Abstract
Rice functional genomics is a scientific approach that seeks to identify and define the function of rice genes, and uncover when and how genes work together to produce phenotypic traits. Rapid progress in rice genome sequencing has facilitated research in rice functional genomics in China. The Ministry of Science and Technology of China has funded two major rice functional genomics research programmes for building up the infrastructures of the functional genomics study such as developing rice functional genomics tools and resources. The programmes were also aimed at cloning and functional analyses of a number of genes controlling important agronomic traits from rice. National and international collaborations on rice functional genomics study are accelerating rice gene discovery and application.
Keywords: rice, Oryza sativa, functional genomics, agronomic traits, genome sequencing, microarray
1. Introduction
Rice is the principal food for over half of the population of the world. With a compact genome spanning approximately 430 megabase (Mb) pairs, an extensive genetic map (Harushima et al. 1998) and established synteny with other cereal crops (Gale & Devos 1998), the cultivated rice species Oryza sativa represents a model for cereals as well as other monocot plants (Shimamoto & Kyozuka 2002). The completion of the genome sequence of rice, in December 2004, opens a new and exciting chapter in our quest to functionally characterize all of the approximately 50 000 annotated genes in rice (Feng et al. 2002; Sasaki et al. 2002; Rice Chromosome 10 Sequencing Consortium 2003). A systematic approach to characterizing these genes will allow us to dissect and understand the regulatory networks and evolutionary selection controlling such complex traits as yield, grain quality, biotic and abiotic stresses, reproductive barriers, epigenetics and flowering time. The next essential steps towards deciphering the sequenced genome are to develop complete and accurate maps of actively transcribed regions during rice development, and to generate more and more genome-wide rice mutant resources. These will facilitate the identification of all the genes and proteins encoded in the DNA sequence. Such information will allow further analysis of their function, regulation and how they cooperate in complex biological processes in a systematic manner.
In China, the Ministry of Science and Technology has funded two rice functional genomics programmes since 1999 under the National Basic Sciences Initiatives (Xue & Xu 2002; Xue et al. 2003) and the ‘863’ High-Tech Development Plan. The 863 programme was started in 2002. The Chinese Rice Functional Genomics Programme is composed of three parts: (i) development of technical platforms, (ii) functional genomics of agriculturally important traits, and (iii) molecular cloning and functional analysis of agronomic genes. The platforms are aimed at enabling high-throughput analyses and effective determination of gene functions, which consist of three major components: generation and characterization of a large mutant library; expression profiling of the predicted exons and expressed sequence tags (ESTs) of the entire genome; and isolation of full-length complementary DNAs (cDNAs). The traits targeted for functional genomics studies in this programme include grain quality, yield potential, architecture, stress tolerance, disease and insect resistance, and nutrient efficiency, which are also the traits targeted for gene isolation. The organized national consortium for rice functional genomics research has been established in China. This consortium includes a number of institutes and universities in Beijing, Shanghai, Wuhan, Hangzhou, Guangzhou and Yangzhou. Here, we report the progress that has been achieved in the last several years in these programmes.
2. Rapid progress in rice genome sequencing
Domesticated Asian rice, O. sativa, which consists of two major subspecies indica and japonica partially isolated in terms of sexual reproduction, has become the most important rice crop in the world (Goff et al. 2002; Linares 2002). Indica–japonica genome comparison has been chosen as a model system for understanding the origin, speciation, domestication and genome evolution of rice. Indica and japonica cultivars can be classified based on their agronomic traits (Oka & Morishima 1997) and the use of molecular markers (Glaszmann 1987; Cheng et al. 2003). Rapid progress in both indica and japonica genome sequencing has been made in China (Yu et al. 2002, 2005). Here, we describe the completed sequence of rice chromosome 4 and whole-genome shotgun sequencing of an indica variety 93-11.
(a) Rice Oryza sativa japonica chromosome 4 sequencing as part of international efforts to sequence the entire genome
The International Rice Genome Sequencing Project (IRGSP) has adopted the clone-by-clone approach for obtaining a finished rice genome sequence, because it is modular, allows efficient gap filling, avoids problems arising from distant repetitive sequences and results in the early completion of larger contiguous segments of a genome (Sasaki & Burr 2000). As part of international efforts to completely sequence the entire rice genome, the National Center for Gene Research (NCGR) of the Chinese Academy of Sciences has finished the sequence of rice japonica chromosome 4 through a map-based strategy (Feng et al. 2002; Zhao et al. 2002; IRGSP 2005). This was one of the first two rice chromosomes to have been completely sequenced. The finished sequence spans 34.6 Mb and represents 97.3% of the chromosome. To characterize the blueprint of intraspecific DNA-sequence variations between indica and japonica subspecies, NCGR has conducted an indica–japonica chromosome 4 comparison at the nucleotide sequence level. A 22.1 Mb of the indica Guangluai 4 (GLA4) chromosome 4 has been sequenced, the largest map-based high-quality indica sequence, and compared with 23.2 Mb of the japonica orthologous chromosome 4 (Han et al. 2007 unpublished data). This comparison provided a fine-scale indica–japonica comparison. A sequence comparison of the collinear regions from indica and japonica chromosome 4 has revealed not only their extensive micro-collinearity in gene order and content, but also the intraspecific sequence polymorphisms in both coding and non-coding regions (Feng et al. 2002; Han & Xue 2003; Li et al. 2004). Comparative genome analysis between cultivated rice subspecies shows that there is an overall syntenic relationship between the chromosomes and the divergence at the level of single-nucleotide polymorphisms (SNPs) and insertions and deletions. The rice genome has been well mapped both genetically and physically and has a syntenic relationship with other cereals. Identification of the sequence polymorphisms of the two subspecies has produced the greatest density of polymorphic sequence maps of the indica and japonica chromosome 4. Importantly, this comparison revealed the dynamic changes that may reflect major evolutionary events occurring throughout the domestication and natural selection of rice.
A complete sequence of a chromosome centromere is necessary to fully understand centromere function. The rice chromosome 4 centromere sequence has been completely determined and its structures have been analysed. Complete sequencing of the 124 kb rice chromosome 4 centromere revealed that it consisted of 18 tracts of 379 tandemly arrayed repeats known as CentO and a total of 19 centromeric retroelements (CRs), but no unique sequences were detected. The preferential insert of the CRs among CentO repeats indicated that the centromere-specific retroelements may contribute to centromere expansion during evolution. The presence of three intact retrotransposons in the centromere suggests that they may be responsible for functional centromere initiation through a transcription-mediated mechanism (Zhang et al. 2004).
(b) Whole-genome shotgun sequencing of indica 93-11
A whole-genome shotgun sequencing approach was carried out for sequencing an indica variety 93-11 genome by the Beijing Genomics Institute of the Chinese Academy of Sciences (Yu et al. 2002, 2005). The improved whole-genome shotgun sequences for the genomes of indica and japonica rice, both with multimegabase contiguity, gave an almost 1000-fold improvement over the drafts of 2002 (Yu et al. 2005). Tested against a non-redundant collection of 19 079 full-length cDNAs, 97.7% of the genes are aligned, without fragmentation, to the mapped superscaffolds of one or the other genome. Yu et al. (2005) introduced a gene identification procedure for plants that do not rely on similarity to known genes to remove erroneous predictions resulting from transposable elements. At least a quarter of the two sequences could not be aligned, and where they could be aligned, SNP rates varied from as little as 3.0 SNP kb−1 in the coding regions to 27.6 SNP kb−1 in the transposable elements. These results have revealed ancient whole-genome duplication, a recent segmental duplication on chromosomes 11 and 12, and massive ongoing individual gene duplications. More importantly, ongoing individual gene duplications provide a never-ending source of raw materials for gene genesis and are major contributors to the differences between members of the grass family.
