number of genes involved in the utilisation of alternative or non-fermentative carbon sources are repressed in the presence of glucose and derepressed in its absence [60]. Glucose sensing, signalling, and repression are thus closely linked to respirative and fermentative modes of metabolism. Accordingly, the way the cell recognises, senses, and signals the presence of xylose must affect the efficiency of its utilisation and fermentation. Previous studies of xylose-metabolising S. cerevisiae have suggested that xylose has a repressing effect on gene expression [21,61,62]. The results of the present study show that the expression of several genes repressed via the Snf1p/Mig1p glucose repression pathway i.e. genes encoding respiratory, TCA, glyoxylate cycle and gluconeogenic enzymes was lower during growth on xylose than in derepressed glucose-grown cells (24 h in Fig. 1). However, expression of these genes was higher than observed in glucose repressed cells (5 h in Fig. 1). On the other hand, some genes of the PPP and glycolysis and those typically expressed on non-fermentable carbon sources (e.g. HXK1, SUC2, PFK26 and some MAL-genes) showed increased expression in the cells growing on xylose. Thus, during xylose metabolism a mixed profile of gene expression was observed, suggesting that xylose does not activate all the glucose repression pathways in the same way as glucose.
The mechanisms of signal transduction by xylose are still unknown. It has been suggested that some hexose or triose phosphates derived from glycolysis [63,64] are involved in the regulation of gene expression, at least in lower glycolysis. The cellular concentration of hexose phosphates depends on the rate of glycolysis [65], but a correlation between the repression of genes and the concentration of hexose or triose phosphates on different carbon sources has not been shown [62]. When xylose is the main carbon source glycolytic flux to pyruvate is lower than for cells growing on glucose [14,66]. This may contribute to the observed higher expression of genes repressed via Snf1p/Mig1p in the xylose-grown cells, as Mig1p is expected to be totally dephosphorylated (and thus to repress its target genes) only at high glycolytic rates [67].
In recent studies, growth rate has been shown to affect the levels of transcripts, proteins and metabolites [68,69]. It could thus be argued that the intermediate expression level of many glucose repressed genes in the xylose-grown cells, compared with the glucose repressed and derepressed cells, is due to the different growth rate on xylose. Comparison of the present data with the data of growth rate regulated genes and proteins described in studies of Regenberg et al., 2006 and Castrillo et al., 2007, showed, however, that a majority (roughly 90%) of these genes and proteins (in clusters 1 and 2, Fig. 2) were with unknown function or associated with processes such as translation, RNA metabolism, ribosome biogenesis, amino acid metabolism, and stress, and not with the central pathways of carbon metabolism or glucose sensing and repression [68,69]. Additionally, in our previous chemostat studies, TCA cycle genes had a lower expression level on a mixture of xylose and glucose (27 and 3 g l-1, respectively) than under glucose limitation (10 g l-1) at the same specific growth rate suggesting that xylose had a repressive effect on the expression of these genes [21]. This supports the assumption that the lower growth rate is not the sole explanation for the only partial repression of the glucose repressed genes on xylose. The intermediate expression on xylose of respiratory, TCA and glyoxylate cycle genes was probably neither due to the lower concentration of ethanol on xylose compared with the glucose derepressed cells since Belinchón and Gancedo, (2003) showed that in a recombinant, xylose-metabolising S. cerevisiae strain, even in the presence of ethanol, xylose decreased the expression of ICL1 and FBP1 compared with derepressed, ethanol-grown cells [62]. Contradictorily, Jin et al., (2004), comparing the transcriptome of xylose-grown cells to that of glucose repressed cells, stated that xylose is not a repressive carbon source [17]. Their study, however, lacked the comparison of xylose-grown cells to fully derepressed cells, whereas comparison of the xylose-grown cells to the derepressed cells in the present study showed that xylose decreased the expression of several genes repressed by glucose via the Snf1p/Mig1p-pathway.
Yet another possibility for the intermediate expression in xylose-grown cells may be the way cAMP-PKA mediated pathways function on xylose. In the present study, over 50% of the genes repressible via this pathway [70] were expressed in xylose-grown cells at levels lower than those in glucose derepressed cells but above those in glucose repressed cells [Additional file 16]. Glucose is sensed by the receptor Gpr1p that initiates a signalling cascade leading to stimulation of fermentation. In the study of Rolland et. al., (2000) xylose (100 mM) was not sensed by Gpr1p [71]. In the present work, xylose was 210 mM at the time of sampling, and possibly even a higher concentration is needed for Gpr1p to sense this sugar. Furthermore, full activation of cAMP synthesis by glucose requires phosphorylation of glucose by Hxk1p, Hxk2p or Glk1p [71], which does not occur on xylose.