3. Infrastructures of rice functional genomics
(a) Rice chips
As a first attempt to decipher the rice genome, computational annotation has been successful, although improvements are needed (Yuan et al. 2003). Recent efforts to verify experimentally the gene model structure by sequencing cDNA and ESTs have provided valuable information towards our understanding of gene structure and genome-coding capacity (Wu et al. 2002; Rice Full-Length cDNA Consortium 2003; Rensink & Buell 2004). An essential and necessary step in this effort is the determination of the coding information and expression patterns of each sequenced chromosome. However, until now only about half of the predicted genome-coding capacity had any cDNA or EST expression support. Clearly, experimental approaches complementary to computation-based genome annotation are essential for an understanding of genome structures. Owing to the presence of large amounts of unfinished sequence data, unusual compositional gradients in genes and the large size of the rice genome (Rensink & Buell 2004), there is even greater need for experimental approaches in rice genome annotation.
(i) Rice chromosome 4 tilling chip: chromosomal level regulation
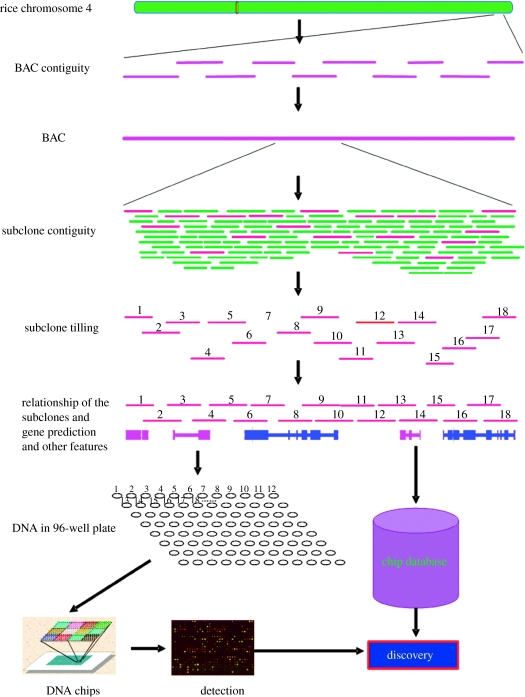
A chromosome-scale transcriptional analysis will expand our knowledge of possible chromosomal level transcriptional regulation. One prominent feature of eukaryotic chromosomes is their organization into heterochromatic and euchromatic regions. Heterochromatin was first distinguished from euchromatin cytologically as more intensely staining nuclear material throughout the cell cycle in Bryophyta. In fact, heterochromatin has emerged as a key regulator in the epigenetic control of gene expression, chromosome behaviour and evolution. Using tilling path microarray analysis as a tool, it is now possible to perform high-throughput profiling of the transcriptional activities along an entire sequenced chromosome to examine potential connections between transcription and cytologically defined chromatin organization. Xing-Wang Deng's and Bin Han's laboratories have developed a tilling path DNA microarray consisting of overlapping PCR-amplified genomic fragments covering over 33 Mb (95.5%) of japonica rice chromosome 4 (figure 1). Using this array, they analysed the transcriptional activity of chromosome 4 in six representative rice chromosome 4 transcriptome organs or tissues. Chromosomal-scale transcription patterns were analysed in comparison with cytologically observed chromatin organization and the distribution of transposon-related and various other gene model groups (Jiao et al. 2005). Six representative rice organ types were examined using this microarray to catalogue the transcribed regions of rice chromosome 4 and to reveal organ- and developmental stage-specific transcription patterns. This analysis provided expression support for 82% of the gene models in the chromosome. Transcriptional activities in 1643 non-annotated regions were also detected. Comparison with cytologically defined chromatin features indicated that in juvenile stage rice the euchromatic region is more actively transcribed than the transposon-rich heterochromatic portion of the chromosome. Interestingly, elevated transcription of transposon-related gene models in certain heterochromatic regions was observed in mature stage rice organs and in suspension cultured cells. These results suggest a close correlation between transcriptional activity and chromosome organization and developmental regulation of transcription activity at the chromosomal level (Jiao et al. 2005; Li et al. 2005).
Figure 1.
Diagram of rice chromosome 4 tilling chips. BAC, bacterial artificial chromosome.
(ii) Rice cDNA chips
A 10 K cDNA microarray was developed and used in an analysis of genetic programmes controlling pollination (fertilization and stress responses) in rice (Lan et al. 2004, 2005). Yongbiao Xue and his colleagues have identified 253 cDNAs that are regulated by pollination/fertilization in rice by using this cDNA microarray. To investigate this relationship, they obtained the expression profiles after dehydration and wounding treatments in this study. Venn diagram analysis indicated that 53.8% (136/253) and 21% (57/253) of the pollination/fertilization-related genes are indeed regulated by dehydration and wounding, respectively, and nearly half of the genes expressed preferentially in unpollinated pistils (UP) are responsive to dehydration. These results indicated that an extensive gene set is shared among these responses, suggesting that the genetic programmes regulating them are probably related. Among them, the genetic network of the water stress control may be a key player in pollination and fertilization. Additionally, 39.5% (100/253) cDNAs that are related to pollination/fertilization appear not to be regulated by the stress treatments (dehydration and wounding), suggesting that the existence of additional genetic networks is involved in pollination/fertilization. Furthermore, comparative analysis of the expression profiles of the 253 cDNAs under 18 different conditions (various tissues, treatments and developmental status) revealed that the genetic networks regulating photosynthesis, starch metabolisms, gibberelin and defence responses are involved in pollination and fertilization. Taken together, these results provided some clues to elucidate the molecular mechanisms of pollination and fertilization in rice.
(iii) Development of a rice whole-genome oligonucleotide microarray
With the complete genome sequence available, it is possible to design a microarray for expression profiling of all known and predicted genes in the rice genome. For this purpose, a rice 70-mer oligonucleotide microarray was designed during the period October–December 2002, in collaboration with the Beijing Genome Institute and Peking–Yale Joint Center of Plant Molecular Genetics and Agrobiotechnology. The oligonucleotide set was based on a combination of FGENESH-predicted gene models from an indica rice draft sequence (Yu et al. 2002) and the available 28 444 full-length cDNAs (Rice Full-Length cDNA Consortium 2003), 15 558 public EST-derived unigenes (Rensink & Buell 2004) and 31 776 ESTs derived from indica rice (Zhao et al. 2004) at the time. The following two criteria were used to remove redundancy. First, for gene models overlapping with cDNAs with open reading frame (ORF), cDNA sequences instead of predicted gene models were used for oligonucleotides' design. Second, FGENESH-predicted gene models were used with or without partially overlapping EST supports. In total, a set of 61 123 non-redundant protein-coding genes or gene models were generated and used for oligonucleotide design. The design of oligonucleotide optimized cross-hybridization, melting temperature and GC-content minimized the potential of hairpin stem formation and biased for 3′ ends of the coding region of annotated genes (Wang et al. 2003). A total of 58 404 oligonucleotides were designed and custom synthesized. After the recent release of the improved indica rice genome sequence and corresponding annotation (Yu et al. 2005), we aligned oligonucleotides to updated rice gene annotation. This oligonucleotide set matched 41 754 known and predicted gene models (excluding transposon-related models), including those located in the physically unassembled rice DNA sequences (Yu et al. 2005). Among this group, 70-mer oligonucleotides for 4622 gene models could match another one or more gene models with a 70% identity threshold and have potential cross-hybridization. Therefore, this 70-mer oligonucleotide set covers 37 132 known and predicted genes of the most current version of the indica genome (Yu et al. 2005) with one-to-one specificity. Oligonucleotides' annotation information is hosted on BGI-RIS databases (http://rise.genomics.org.cn). A further alignment of these 37 132 unique oligos to the Syngenta O. sativa L. ssp. japonica (Goff et al. 2002) and the IRGSP O. sativa L. ssp. japonica (http://rgp.dna.affrc.go.jp/IRGSP/Build2/build2.html) sequences indicated that 92% (34 248) of them matched to the O. sativa L. ssp. japonica genome sequences.
(iv) Unbiased transcriptional analysis of rice genome using high-density oligonucleotide tilling microarray
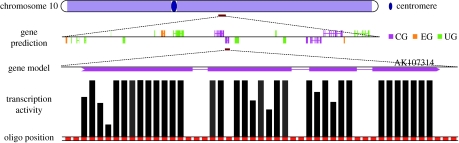
Recent completion of both indica and japonica genome sequence analysis now allowed an unbiased interrogation of transcriptional activity of the whole genome and experimental verification of computational genome annotation. The Maskless Array Synthesizer (MAS) platform was chosen for a high-density oligonucleotide microarray analysis of the complete genomic sequences. Xing-Wang Deng and his colleagues interrogated the non-repetitive sequence of the indica and japonica genomes using sets of 34 or 32 MAS microarrays probed by labelled cDNA from representative tissues to detect transcriptional activity without any presumption of gene structures. In a pilot test, one of the arrays containing approximately 390 000 individual probes representing a portion of the last 11.2 Mb sequence of chromosome 10 was analysed in detail based on a mathematical model developed (Stolc et al. 2005). More recently, a comparative tilling analysis of the entire chromosome 10 of both indica and japonica genomes has been completed (figure 2; Li et al. 2005). This analysis detected expression of about three quarters of the unsupported gene models without previous experimental evidence in both subspecies. This tilling analysis also identified 549 new models for the japonica chromosome, representing an 18% increase in the annotated protein-coding capacity. This analysis also revealed that a chromosomal level transcriptional regulation largely coincides with the heterochromatin and euchromatin domains.
Figure 2.
A transcriptional analysis of the entire rice chromosome 10 genome using high-density oligonucleotide tilling microarray.
(b) Development of a T-DNA tagging system for functional analysis of the rice genome
Reverse genetics aims to determine the functions of genes. One of the best strategies for reverse genetics is based on insertional mutagenesis using T-DNA or transposon as a mutagen (Ito et al. 2002). As a powerful strategy for functional genomics study, T-DNA tagging was widely used in many species. The main advantages of using T-DNA tags are that T-DNA insertions are stable and their copy number is low (Delseny et al. 2001). Several strategies such as inverse PCR and thermal asymmetric interlaced-PCR (TAIL-PCR) have been used to obtain the flanking sequences of T-DNA or transposons inserted in a plant genome. With their flanking sequences, T-DNA tags can be mapped in silico to the chromosomes to reveal the distribution pattern in the tagged genome (Chen et al. 2003b; Yang et al. 2004).
The Chinese Rice Functional Genomics Programme aimed at a T-DNA insertion mutant library of 200 000 independent transformants. The T-DNA vector adopted three strategies for analysing the rice genome: insertional mutation, enhancer trapping and ectopic expression (Wu et al. 2003). Three major groups in Beijing, Shanghai and Wuhan have been involved in this work, and collectively these groups have generated approximately 270 000 independent transformants using a japonica cultivar Zhonghua 11. With an average of 2.1 copies of T-DNA insertions per genome, this amounts to a total of over 500 000 independent insertions, which is 10 times the predicted 50 000 genes. A total of 20 000 flanking sequences have been isolated currently. Large-scale screening of mutations is in progress for agronomical important traits including stress tolerance/resistance (rice blast, bacterial blight, temperature, drought and salinity), grain quality, plant architecture, fertility, tillering, nutrition efficiency, photosynthesis, etc. (figure 3). High-throughput platform for quality assays via infrared, in which amount of starch, amylase, protein and fat could be quantitatively measured, has been established. Expression profiling of the enhancer trap lines has also been conducted by detecting the reporter gene expression in various tissues of the lines in the mutant library. Such screening has produced a large number of mutants that are now tagged for gene isolation and lines with specific expression patterns for identification of regulatory elements such as promoters and enhancers. A centralized database (http://rmd.ncpgr.cn) has been developed for this mutant library (Zhang et al. 2006).
Figure 3.
Phenotypes of the various mutants observed in the T-DNA insertion library during field planting (photos provided by Changyin Wu's group): (a) albino seedling, (b) semi-dwarf, fewer tillers with branches on the culm, (c) softer stems, (d) semi-dwarf and later flowering, (e) bushy tillers, (f) more sensitive to cold stress, (g) smaller number of grains per panicle, (h) change in panicle morphology, (i) thinner grains and (j) thicker and shorter grains.
(c) Full-length cDNA cloning
Full-length cDNAs are essential for the correct annotation of genomic sequences and the functional analysis of genes at both transcriptional and translational levels. Although over 28 000 japonica full-length cDNAs have been identified (Rice Full-Length cDNA Consortium 2003), more cDNAs are needed to be cloned since there are approximately 50 000 genes predicted in rice. Additional indica cDNA clones may be particularly useful for comparative analysis between indica and japonica subspecies, improvement of genome sequence annotation and identification of lineage-specific genes because indica-specific genes have been found (Zhang et al. 2005). The NCGR has built-up full-length cDNA libraries from O. sativa indica Guangluai 4 for large-scale collection of indica rice full-length expression sequences. Now, five full-length cDNA libraries have been constructed from the germinated seed, seedling, root, filling, etiolated seedling and plants grown under various stresses, respectively (X. Liu et al. 2006, unpublished data). The 5′-end ESTs of 100 000 cDNAs have been clustered into 16 000 non-redundant cDNA groups. The 3′ end of the 16 000 non-redundant cDNA is under sequencing. First, the highest quality RNAs were prepared in their full-length form, not in truncated form. Second, Superscript II reverse transcriptase, which has a good rate of processing activity, was used in cDNA synthesis in order to obtain cDNAs in their full-length forms, but not in truncated forms. Full-length mRNA and cDNA were selected by the 5′ caps. Ligation, transformation and propagation are more efficient for shorter cDNAs than longer ones, thus introducing size bias in cDNA library generation. In our approach, duplex cDNA was size fractioned. Different cDNAs with size ranges greater than 2 kb, 1–2 kb, 0.5–1 kb and 250–500 bp were ligated to the cloning vector, respectively, in order to reduce size bias and improve clone representation. Each full-length cDNA library from plants at various developmental stages and from various plant tissues contains over 106 primary non-amplified clones. Over 90% of the clones are full-length cDNAs containing the complete protein-coding region as well as the 5′- and 3′-untranslated regions (UTRs). The promoter sequences can be obtained by comparing the 5′-end sequences of cDNAs with the rice genome sequences.
In addition, 10 828 full-length cDNAs from another indica cultivar Minghui 63 have been characterized (Xie et al. 2005). Comparison with the databases showed that 780 of them are new rice cDNAs with no match in japonica cDNA database. Totally, 9078 of the full-length cDNAs contained predicted ORFs matching with japonica full-length cDNAs, with 6543 finding homologous proteins. Moreover, 53% of the matched full-length cDNAs had longer 5′ UTR than the japonica homologues. In silico mapping showed that 9776 (90.28%) of them had matched genomic sequences in the japonica genome and 10 046 (92.78%) had matched genomic sequences in the indica genome. The average nucleotide sequence identity between the two subspecies is 99.2%. More than 60% of the new cDNAs isolated in this study had no homology to known proteins.