Glucose is also sensed at high and low concentrations, respectively by Rgt2p and Snf3p sensors. In glucose-grown cells the signal leads to induction of the HXT genes encoding glucose transporters by relieve of repression by Rgt1p through degradation of its co-repressors Mth1p and Std1p [43,72]. The Snf1p/Mig1p repression pathway contributes to glucose induction of HXT genes by repressing SNF3 and MTH1 [46]. The induction of MTH1 and only partial repression of SNF3 in the xylose-grown cells shows again that the glucose repression pathway via Snf1p/Mig1p does not operate to the same extent on xylose as it does on glucose. However, the expression profile of SNF3 and the highest expression of RGT2 in xylose-grown cells suggest that these sensors detect extracellular xylose.
Changes were observed in the expression of genes of acetaldehyde and acetyl-CoA metabolism. The increased expression of ACS1 and ALD5, and increased abundance of Ald4p suggest that acetaldehyde was utilised for growth instead of ethanol production during metabolism of xylose. The observed higher expression of ACH1, encoding the glucose repressed mitochondrial acetyl-CoA hydrolase, suggests that mitochondrial acetate concentration was perhaps elevated in xylose-grown cells compared with the glucose repressed cells. The changes may also reflect regulation of intracellular redox balance between the cytosol and mitochondria, since oxidation of acetaldehyde to acetate by Ald6p or Ald4/5p provides cytosolic and mitochondrial NADPH, respectively [73].
Jin et al., (2004) observed no change in the expression levels of genes encoding glycolytic enzymes in a xylose-utilising S. cerevisiae strain when xylose was provided as a carbon source instead of glucose in shake flask cultures [17]. In the present study, some of these genes had increased expression, and the proteome showed higher abundance of several glycolytic enzymes on xylose compared with either the glucose repressed or derepressed cells. Several recent studies suggest that regulation of glycolysis in S. cerevisiae occurs mainly at post-translational levels at least when aerobic and anaerobic conditions are compared, or ethanol and acetate are supplied instead of glucose [74-76]. Supporting these findings, the proteome analysis of the present study revealed that the relative abundance of different isoforms of some glycolytic enzymes correlated with the carbon source. Distinct patterns of phosphorylation of hexokinase 2, glucokinase and enolase in cells grown on xylose or glucose suggested that these enzymes were regulated differently by phosphorylation in the glucose repressed and derepressed cells, and in the cells grown on xylose. Apart from Hxk2p, pyruvate kinase (Pyk1/2p), and the regulatory Pfk26/27p [77,78] there is little information on the phosphorylation of glycolytic enzymes. It has been shown that Hxk2p is dephosphorylated on fermentable carbon sources, whereas both phosphorylated and dephosphorylated forms exist on poorly fermentable carbon sources [79]. The present data showed that only one of the three phosphorylated forms of Hxk2p was present in glucose repressed cells whereas the other two were present only in glucose derepressed cells and cells grown on xylose. This suggests that xylose was sensed rather as a non-fermentable carbon source. It is tempting to speculate that phosphorylation of also other glycolytic enzymes than the three kinases would be a yet poorly understood means to modulate the activity of glycolysis. However, phosphorylation may also relate to the role of these proteins in other cellular processes [80].
The present study shows that several genes that are subject to glucose repression were expressed during batch growth on xylose at levels intermediate between those in glucose repressed and derepressed cells. This suggests that cells growing on xylose do not have the appropriate expression levels of certain genes that may be one reason why they do not ferment xylose as fast as glucose. The results are consistent with the hypothesis that xylose does not activate all the glucose repression pathways in the same way as glucose, clearly indicating that this is an interesting area for further research towards improving xylose fermentation.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LS participated in the design of the study, performed the batch fermentations, RNA isolations and 2-D gel based proteome analyses, analysed the data and wrote the paper together with LR. MK carried out the statistical analysis of transcription and proteome data. RS carried out the mass spectrometric identification of proteins. JPP carried out the analysis presented in Figure 3 and commented the paper. MP participated in the design of the study. LR participated in the design of the study and wrote the paper together with LS. All authors have read and approved the final manuscript.