Depending on the expression of the cellular mRNA, mRNA can be defined as abundant, intermediate and rare. In a typical cell, 5–10 species of abundant mRNA comprise at least 20% of the mass mRNA, 500–2000 species of intermediately expressed mRNA comprise 40–60% of the mass mRNA and 10 000–20 000 rare mRNA may account for less than 20–40% of the mRNA mass. Sequencing cDNAs from standard cDNA libraries is ineffective for discovering rarely expressed genes, when highly and intermediately expressed cDNAs would be sequenced redundantly. Normalization and subtraction procedures were introduced into the construction of the full-length cDNA libraries to reduce the frequency of highly expressed mRNA in the library and to enhance the discovery of new cDNAs. Subtraction procedures were based on hybridization of the first-strand cDNA with RNA drivers from highly expressed cDNAs and previously sequenced cDNAs or DNA primers designed from already known cDNA of japonica. Large-scale new gene discovery was accelerated by using these procedures.
4. Cloning and functional identification of genes of agronomic significance
Cloning and functional analysis of the agronomic important traits including plant architecture, stress tolerance/resistance (bacteria blight and salinity), grain quality, fertility and nutrition efficiency are important targets for China Rice Functional Genomics Programmes. Here, we highlight recent progresses in these studies.
(a) A gene controlling rice tillering
Tillering in rice is an important agronomic trait for grain production, and also a model system for the study of branching in monocotyledonous plants. Rice tiller is a specialized grain-bearing branch that is formed on the unelongated basal internodes and grows independently of the mother stem (culm) by means of its own adventitious roots. Rice tillering occurs in a two-stage process: the formation of an axillary bud at each leaf axil and its subsequent outgrowth. Although the morphology and histology and some mutants of rice tillering have been well described, the molecular mechanism of rice tillering remains to be elucidated. Jiayang Li and his colleagues reported the isolation and characterization of MONOCULM 1 (MOC1), a gene that is important in the control of rice tillering (Li et al. 2003a,b). The moc1 mutant plants have only a main culm without any tillers owing to a defect in the formation of tiller buds. MOC1 encodes a putative GRAS family nuclear protein that is expressed mainly in the axillary buds and functions to initiate the axillary buds and promote their outgrowth. To identify genes involved in the control of rice tillering, they have screened for mutants with altered tiller numbers from collections derived from spontaneous mutations or γ-ray radiation and ethyl methanesulphonate (EMS) mutagenesis. A spontaneous moc1 mutant is of particular interest, because moc1 plants nearly completely lose their tillering ability, producing only one main culm, in contrast to the multiple tillers in wild-type plants. Genetic analysis with reciprocal crosses between moc1 and wild-type plants revealed that moc1 possesses a recessive mutation in a single nuclear locus. Allelic tests between the moc1 mutant and five recessive tillering mutants with reduced culm number (rcn1–rcn5) indicated that MOC1 is a previously unknown locus that is involved in the control of rice tillering.
(b) Cloning of GS3, a major quantitative trait locus (QTL) for grain length and weight and a minor QTL for grain width and thickness
Grain size is a major determinant of grain weight, one of the three components (number of panicles per plant, number of grains per panicle and grain weight) of grain yield. Grain size is also a highly important quality trait in rice (figure 4). Although the preference for rice grain characteristics varies with consumer groups, long and slender grain is generally preferred for indica rice by the majority of consumers in China, the United States and most Asian countries. For example, a length/width ratio of 2.8 is adopted as an enforced threshold for a national quality standard of indica rice in China (http://www.knowledgebank.irri.org/regionalSites/china/10_CodesStandards/default.htm). Thus, understanding the genetic and molecular basis of grain size is extremely important for rice improvement programmes.
Figure 4.
Grains of six cultivars, showing great diversity in grain length, width, thickness and overall size.
Many QTLs associated with rice grain size were identified in the last decade. Among them, a QTL with major effect on grain size was consistently detected around the centromeric region of chromosome 3 in numerous studies across different genetic backgrounds and environments (Yu et al. 1997; Tan et al. 2000; Xing et al. 2002). Thus, this gene, referred to as GS3 hereafter, can be very useful for improvement of both yield and quality in rice breeding programmes.
Fan et al. (2006) isolated GS3 by map-based cloning. Near isogenic lines of GS3 were developed by successive crossing and backcrossing of Minghui 63 (large grain) with Chuan 7 (small grain), using Minghui 63 as the recurrent parent. Analysis of a random population of 201 individuals from the BC3F2 progeny confirmed that the GS3 locus explained 80–90% of the variation for grain weight and length in this population. In addition, this locus was resolved as a minor QTL for grain width and thickness. Using 1384 individuals with recessive phenotype (large grain) from a total of 5740 BC3F2 plants and 11 molecular markers based on sequence information, GS3 was mapped to a DNA fragment approximately 7.9 kb in length. A full-length cDNA corresponding to the target region was identified, which provided complete sequence information for the GS3 candidate. This gene consists of five exons and encodes 232 amino acids with a putative PEBP-like domain, a transmembrane region, a putative TNFR/NGFR family cysteine-rich domain and a VWFC module. Comparative sequencing analysis identified a nonsense mutation, shared among all the large-grain varieties tested in comparison with the small grain varieties, in the second exon of the putative GS3 gene. This mutation causes a 178-amino acid truncation in the C-terminus of the predicted protein, suggesting that GS3 may function as a negative regulator for grain size. Cloning of such a gene provided the opportunity for fully characterizing the regulatory mechanism and related processes during grain development.
(c) Control of rice root formation
Adventitious root formation in rice is involved in the whole development of rice root. Ping Wu and his colleagues identified a novel rice mutant, arl1 (adventitious rootless1), which completely lacks adventitious root during the whole developmental period (Liu et al. 2005). They have done cross-sections at the base of the stem of 5-day-old seedlings and found that formation of adventitious primordium is impaired in the arl1 mutant. Both α-naphthylacetic acid and ethephon at the optimal concentrations to promote adventitious root growth cannot rescue the defect of arl1. They cloned and demonstrated that the ARL1 gene was responsible for the arl1 mutant phenotype. Subcellular localization of ARL1 suggests that ARL1 is a nuclear protein. Yeast two-hybrid experiments suggest that as a nucleic protein, ARL1 may homodimerize and activate reporter gene. In addition to the characterization of the expression pattern of ARL1 gene, the response of ARL1 gene expression to different hormones was also investigated in this study (Liu et al. 2005).
(d) Genes related to rice mechanical strength
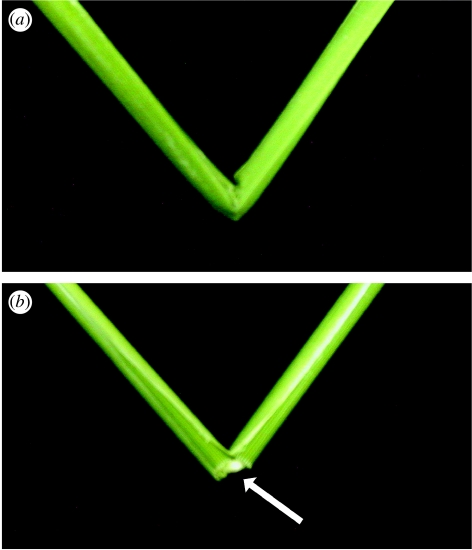
Plant mechanical strength is an important agronomic trait. To understand the molecular mechanism that controls the plant mechanical strength of crops, Jiayang Li and his colleagues characterized the classic rice mutant brittle culm1 (bc1) and isolated BC1 using a map-based cloning approach (figure 5). BC1, which encodes a COBRA-like protein, is expressed mainly in developing sclerenchyma cells and in vascular bundles of rice. In these types of cells, mutations in BC1 cause not only a reduction in cell wall thickness and cellulose content, but also an increase in lignin level, suggesting that BC1, a gene that controls the mechanical strength of monocots, plays an important role in the biosynthesis of the cell walls of mechanical tissues. The plant cell wall, a strong fibrillar network that provides mechanical support to cells, tissues and the entire plant body, is a highly organized composite that may contain many different polysaccharides, aromatic substances and proteins. The structure and composition of plant cell walls are ideally suited to the functions they perform. For example, parenchyma cells, which consist of primary walls, provide the main structural support in growing regions of the plant body. Sclerenchyma cells, which have both primary walls and thick secondary walls, provide the major mechanical support in non-elongating regions of the plant body (Carpita & McCann 2000). Cellulose usually constitutes 20–30% of the dry weight of the primary walls and 40–90% of the secondary walls, depending on the cell type (Taylor et al. 1999). In some cells, lignin may be incorporated into the cell wall, enhancing its mechanical strength. Despite extensive descriptions of the chemical and physical structures of cell walls, the mechanisms that regulate the deposition of cell wall materials and that determine cell wall strength remain to be elucidated. To understand the mechanisms that regulate the mechanical strength of the plant body and the biosynthesis of plant cell walls, mutants defective in stem strength have been isolated and characterized.
Figure 5.
Phenotypes and physical properties of wild-type and bc1-2 mutant rice plants. (a) A wild-type culm. (b) An easily broken bc1-2 culm, as indicated by the arrow.
(e) Rice bacterial blight resistance gene
Rice bacterial blight, caused by Xanthomonas oryzae pv. oryzae (Xoo), is one of the most serious rice diseases worldwide. It is not only widespread throughout Asia, but is also reported to occur in Australia, the United States and several rice-growing countries of Africa and Latin America. Yield loss due to the disease ranges from 2 to 74%. A rice gene, Xa26, conferring resistance against Xoo at both seedling and adult stages was isolated from the rice cultivar Minghui 63 using a map-based cloning approach by Shiping Wang's group (Yang et al. 2003; Sun et al. 2004). Xa26 encodes a leucine-rich repeat receptor kinase-like protein consisting of 1103 amino acids. The resistance conferred by Xa26 is influenced by the genetic background such that transgenic plants with a Mudanjiang 8 background carrying the Xa26 gene had broader spectrum and higher level of resistance than the donor parent. The function of Xa26 is also dosage dependent. Overexpression of Xa26 could enlarge the resistance spectrum of rice to different Xoo strains. Sequence analysis showed that Xa26 belongs to a multigene family. Another rice gene, Xa4 for bacterial blight resistance, is likely to be a member of the Xa26 family. Comparative sequence analysis of four rice varieties from the two subspecies of cultivated rice, indica and japonica, showed that the Xa26 family consists of 4–12 members in different rice haplotypes. This family is formed by tandem duplication followed by diversification through recombination, deletion and point mutation during evolution. The haplotype polymorphism and positive selection for diversification of the Xa26 family provide sequence reservoir and evolutionary force to generate rapid variation for novel resistance specificities.
(f) Functional analysis of a rice cytoplasmic male sterility/restorer system
Cytoplasmic male sterility (CMS) is a widespread phenomenon observed in over 150 flowering plant species (Laser & Lersten 1972). CMS is a maternally inherited trait and often associated with unusual ORFs in the mitochondrial genomes; in many instances, the male fertility of CMS plants can be restored by specific nuclear genes, called fertility restorer (Rf) genes (Schnable & Wise 1998). Therefore, the CMS–restoration interaction represents a striking example of the genetic interaction and cooperative function of the mitochondrial and nuclear genomes in plants. CMS and nucleus-controlled fertility restoration are widespread plant reproductive features which provide useful tools to exploit hybrid vigour in agriculture. In rice, several CMS/restorer systems defined by the different origins of CMS cytoplasm with distinct genetic features have been identified, including CMS-BT (Boro II), CMS-WA (wild abortive) and CMS-HL (Honglian; Zhu 2000). These systems have been widely used for hybrid rice in China and other Asian countries since the latter half of the 1970s. Hybrid rice usually has yields 20–30% higher than inbred varieties, greatly contributing to the increase in rice production (Li & Yuan 2000). Recently, four Rf genes have been cloned from rice (Komori et al. 2004). However, very few CMS gene candidates have been functionally tested. The rice male sterile BT-cytoplasm has been primarily identified in a variety, Chinsurash Boro II, of the indica subspecies and transferred into a number of japonica and indica varieties by backcrossing. The mitochondrial genome of the BT-cytoplasm contains two duplicated copies of the atp6 gene encoding a subunit of the ATPase complex, N-atp6 and B-atp6, and they are transcribed constitutively to produce different lengths of mRNAs, of which B-atp6 mRNA has an additional sequence downstream of the atp6-coding sequence (Iwabuchi et al. 1993). In the downstream sequence of B-atp6, an ORF, orf79, was predicted (Akagi et al. 1994). The roles of B-atp6 and orf79 in the CMS and the mechanistic connection between the Rf gene's function and the fertility restoration are unclear, although it was proposed that the editing extents of B-atp6 mRNA or lower levels of the antisense transcripts might be related to the CMS and restoration.
Yaoguang Liu and his colleagues identified that, in the BT-type of CMS rice, an abnormal mitochondrial ORF, orf79, which is cotranscribed with a duplicated copy of atp6 (B-atp6), encodes a cytotoxic peptide and confers gametophytic CMS (Wang et al. in press). Two fertility restorer genes, Rf1a and Rf1b, are identified at the classical locus Rf-1 as members of a multigene cluster encoding pentatricopeptide repeat (PPR) proteins. The restorer genes function to restore the male fertility by silencing orf79 via either endonucleolytic cleavage or degradation of the dicistronic B-atp6/orf79 mRNA. They further demonstrated that Rf1a has a role in promoting the RNA editing of atp6 independent of the cleavage activity. The results revealed a novel binary post-transcriptional controlling mechanism in plant CMS/restorer systems and have general implications on the function of PPR proteins. Mitochondrial genomes encode only a fraction of the genetic information required for their biogenesis and function. Consequently, a large number of genetic and biochemical features displayed in plant mitochondria arose in the context of coevolution and coordinated gene functions between the mitochondrial and nuclear genomes (Wang et al. 2006).
(g) Waxy gene and its regulation
Starch is the major constituent and energy reserve in many important foods, including the seeds of cereal grains and the potato tuber. Starch consists of two types of polysaccharides: amylose and amylopectin. Amylose makes up 0–30% of the starch in the cereal grains. The amylose content varies widely in different plant species and even in different cultivars of same species. Studies on starch mutants of maize, rice and barley have shown that the enzyme granule-bound starch synthase, encoded by the Waxy (Wx) gene, catalyses the synthesis of amylose. Mengmin Hong and Zongyang Wang have identified that Wx transcripts could be detected only in the endosperm, pollen and embryo sac, indicating that the expression of the Wx gene is probably regulated at the level of transcription. They previously cloned and sequenced the Wx gene of rice and demonstrated that the sequences within the first intron of the gene enhance its expression. Additional studies revealed that post-transcriptional events play important roles in regulating the concentrations of Wx protein and the amylose in rice seeds. It was also demonstrated that a sequence (860–640) upstream from the rice Wx gene is a positive regulator of its expression. The results of electrophoretic mobility shift assays showed that a 31 bp nucleotide sequence (840–810) located within the 860–640 region is the binding site for nuclear proteins isolated from developing rice seeds. Furthermore, experiments performed on transgenic rice plants containing a 3-glucuronidase (GUS) reporter gene controlled by the Wx promoter with the 31 bp sequence present synthesized 2–3 times the amount of GUS as plants carrying the reporter gene and a Wx promoter with the 31 bp sequence deleted. These results indicated that the 31 bp sequence plays an important role in the transcriptional regulation of the rice Wx gene (Chen et al. 2003a; Yang et al. 2002; Zhu et al. 2003).