Supplementary Material
Pearson correlation coefficient values between the biological and technical replicate arrays from samples of glucose fermentations. The data provided shows the Pearson correlation coefficient values between the biological and technical replicate arrays of from samples of glucose fermentations (Glc5h and Glc24h).
Pearson correlation coefficient values between the biological and technical replicate arrays of samples from xylose fermentations. The data provided shows the Pearson correlation coefficient values between the biological and technical replicate arrays of xylose samples from fermentations (Xyl72h).
Scatterplots of RMA pre-processed arrays from cells grown on glucose for 5 h. The figure provided represents the scatterplots of the expression values of the replicate microarrays hybridised with the samples derived from cells grown on glucose for 5 h.
Scatterplots of RMA pre-processed arrays from cells grown on glucose for 24 h. The figure provided represents the scatterplots of the expression values of the replicate microarrays hybridised with the samples derived from cells grown on glucose for 24 h.
Scatterplots of RMA pre-processed arrays from cells grown on xylose for 72 h. The figure provided represents the scatterplots of the expression values of the replicate microarrays hybridised with the samples derived from cells grown on xylose for 72 h.
Cluster 1. List of open reading frames in cluster 1 shown in Fig. 2 of the paper.
Cluster 2. List of open reading frames in cluster 2 shown in Fig. 2 of the paper.
Cluster 3. List of open reading frames in cluster 3 shown in Fig. 2 of the paper.
Cluster 4. List of open reading frames of in cluster 4 shown in Fig. 2 of the paper.
Cluster 5. List of open reading frames in cluster 5 shown in Fig. 2 of the paper.
Cluster 6. List of open reading frames of in cluster 6 shown in Fig. 2 of the paper.
Cluster 7. List of open reading frames in cluster 7 shown in Fig. 2 of the paper.
Cluster 8. List of open reading frames in cluster 8 shown in Fig. 2 of the paper.
Image of the 11% SDS-PAGE 2-DE-gel. The image of the 2-DE-gel showing the locations of the seventy protein spots, which had different abundance in cells growing on glucose or xylose.
Clustering of the proteome data. The figure shows seventy proteins, which were differentially translated in the glucose repressed, glucose derepressed and xylose-grown cells and clustered by using hierarchical clustering with Euclidean distance and average linkage.
Expression profiles of genes involved in cyclic AMP – phosphokinase A pathway (cAMP-PKA) of S. cerevisiae. The figure provided shows the expression trend of genes involved in cyclic AMP – phosphokinase A pathway in cells grown on glucose for 5 or 24 h or on xylose for 72 h.
Images of 2-DE gels with phosphorylated proteins. Images of 2-DE gels showing the locations of Hxk2p, Glk1p, Eno2p and Eno1p in samples from cells grown for 72 h on xylose and for 5 h on glucose and stained either with phosphoprotein specific Pro-Q Diamond or Sypro Ruby.
Acknowledgments
Acknowledgements
We thank Marilyn Wiebe, Mervi Toivari and John Londesborough for fruitful discussions and valuable comments on the manuscript, Marc Baumann from the University of Helsinki for collaboration in the proteome analyses, and Eila Leino and Pirjo Tähtinen for excellent technical assistance. The microarray analyses were carried out at the Finnish DNA Microarray Centre at Turku Centre for Biotechnology. This work was financed by the Finnish Funding Agency for Technology and Innovation (Project number Tekes 40416/01). The work was part of the research programme "VTT Industrial Biotechnology" (Academy of Finland; Finnish Centre of Excellence programme, 2000–2005, Project number 64330).
Contributor Information
Laura Salusjärvi, Email: laura.salusjarvi@vtt.fi.
Matti Kankainen, Email: matti.kankainen@helsinki.fi.
Rabah Soliymani, Email: rabah.soliymani@helsinki.fi.
Juha-Pekka Pitkänen, Email: juha-pekka.pitkanen@vtt.fi.
Merja Penttilä, Email: merja.penttila@vtt.fi.
Laura Ruohonen, Email: laura.ruohonen@vtt.fi.