(h) Genes involved in tolerance to phosphate starvation in rice
Ping Wu reported that a novel transcription factor was involved in tolerance to phosphate (Pi) starvation in rice (Yi et al. 2005). This transcription factor contains a bHLH domain and was designated as OsPTF1. The expression of OsPTF1 is phosphate (Pi) starvation induced in roots while it is constitutively expressed in shoots. Transformation of promoter : GUS fusion construct shows that OsPTF1 is expressed in phloem cells of primary root and in all cells of lateral roots. Overexpression of OsPTF1 enhanced tolerance to Pi starvation in transgenic rice. Tillering ability, root and shoot biomass, and P content of transgenic rice plants were approximately 30% higher than those of the wild-type plants under Pi-deficient conditions in the hydroponic experiment. In soil pot and field experiments, approximately 20% increase in tiller number, panicle weight and P content was observed in transgenic plants compared with the wild-type plants under Pi-deficient conditions. Under Pi-deficient conditions, transgenic rice plants showed significantly higher instantaneous Pi uptake rate over their wild-type counterparts. Microarray result demonstrates that OsPTF1 is involved in the efficient Pi recycling under Pi-deficient conditions, which is important in alleviating phosphorus deficiency in rice under Pi-starvation conditions.
(i) Mapping and map-based cloning of QTL for rice salt tolerance
Soil salinity is a major abiotic stress in crop productivity worldwide (Zhu 2001). It is estimated that saline soils cover from 400 to 950 million hectares of Earth's surface. Improving the salt tolerance in rice is one of the most important objectives of rice breeding programmes in coastal areas. To facilitate the development of new varieties with high level of salinity tolerance, understanding of the genetic control mechanisms for salt tolerance is required. Salt tolerance is a complex trait controlled by QTLs and is the final manifestation of several components, such as Na+ uptake, K+ uptake, ions balance, ions compartmentation, etc. To map QTLs for rice salt tolerance, we constructed an F2 and an equivalent F3 population derived from a cross between a high salt tolerance indica variety, Nona Bokra, and a susceptible elite japonica variety, Koshihikari (Lin et al. 2004). Eight QTLs responsible for the variations in their K+ or Na+ content under salt stress were mapped. Out of these QTLs, a major QTL, SKC1, that explained 40% of the phenotypic variation in shoot K+ content under salt stress was mapped on chromosome 1. To understand the molecular basis for QTL related to ion transport or salt tolerance, we cloned the SKC1 by using advanced backcross progeny and map-based cloning. It showed that SKC1 encodes an ion transporter (Ren et al. 2005). The complementation test of the candidate SKC1 gene was successful (figure 6). Physiological analysis suggested that SKC1 is involved in the long-distance K+/Na+ transport (from the root to the shoot) thereby regulating K+/Na+ homeostasis in the shoots under salt stress, suggesting a potential gene for improving salt tolerance in rice.
Figure 6.
NIL (SKC1) seedlings (right) are more tolerant to salt than Koshihikari (left) under salt stress (125 mM NaCl for 32 days). It indicated that the QTL SKC1 is involved in salt tolerance.
Trehalose is a non-reducing disaccharide of glucose that functions as a compatible solute in the stabilization of biological structures under abiotic stress in bacteria, fungi and invertebrates. In plants, it also plays important roles in the development and tolerance against various abiotic stresses. Trehalose-6-phosphate phosphatase catalyses the final step in the trehalose synthesis pathway, thereby contributing much to abiotic stress tolerance. We cloned a novel gene for coding trehalose-6-phosphate phosphatase (OsTPPA), which is transiently and remarkably induced immediately after 140 mM NaCl treatment (Chao et al. 2005). Analysing cDNA and genomic sequences of OsTPPA shows that it contains 12 exons, and an initial codon is located in the third exon. Microarray, RT-PCR and northern blotting results show that OsTPPA was rapidly upregulated under salt, drought and cold stress in roots and shoots of the salt tolerance variety. To further study its functions and exploit its application potential, we constructed 35S-OsTPPA-sense and 35S-OsTPPA-RNAi vectors and transferred them into rice. Salt tolerance of T1 progeny containing 35S-OsTPPA-sense transgene significantly increased compared with the vector control under salt stress. Drought and cold tolerance of T1 progeny containing 35S-OsTPPA-RNAi transgene significantly reduced compared with the vector control. These results indicated that OsTPPA plays important roles in rice abiotic stress tolerance.
5. Conclusion
Rice has become an important model flowering plant for studying many aspects of plant biology. The completion of the rice genome has afforded an unprecedented opportunity for systematic studies of plant gene function. The rice genome sequence provides a complete catalogue of genes that are important for improving not only rice but also other cereals, as functionally important sequences are conserved and may be identified by their similarity. The high-throughput tools and approaches have been developed and used in rice functional genomics research. National and international collaborations (Fischer et al. 2000) on rice functional genomics study have accelerated the gene discovery and sharing of the rice genomic resources. There is no doubt that rice functional genomics research will impact on development of a sustainable agriculture in China and worldwide.
Acknowledgments
The China Rice Functional Genomics Programmes are funded by the Ministry of Science and Technology of China (G19990116600, 2002AA2Z1001, 2002AA2Z1002 and 2002AA2Z1003), and are also supported by the Chinese Academy of Sciences (038019315). We thank Mengmin Hong, Hongxuan Lin, Ping Wu, Zongyang Yaoguang Liu, Chengcai Chu, Wang, Shiping Wang, Tiegang Lu, Hongwei Xue, Yuezhi Tao and Qian Qian for their assistance in preparation of this manuscript and for communicating their unpublished results.
Footnotes
One contribution of 14 to a Theme Issue ‘Biological science in China’.
References
- Akagi H, Sakamoto M, Shinjyo C, Shimada H, Fujimura T. A unique sequence located downstream from the rice mitochondrial apt6 may cause male sterility. Curr. Genet. 1994;25:52–58. doi: 10.1007/BF00712968. doi:10.1007/BF00712968 [DOI] [PubMed] [Google Scholar]
- Carpita N, McCann M. The cell wall. In: Buchanan B.B, Gruissem W, Jones R.L, editors. Biochemistry and molecular biology of plants. American Society of Plant Physiologists; Rockville, MD: 2000. pp. 52–108. [Google Scholar]
- Chao D.Y, Luo Y.H, Shi M, Luo D, Lin H.X. Salt-responsive genes in rice revealed by cDNA microarray analysis. Cell Res. 2005;15:796–810. doi: 10.1038/sj.cr.7290349. doi:10.1038/sj.cr.7290349 [DOI] [PubMed] [Google Scholar]
- Chen J, Tang W, Hong M, Min. Wang Z.Y. OsBP-73, a rice gene, encodes a novel DNA-binding protein with a SAP-like domain and its genetic interference by double-stranded RNA inhibits rice growth. Plant Mol. Biol. 2003a;52:579–590. doi: 10.1023/a:1024854101965. doi:10.1023/A:1024854101965 [DOI] [PubMed] [Google Scholar]
- Chen S, Jin W, Wang M, Zhang F, Zhou J, Jia Q, Wu Y, Liu F, Wu P. Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J. 2003b;36:105–113. doi: 10.1046/j.1365-313x.2003.01860.x. doi:10.1046/j.1365-313X.2003.01860.x [DOI] [PubMed] [Google Scholar]
- Cheng C, Motohashi R, Tsuchimoto S, Fukuta Y, Ohtsubo H, Ohtsubo E. Polyphyletic origin of cultivated rice: based on the interspersion pattern of SINEs. Mol. Biol. Evol. 2003;20:67–75. doi: 10.1093/molbev/msg004. doi:10.1093/molbev/msg004 [DOI] [PubMed] [Google Scholar]
- Delseny M, et al. Rice genomics: present and future. Plant Physiol. Biochem. 2001;39:323–334. doi:10.1016/S0981-9428(01)01245-1 [Google Scholar]
- Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Zhang Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. doi:10.1007/s00122-006-0218-1 [DOI] [PubMed] [Google Scholar]
- Feng Q, et al. Sequence and analysis of rice chromosome 4. Nature. 2002;420:316–320. doi: 10.1038/nature01183. doi:10.1038/nature01183 [DOI] [PubMed] [Google Scholar]
- Fischer K.S, Barton J, Khush G.S, Leung H, Cantrell R. Collaborations in rice. Science. 2000;290:279–280. doi: 10.1126/science.290.5490.279. doi:10.1126/science.290.5490.279 [DOI] [PubMed] [Google Scholar]
- Gale M.D, Devos K.M. Comparative genetics in the grasses. Proc. Natl Acad. Sci. USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. doi:10.1073/pnas.95.5.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaszmann J.C. Isozymes and classification of Asian rice varieties. Theor. Appl. Genet. 1987;74:21–30. doi: 10.1007/BF00290078. doi:10.1007/BF00290078 [DOI] [PubMed] [Google Scholar]
- Goff S.A, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. doi:10.1126/science.1068275 [DOI] [PubMed] [Google Scholar]
- Han B, Xue Y. Genome-wide intraspecific DNA-sequence variations in rice. Curr. Opin. Plant Biol. 2003;6:134–138. doi: 10.1016/s1369-5266(03)00004-9. doi:10.1016/S1369-5266(03)00004-9 [DOI] [PubMed] [Google Scholar]
- Harushima Y, et al. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. doi:10.1038/nature03895 [DOI] [PubMed] [Google Scholar]
- Ito T, Motohashi R, Kuromori T, Mizukado S, Sakurai T, Kanahara H, Seki M, Shinozaki K. A new resource of locally transposed dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome. Plant Physiol. 2002;129:1695–1699. doi: 10.1104/pp.002774. doi:10.1104/pp.002774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi M, Kyozuka J, Shimamoto K. Processing followed by complete editing of an altered motochondrial apt6 RNA restores fertility of cytoplasmic male sterile rice. EMBO J. 1993;12:1437–1446. doi: 10.1002/j.1460-2075.1993.tb05787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, et al. A tiling microarray expression analysis of rice chromosome 4 suggests a chromosomal level regulation of transcription. Plant Cell. 2005;17:1641–1657. doi: 10.1105/tpc.105.031575. doi:10.1105/tpc.105.031575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Ohta S, Murai N, Takakura Y, Kuraya Y, Suzuki S, Hiei Y, Imaseki H, Nitta N. Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.) Plant J. 2004;37:315–325. doi: 10.1046/j.1365-313x.2003.01961.x. doi:10.1046/j.1365-313X.2003.01961.x [DOI] [PubMed] [Google Scholar]
- Lan L, et al. Monitoring of gene expression profiles and isolation of candidate genes involved in pollination and fertilization in rice (Oryza sativa L.) with a 10 K cDNA microarray. Plant Mol. Biol. 2004;54:471–487. doi: 10.1023/B:PLAN.0000038254.58491.c7. doi:10.1023/B:PLAN.0000038254.58491.c7 [DOI] [PubMed] [Google Scholar]
- Lan L, Li M, Lai Y, Xu W, Kong Z, Ying K, Han B, Xue Y. Microarray analysis reveals similarities and variations in genetic programs controlling pollination/fertilization and stress responses in rice (Oryza sativa L.) Plant Mol. Biol. 2005;59:151–164. doi: 10.1007/s11103-005-3958-4. doi:10.1007/s11103-005-3958-4 [DOI] [PubMed] [Google Scholar]
- Laser K.D, Lersten N.R. Anatomy and cytology of microsporogenesis in cytoplasmic male sterile angiosperms. Bot. Rev. 1972;38:425–454. [Google Scholar]
- Li J.M, Yuan L.P. Hybrid rice: genetics, breeding, and seed production. Plant Breeding Rev. 2000;17:15–158. [Google Scholar]
- Li X, et al. Control of tillering in rice. Nature. 2003a;422:618–621. doi: 10.1038/nature01518. doi:10.1038/nature01518 [DOI] [PubMed] [Google Scholar]
- Li Y, et al. BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell. 2003b;15:2020–2031. doi: 10.1105/tpc.011775. doi:10.1105/tpc.011775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhang Y, Ying K, Liang X, Han B. Sequence variations of simple sequence repeats on chromosome-4 in two subspecies of the Asian cultivated rice. Theor. Appl. Genet. 2004;108:392–400. doi: 10.1007/s00122-003-1457-z. doi:10.1007/s00122-003-1457-z [DOI] [PubMed] [Google Scholar]
- Li L, et al. Tiling microarray analysis of rice chromosome 10 to identify the transcriptome and relate its expression to chromosomal architecture. Genome Biol. 2005;6:R52. doi: 10.1186/gb-2005-6-6-r52. doi:10.1186/gb-2005-6-6-r52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.X, et al. QTLs for Na+, K+ uptake controlling rice salt tolerance of shoot and root. Theor. Appl. Genet. 2004;108:253–260. doi: 10.1007/s00122-003-1421-y. doi:10.1007/s00122-003-1421-y [DOI] [PubMed] [Google Scholar]
- Linares O.F. African rice (Oryza glaberrima): history and future potential. Proc. Natl Acad. Sci. USA. 2002;99:16 360–16 365. doi: 10.1073/pnas.252604599. doi:10.1073/pnas.252604599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang S, Yu X, Yu J, He X, Zhang S, Shou H, Wu P. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005;43:47–56. doi: 10.1111/j.1365-313X.2005.02434.x. doi:10.1111/j.1365-313X.2005.02434.x [DOI] [PubMed] [Google Scholar]
- Oka H, Morishima H. Wild and cultivated rice. In: Matsuo T, Futsuhara Y, Kikushi F, Yamaguchi H, editors. Science of the rice plant. Genetics. vol. 3. Food and Agriculture Policy Research Center; Tokyo, Japan: 1997. pp. 88–111. [Google Scholar]
- Ren Z.H, et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005;37:1141–1146. doi: 10.1038/ng1643. doi:10.1038/ng1643 [DOI] [PubMed] [Google Scholar]