References
- Barnett JA, Payne RW, Yarrow D, Yeasts . Characteristics and identification. Cambridge, UK: Cambridge Univ Press; 1991. [Google Scholar]
- Attfield PV, Bell PJ. Use of population genetics to derive nonrecombinant Saccharomyces cerevisiae strains that grow using xylose as a sole carbon source. FEMS Yeast Res. 2006;6:862–868. doi: 10.1111/j.1567-1364.2006.00098.x. [DOI] [PubMed] [Google Scholar]
- van Maris AJ, Abbott DA, Bellissimi E, Brink J van den, Kuyper M, Luttik MA, Wisselink HW, Scheffers WA, van Dijken JP, Pronk JT. Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Antonie Van Leeuwenhoek. 2006;90:391–418. doi: 10.1007/s10482-006-9085-7. [DOI] [PubMed] [Google Scholar]
- Jeffries TW. Engineering yeasts for xylose metabolism. Curr Opin Biotechnol. 2006;17:320–326. doi: 10.1016/j.copbio.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Kötter P, Ciriacy M. Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1993;38:776–783. doi: 10.1007/BF00167144. [DOI] [Google Scholar]
- Walfridsson M, Anderlund M, Bao X, Hahn-Hägerdal B. Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilisation. Appl Microbiol Biotechnol. 1997;48:218–224. doi: 10.1007/s002530051041. [DOI] [PubMed] [Google Scholar]
- Eliasson A, Hofmeyr JH, Pedler S, Hahn-Hägerdal B. The xylose reductase/xylitol dehydrogenase/xylulokinase ratio affects product formation in recombinant xylose-utilising Saccharomyces cereviciae. Enzyme Microb Techno. 2001;29:288–297. doi: 10.1016/S0141-0229(01)00386-6. [DOI] [Google Scholar]
- Toivari MH, Aristidou A, Ruohonen L, Penttilä M. Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae: importance of xylulokinase (XKS1) and oxygen availability. Metab Eng. 2001;3:236–249. doi: 10.1006/mben.2000.0191. [DOI] [PubMed] [Google Scholar]
- Kuyper M, Winkler AA, van Dijken JP, Pronk JT. Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res. 2004;4:655–664. doi: 10.1016/j.femsyr.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Kuyper M, Hartog MM, Toirkens MJ, Almering MJ, Winkler AA, van Dijken JP, Pronk JT. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 2005;5:399–409. doi: 10.1016/j.femsyr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Kuyper M, Toirkens MJ, Diderich JA, Winkler AA, van Dijken JP, Pronk JT. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res. 2005;5:925–934. doi: 10.1016/j.femsyr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Gárdonyi M, Jeppsson M, Liden G, Gorwa-Grauslund MF, Hahn-Hägerdal B. Control of xylose consumption by xylose transport in recombinant Saccharomyces cerevisiae. Biotechnol Bioeng. 2003;82:818–824. doi: 10.1002/bit.10631. [DOI] [PubMed] [Google Scholar]
- Karhumaa K, Hahn-Hägerdal B, Gorwa-Grauslund MF. Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast. 2005;22:359–368. doi: 10.1002/yea.1216. [DOI] [PubMed] [Google Scholar]
- Pitkänen J-P, Aristidou A, Salusjärvi L, Ruohonen L, Penttilä M. Metabolic flux analysis of xylose metabolism in recombinant Saccharomyces cerevisiae using continuous culture. Metab Eng. 2003;5:16–31. doi: 10.1016/S1096-7176(02)00012-5. [DOI] [PubMed] [Google Scholar]
- Salusjärvi L, Poutanen M, Pitkänen J-P, Koivistoinen H, Aristidou A, Kalkkinen N, Ruohonen L, Penttilä M. Proteome analysis of recombinant xylose-fermenting Saccharomyces cerevisiae. Yeast. 2003;20:295–314. doi: 10.1002/yea.960. [DOI] [PubMed] [Google Scholar]
- Wahlbom CF, Cordero Otero RR, van Zyl WH, Hahn-Hägerdal B, Jonsson LJ. Molecular analysis of a Saccharomyces cerevisiae mutant with improved ability to utilize xylose shows enhanced expression of proteins involved in transport, initial xylose metabolism, and the pentose phosphate pathway. Appl Environ Microbio. 2003;69:740–746. doi: 10.1128/AEM.69.2.740-746.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YS, Laplaza JM, Jeffries TW. Saccharomyces cerevisiae engineered for xylose metabolism exhibits a respiratory response. Appl Environ Microbiol. 