- Rensink W.A, Buell C.R. Arabidopsis to rice. Applying knowledge from a weed to enhance our understanding of a crop species. Plant Physiol. 2004;135:622–629. doi: 10.1104/pp.104.040170. doi:10.1104/pp.104.040170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Chromosome 10 Sequencing Consortium. In-depth view of structure, activity, and evolution of rice chromosome 10. Science. 2003;300:1566–1669. doi: 10.1126/science.1083523. doi:10.1126/science.1083523 [DOI] [PubMed] [Google Scholar]
- Rice Full-Length cDNA Consortium. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. doi:10.1126/science.1081288 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Burr B. International rice genome sequencing project: the effort to completely sequence the rice genome. Curr. Opin. Plant Biol. 2000;3:138–141. doi: 10.1016/s1369-5266(99)00047-3. doi:10.1016/S1369-5266(99)00047-3 [DOI] [PubMed] [Google Scholar]
- Sasaki T, et al. The genome sequence and structure of rice chromosome 1. Nature. 2002;420:312–316. doi: 10.1038/nature01184. doi:10.1038/nature01184 [DOI] [PubMed] [Google Scholar]
- Schnable P.S, Wise R.P. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 1998;3:175–180. doi:10.1016/S1360-1385(98)01235-7 [Google Scholar]
- Shimamoto K, Kyozuka J. Rice as a model for comparative genomics of plants. Annu. Rev. Plant Biol. 2002;53:399–419. doi: 10.1146/annurev.arplant.53.092401.134447. doi:10.1146/annurev.arplant.53.092401.134447 [DOI] [PubMed] [Google Scholar]
- Stolc V, et al. A pilot study of transcription unit analysis in rice using oligonucleotide tiling-path microarray. Plant Mol. Biol. 2005;59:137–149. doi: 10.1007/s11103-005-6164-5. doi:10.1007/s11103-005-6164-5 [DOI] [PubMed] [Google Scholar]
- Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes a LRR receptor kinase-like protein. Plant J. 2004;37:517–527. doi: 10.1046/j.1365-313x.2003.01976.x. doi:10.1046/j.1365-313X.2003.01976.x [DOI] [PubMed] [Google Scholar]
- Tan Y, Xing Y, Li J, Yu S, Xu C, Zhang Q. Genetic bases of appearance quality of rice grains in Shanyou 63, an elite rice hybrid. Theor. Appl. Genet. 2000;101:823–829. doi: 10.1007/s001220051279. doi:10.1007/s001220051549 [DOI] [PubMed] [Google Scholar]
- Taylor N.G, Scheible W, Cutler S, Somerville C.R, Turner S.R. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell. 1999;11:769–779. doi: 10.1105/tpc.11.5.769. doi:10.1105/tpc.11.5.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.Y, Malek R.L, Kwitek A.E, Greene A.S, Luu T.V, Behbahani B, Frank B, Quackenbush J, Lee N.H. Assessing unmodified 70-mer oligonucleotide probe performance on glass-slide microarrays. Genome Biol. 2003;4:R5. doi: 10.1186/gb-2003-4-1-r5. doi:10.1186/gb-2003-4-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell. 2006;18:676–687. doi: 10.1105/tpc.105.038240. doi:10.1105/tpc.105.038240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, et al. A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell. 2002;14:525–535. doi: 10.1105/tpc.010274. doi:10.1105/tpc.010274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, et al. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 2003;35:418–427. doi: 10.1046/j.1365-313x.2003.01808.x. doi:10.1046/j.1365-313X.2003.01808.x [DOI] [PubMed] [Google Scholar]
- Xie K, Zhang J, Xiang Y, Feng Q, Han B, Chu Z, Wang S, Zhang Q, Xiong L. Isolation and annotation of 10,828 putative full length cDNAs from indica rice. Science in China (ser C) 2005;35:6–12. doi: 10.1360/062004-90. [DOI] [PubMed] [Google Scholar]
- Xing Z, Tan Y, Hua J, Sun X, Xu C, Zhang Q. Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor. Appl. Genet. 2002;105:248–257. doi: 10.1007/s00122-002-0952-y. doi:10.1007/s00122-002-0952-y [DOI] [PubMed] [Google Scholar]
- Xue Y, Xu Z. An introduction to the China rice functional genomics program. Comp. Funct. Genomics. 2002;3:161–163. doi: 10.1002/cfg.147. doi:10.1002/cfg.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Li J, Xu Z. Recent highlights of the China rice functional genomics program. Trends Genet. 2003;19:390–394. doi: 10.1016/S0168-9525(03)00141-0. doi:10.1016/S0168-9525(03)00141-0 [DOI] [PubMed] [Google Scholar]
- Yang H, Shen H, Chen L, Xing Y.Y, Wang Z.Y, Zhang J.L, Hong M.M. The OsEBP-89 gene of rice encodes a putative EREBP transcription factor and is temporally expressed in developing endosperm and intercalary. Plant Mol. Biol. 2002;50:379–391. doi: 10.1023/a:1019859612791. doi:10.1023/A:1019859612791 [DOI] [PubMed] [Google Scholar]
- Yang Z, Sun X, Wang S, Zhang Q. Genetic and physical mapping of a new gene for bacterial blight resistance in rice. Theor. Appl. Genet. 2003;106:1467–1472. doi: 10.1007/s00122-003-1205-4. [DOI] [PubMed] [Google Scholar]
- Yang Y, Peng H, Huang H, Wu J, Jia S, Huang D, Lu T. Large-scale production of enhancer trapping lines for rice functional genomics. Plant Sci. 2004;167:281–288. doi:10.1016/j.plantsci.2004.03.026 [Google Scholar]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P. OsPTF1, a novel transcription factor involved in tolerance to phosphate-starvation in rice (Oryza sativa L.) Plant Physiol. 2005;138:2087–2096. doi: 10.1104/pp.105.063115. doi:10.1104/pp.105.063115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Li J, Xu C, Tan Y, Gao Y, Li X, Zhang Q, Saghai Maroof M.A. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc. Natl Acad. Sci. USA. 1997;94:9226–9231. doi: 10.1073/pnas.94.17.9226. doi:10.1073/pnas.94.17.9226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, et al. A draft sequence of the rice genome (Oryza sativa ssp. indica) Science. 2002;296:79–91. doi: 10.1126/science.1068037. doi:10.1126/science.1068037 [DOI] [PubMed] [Google Scholar]
- Yu J, et al. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. doi:10.1371/journal.pbio.0030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Liu J, Suh B, Cheung F, Sultana R, Lee D, Quackenbush J, Buell C.R. The TIGR rice genome annotation resource: annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res. 2003;31:229–233. doi: 10.1093/nar/gkg059. doi:10.1093/nar/gkg059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Structural features of the rice chromosome 4 centromere. Nucleic Acids Res. 2004;32:2023–2030. doi: 10.1093/nar/gkh521. doi:10.1093/nar/gkh521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. Features of the expressed sequences revealed by a large-scale analysis of ESTs from a normalized cDNA library of an elite indica rice cultivar Minghui 63. Plant J. 2005;42:772–780. doi: 10.1111/j.1365-313X.2005.02408.x. doi:10.1111/j.1365-313X.2005.02408.x [DOI] [PubMed] [Google Scholar]
- Zhang J, Li C, Wu C, Xiong L, Chen G, Zhang Q, Wang S. RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Res. 2006;34:D745–D748. doi: 10.1093/nar/gkj016. doi:10.1093/nar/gkj016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, et al. A fine physical map of the rice chromosome 4. Genome Res. 2002;12:817–823. doi: 10.1101/gr.48902. doi:10.1101/gr.48902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, et al. BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res. 2004;32:D377–D382. doi: 10.1093/nar/gkh085. doi:10.1093/nar/gkh085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, editor. Biology of the male sterility in rice. Wuhan University Press; Wuhan, China: 2000. [Google Scholar]
- Zhu J.K. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. doi:10.1016/S1360-1385(00)01838-0 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Cai X, Wang Z.Y, Hong M.M. An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. J. Biol. Chem. 2003;278:47 803–47 811. doi: 10.1074/jbc.M302806200. doi:10.1074/jbc.M302806200 [DOI] [PubMed] [Google Scholar]