2004;70:6816–6825. doi: 10.1128/AEM.70.11.6816-6825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonderegger M, Jeppsson M, Hahn-Hägerdal B, Sauer U. Molecular basis for anaerobic growth of Saccharomyces cerevisiae on xylose, investigated by global gene expression and metabolic flux analysis. Appl Environ Microbio. 2004;70:2307–2317. doi: 10.1128/AEM.70.4.2307-2317.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotkjaer T, Christakopoulos P, Nielsen J, Olsson L. Comparative metabolic network analysis of two xylose fermenting recombinant Saccharomyces cerevisiae strains. Metab Eng. 2005;7:437–444. doi: 10.1016/j.ymben.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Pitkänen J-P, Rintala E, Aristidou A, Ruohonen L, Penttilä M. Xylose chemostat isolates of Saccharomyces cerevisiae show altered metabolite and enzyme levels compared with xylose, glucose, and ethanol metabolism of the original strain. Appl Microbiol Biotechnol. 2005;67:827–837. doi: 10.1007/s00253-004-1798-9. [DOI] [PubMed] [Google Scholar]
- Salusjärvi L, Pitkänen J-P, Aristidou A, Ruohonen L, Penttilä M. Transcription analysis of recombinant Saccharomyces cerevisiae reveals novel responses to xylose. Appl Biochem Biotechno. 2006;128:237–261. doi: 10.1385/ABAB:128:3:237. [DOI] [PubMed] [Google Scholar]
- Verho R, Londesborough J, Penttilä M, Richard P. Engineering redox cofactor regeneration for improved pentose fermentation in Saccharomyces cerevisiae. Appl Environ Microbiol. 2003;69:5892–5897. doi: 10.1128/AEM.69.10.5892-5897.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles E, Gohlmann HW, Zimmermann FK. Cloning of a second gene encoding 5-phosphofructo-2-kinase in yeast, and characterization of mutant strains without fructose-2,6-bisphosphate. Mol Microbiol. 1996;20:65–76. doi: 10.1111/j.1365-2958.1996.tb02489.x. [DOI] [PubMed] [Google Scholar]
- Ruohonen L, Aalto MK, Keränen S. Modifications to the ADH1 promoter of Saccharomyces cerevisiae for efficient production of heterologous proteins. J Biotechnol. 1995;39:193. doi: 10.1016/0168-1656(95)00024-K. [DOI] [PubMed] [Google Scholar]
- Richard P, Toivari MH, Penttilä M. The role of xylulokinase in Saccharomyces cerevisiae xylulose catabolism. FEMS Microbiol Lett. 2000;190:39–43. doi: 10.1111/j.1574-6968.2000.tb09259.x. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks JB. Methods in Yeast Genetics. A Laboratory Manual. Cold Springs Harbor, N.Y.: Cold Springs Harbor Laboratory; 1983. [Google Scholar]
- Carvalho B, Bengtsson H, Speed TP, Irizarry RA. Exploration, normalization, and genotype calls of high density oligonucleotide SNP array data. Biostatistics. 2007;8:485–485. doi: 10.1093/biostatistics/kxl042. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM 4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Zar JH. Biostatistical Analysis. 4. Prentice Hall, NJ; 1999. ANOVA – One-way Analysis of Variance. [Google Scholar]
- Soukas A, Cohen P, Socci ND, Friedman JM. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 2000;14:963–980. [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder – open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laboratory of Mass Spectrometry and Gaseous Ion Chemistry http://prowl.rockefeller.edu/prowl-cgi/profound.exe
- xProteinProspector http://prospector.ucsf.edu/mshome.htm
- MASCOT http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=PMF
- Hamacher T, Becker J, Gardonyi M, Hahn-Hägerdal B, Boles E. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology. 2002;148:2783–2788. doi: 10.1099/00221287-148-9-2783. [DOI] [PubMed] [Google Scholar]
- Saloheimo A, Rauta J, Stasyk OV, Sibirny AA, Penttilä M, Ruohonen L. Xylose transport studies with xylose-utilizing Saccharomyces cerevisiae strains expressing heterologous and homologous permeases. Appl Microbiol Biotechnol. 2007;74:1041–1052. doi: 10.1007/s00253-006-0747-1. [DOI] [PubMed] [Google Scholar]
- Liang H, Gaber RF. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol Biol Cell. 1996;7:1953–1966. doi: 10.1091/mbc.7.12.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S. Two different signals regulate repression and induction of gene expression by glucose. J Biol Chem. 2002;277:46993–46997. doi: 10.1074/jbc.M208726200. [DOI] [PubMed] [Google Scholar]
- Özcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Dover J, Rosenwald AG, Wolfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H, Johnston M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci USA. 2004;101:1572–1577. doi: 10.1073/pnas.0305901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomino A, Herrero P, Moreno F. Rgt1, a glucose sensing transcription factor, is required for transcriptional repression of the HXK2 gene in Saccharomyces cerevisiae. Biochem. 2005;388:697–703. doi: 10.1042/BJ20050160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Brachet V, Moriya H, Johnston M. Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:167–173. doi: 10.1128/EC.5.1.167-173.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard SL, Oliveira AP, Bro C, Olsson L, Nielsen J. A systems biology approach to study glucose repression in the yeast Saccharomyces cerevisiae. Biotechnol Bioeng. 2007;96:134–145. doi: 10.1002/bit.21135. [DOI] [PubMed] [Google Scholar]
- Johnston M. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 1999;15:29–33. doi: 10.1016/S0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- Entian KD, Meurer B, Kohler H, Mann KH, Mecke D. Studies on the regulation of enolases and compartmentation of cytosolic enzymes in Saccharomyces cerevisiae. Biochim Biophys Acta. 1987;923:214–221. doi: 10.1016/0304-4165(87)90006-7. [DOI] [PubMed] [Google Scholar]
- Young ET, Dombek KM, Tachibana C, Ideker T. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J Biol Chem. 2003;278:26146–26158. doi: 10.1074/jbc.M301981200. [DOI] [PubMed] [Google Scholar]
- Xue Y, Batlle M, Hirsch JP. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Winderickx J, Thevelein JM. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2002;2:183–201. doi: 10.1111/j.1567-1364.2002.tb00084.x. [DOI] [PubMed] [Google Scholar]
- Lutfiyya LL, Johnston M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol Cell Biol. 1996;16:4790–4797. doi: 10.1128/mcb.16.9.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CJ, Olsson L, Nielsen J. Glucose control in Saccharomyces cerevisiae: the role of Mig1 in metabolic functions. Microbiology. 1998;144:13–24. doi: 10.1099/00221287-144-1-13. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, De La Cera T, Herrero P, Moreno F. The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem J. 2001;355:625–631. doi: 10.1042/bj3550625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney RR, Rubenstein EM, Schmidt MC. Snf1 kinase complexes with different beta subunits display stress-dependent preferences for the three Snf1-activating kinases. Curr Genet. 2005;47:335–344. doi: 10.1007/s00294-005-0576-2. [DOI] [PubMed] [Google Scholar]
- Walther K, Schüller HJ. Adr1 and Cat8 synergistically activate the glucose-regulated alcohol dehydrogenase gene ADH2 of the yeast Saccharomyces cerevisiae. Microbiology. 2001;147:2037–2044. doi: 10.1099/00221287-147-8-2037. [DOI] [PubMed] [Google Scholar]
- Saint-Prix F, Bonquist L, Dequin S. Functional analysis of the ALD gene family of Saccharomyces cerevisiae during anaerobic growth on glucose: the NADP+-dependent Ald6p and Ald5p isoforms play a major role in acetate formation. Microbiology. 2004;150:2209–2220. doi: 10.1099/mic.0.26999-0. [DOI] [PubMed] [Google Scholar]
- Buu LM, Chen YC, Lee FJ. Functional characterization and localization of acetyl-CoA hydrolase, Ach1p, in Saccharomyces cerevisiae. J Biol Chem. 2003;278:17203–17209. doi: 10.1074/jbc.M213268200. [DOI] [PubMed] [Google Scholar]
- Gancedo JM. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca C, Haack MB, Olsson L. Engineering of carbon catabolite repression in recombinant xylose fermenting Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2004;63:578–583. doi: 10.1007/s00253-003-1408-2. [DOI] [PubMed] [Google Scholar]
- Belinchón MM, Gancedo JM. Xylose and some non-sugar carbon sources cause catabolite repression in Saccharomyces cerevisiae. Archiv Microbiol. 2003;180:293–297. doi: 10.1007/s00203-003-0593-9. [DOI] [PubMed] [Google Scholar]
- Boles E, Zimmermann FK. Induction of pyruvate decarboxylase in glycolysis mutants of Saccharomyces cerevisiae correlates with the concentrations of three-carbon glycolytic metabolites. Arch Microbiol. 1993;160:324–328. doi: 10.1007/BF00292085. [DOI] [PubMed] [Google Scholar]
- Vincent O, Townley R, Kuchin S, Carlson M. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 2001;15:1104–1114. doi: 10.1101/gad.879301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch D, Johansson M, Ferndahl C, Franzén CJ, Larsson C, Gustafsson L. Characterization of glucose transport mutants of Saccharomyces cerevisiae during a nutritional upshift reveals a correlation between metabolite levels and glycolytic flux. FEMS Yeast Res. 2008;8:10–25. doi: 10.1111/j.1567-1364.2007.00323.x. [DOI] [PubMed] [Google Scholar]
- Wahlbom CF, Eliasson A, Hahn-Hägerdal B. Intracellular fluxes in a recombinant xylose-utilizing Saccharomyces cerevisiae cultivated anaerobically at different dilution rates and feed concentrations. Biotechnol Bioeng. 2001;72:289–296. doi: 10.1002/1097-0290(20010205)72:3<289::AID-BIT5>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Elbing K, Stahlberg A, Hohmann S, Gustafsson L. Transcriptional responses to glucose at different glycolytic rates in Saccharomyces cerevisiae. Eur J Biochem. 2004;271:4855–4864. doi: 10.1111/j.1432-1033.2004.04451.x. [DOI] [PubMed] [Google Scholar]
- Regenberg B, Grotkjaer T, Winther O, Fausboll A, Åkesson M, Bro C, Hansen LK, Brunak S, Nielsen J. Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol. 2006;7:R107. doi: 10.1186/gb-2006-7-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo JI, Zeef LA, Hoyle DC, Zhang N, Hayes A, Gardner DC, Cornell MJ, Petty J, Hakes L, Wardleworth L, Rash B, Brown M, Dunn WB, Broadhurst D, O'Donoghue K, Hester SS, Dunkley TP, Hart SR, Swainston N, Li P, Gaskell SJ, Paton NW, Lilley KS, Kell DB, Oliver SG. Growth control of the eukaryote cell: a systems biology study in yeast. J Biol. 2007;6:4. doi: 10.1186/jbiol54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, Hogan JD, Cusick ME, Davis BP, Fancher AM, Hodges PE, Kondu P, Lengieza C, Lew-Smith JE, Lingner C, Roberg-Perez KJ, Tillberg M, Brooks JE, Garrels JI. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, De Winde JH, Lemaire K, Boles E, Thevelein JM, Winderickx J. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol Microbiol. 2000;38:348–358. doi: 10.1046/j.1365-2958.2000.02125.x. [DOI] [PubMed] [Google Scholar]
- Kim JH, Johnston M. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J Biol Chem. 2006;281:26144–26149. doi: 10.1074/jbc.M603636200. [DOI] [PubMed] [Google Scholar]
- Minard KI, McAlister-Henn L. Sources of NADPH in yeast vary with carbon source. J Biol Chem. 2005;280:39890–39896. doi: 10.1074/jbc.M509461200. [DOI] [PubMed] [Google Scholar]
- Wiebe MG, Rintala E, Tamminen A, Simolin H, Salusjarvi L, Toivari M, Kokkonen JT, Kiuru J, Ketola RA, Jouhten P, Huuskonen A, Maaheimo H, Ruohonen L, Penttilä M. Central carbon metabolism of Saccharomyces cerevisiae in anaerobic, oxygen-limited and fully aerobic steady-state conditions and following a shift to anaerobic conditions. FEMS Yeast Res. 2008;8:140–154. doi: 10.1111/j.1567-1364.2007.00234.x. [DOI] [PubMed] [Google Scholar]
- de Groot MJ, Daran-Lapujade P, van Breukelen B, Knijnenburg TA, de Hulster EA, Reinders MJ, Pronk JT, Heck AJ, Slijper M. Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post-transcriptional regulation of key cellular processes. Microbiology. 2007;153:3864–3878. doi: 10.1099/mic.0.2007/009969-0. [DOI] [PubMed] [Google Scholar]
- Daran-Lapujade P, Jansen MLA, Daran JM, van Gulik WM, de Winde JH, Pronk JT. Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiae. A chemostat culture study. J Biol Chem. 2004;5:9125–9138. doi: 10.1074/jbc.M309578200. [DOI] [PubMed] [Google Scholar]
- Portela P, Howell S, Moreno S, Rossi S. In vivo and in vitro phosphorylation of two isoforms of yeast pyruvate kinase by protein kinase A. J Biol Chem. 2002;277:30477–30487. doi: 10.1074/jbc.M201094200. [DOI] [PubMed] [Google Scholar]
- Goncalves PM, Griffioen G, Bebelman JP, Planta RJ. Signalling pathways leading to transcriptional regulation of genes involved in the activation of glycolysis in yeast. Mol Microbiol. 1997;25:483–493. doi: 10.1046/j.1365-2958.1997.4811847.x. [DOI] [PubMed] [Google Scholar]
- Randez-Gil F, Herrero P, Sanz P, Prieto JA, Moreno F. Hexokinase PII has a double cytosolic-nuclear localisation in Saccharomyces cerevisiae. FEBS Lett. 1998;425:475–478. doi: 10.1016/S0014-5793(98)00289-0. [DOI] [PubMed] [Google Scholar]
- Gancedo C, Flores CL. Moonlighting proteins in yeasts. Microbiol Mol Biol Rev. 2008;72:197–210. doi: 10.1128/MMBR.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeast Proteome Database http://www.proteome.com
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pearson correlation coefficient values between the biological and technical replicate arrays from samples of glucose fermentations. The data provided shows the Pearson correlation coefficient values between the biological and technical replicate arrays of from samples of glucose fermentations (Glc5h and Glc24h).
Pearson correlation coefficient values between the biological and technical replicate arrays of samples from xylose fermentations. The data provided shows the Pearson correlation coefficient values between the biological and technical replicate arrays of xylose samples from fermentations (Xyl72h).
Scatterplots of RMA pre-processed arrays from cells grown on glucose for 5 h. The figure provided represents the scatterplots of the expression values of the replicate microarrays hybridised with the samples derived from cells grown on glucose for 5 h.
Scatterplots of RMA pre-processed arrays from cells grown on glucose for 24 h. The figure provided represents the scatterplots of the expression values of the replicate microarrays hybridised with the samples derived from cells grown on glucose for 24 h.
Scatterplots of RMA pre-processed arrays from cells grown on xylose for 72 h. The figure provided represents the scatterplots of the expression values of the replicate microarrays hybridised with the samples derived from cells grown on xylose for 72 h.
Cluster 1. List of open reading frames in cluster 1 shown in Fig. 2 of the paper.
Cluster 2. List of open reading frames in cluster 2 shown in Fig. 2 of the paper.
Cluster 3. List of open reading frames in cluster 3 shown in Fig. 2 of the paper.
Cluster 4. List of open reading frames of in cluster 4 shown in Fig. 2 of the paper.
Cluster 5. List of open reading frames in cluster 5 shown in Fig. 2 of the paper.
Cluster 6. List of open reading frames of in cluster 6 shown in Fig. 2 of the paper.
Cluster 7. List of open reading frames in cluster 7 shown in Fig. 2 of the paper.
Cluster 8. List of open reading frames in cluster 8 shown in Fig. 2 of the paper.
Image of the 11% SDS-PAGE 2-DE-gel. The image of the 2-DE-gel showing the locations of the seventy protein spots, which had different abundance in cells growing on glucose or xylose.
Clustering of the proteome data. The figure shows seventy proteins, which were differentially translated in the glucose repressed, glucose derepressed and xylose-grown cells and clustered by using hierarchical clustering with Euclidean distance and average linkage.
Expression profiles of genes involved in cyclic AMP – phosphokinase A pathway (cAMP-PKA) of S. cerevisiae. The figure provided shows the expression trend of genes involved in cyclic AMP – phosphokinase A pathway in cells grown on glucose for 5 or 24 h or on xylose for 72 h.
Images of 2-DE gels with phosphorylated proteins. Images of 2-DE gels showing the locations of Hxk2p, Glk1p, Eno2p and Eno1p in samples from cells grown for 72 h on xylose and for 5 h on glucose and stained either with phosphoprotein specific Pro-Q Diamond or Sypro Ruby.