Abstract
In May 2000, the Beijing Institute of Genomics formally announced the launch of a comprehensive crop genome research project on rice genomics, the Chinese Superhybrid Rice Genome Project. SRGP is not simply a sequencing project targeted to a single rice (Oryza sativa L.) genome, but a full-swing research effort with an ultimate goal of providing inclusive basic genomic information and molecular tools not only to understand biology of the rice, both as an important crop species and a model organism of cereals, but also to focus on a popular superhybrid rice landrace, LYP9. We have completed the first phase of SRGP and provide the rice research community with a finished genome sequence of an indica variety, 93-11 (the paternal cultivar of LYP9), together with ample data on subspecific (between subspecies) polymorphisms, transcriptomes and proteomes, useful for within-species comparative studies. In the second phase, we have acquired the genome sequence of the maternal cultivar, PA64S, together with the detailed catalogues of genes uniquely expressed in the parental cultivars and the hybrid as well as allele-specific markers that distinguish parental alleles. Although SRGP in China is not an open-ended research programme, it has been designed to pave a way for future plant genomics research and application, such as to interrogate fundamentals of plant biology, including genome duplication, polyploidy and hybrid vigour, as well as to provide genetic tools for crop breeding and to carry along a social burden—leading a fight against the world's hunger. It began with genomics, the newly developed and industry-scale research field, and from the world's most populous country. In this review, we summarize our scientific goals and noteworthy discoveries that exploit new territories of systematic investigations on basic and applied biology of rice and other major cereal crops.
Keywords: rice, genome, transcriptome, proteome, hybrid vigour
1. Introduction
Most of the genome projects are initially focused on a particular genome, such as the Human Genome Project (International Human Genome Sequencing Consortium 2001, 2004), the Arabidopsis Genome Project (Arabidopsis Genome Initiative 2000), the Drosophila Genome Project (Adams et al. 2000), just to name a few. An immediate goal of these projects is to provide a sequence of the targeted genomes as a reference as well as a set of genes discovered along the way to facilitate genomic and molecular research of the subjects. Genome projects can deliver, with adequate funding, basic genomic information for biological research: physical maps (sequence- and clone-based); genetic maps; genes (sometimes refer as gene models when support evidence is weak); and gene expression information. These goals are2 often achieved in two phases: (i) to assemble a low coverage (accumulated effective sequence length divided by the estimated length of a genome) version of the genome as a ‘draft sequence’ or a ‘working draft of the genome’ so that a particular research community is able to benefit from the immediate release of the data for other research activities, especially for gene discovery and localization; and (ii) to put together a final assembly that integrates, as much as possible, basic genomic information ranging from clone-based physical maps to genetic polymorphisms. For instance, the International Human Genome Sequencing Consortium has published two major papers: the working draft sequence and the completed human genome. Occasionally, another phase of the project can also be executed, a survey sequencing or a genome survey with a genome coverage less than onefold (e.g. Wernersson et al. 2005). This particular phase is always important for large and unknown genomes to work out solid sequencing plans. Although the list of targeted genomes has extended horizontally to closely related genomes within a species or genus, such as the recent extension of Drosophila genome studies, the Drosophila Population Genomic Project (Matthews et al. 2005), and vertically to include species within lineages, such as primates and vertebrates, the purpose of these projects is largely human-centric, to annotate and understand human genes and their functions, or model organism-centric, to understand evolution and particular biological characteristics of a given organism, and to satisfy the basic needs of these research communities. However, for a crop species, especially for crops where most of their landraces are F1 hybrid with infertile progenies, it is highly desirable that a detailed comparative study can be carried out on the distant parental lines at molecular details as the project itself or immediately after the genome sequence becomes available. The Chinese Superhybrid Rice (Oryza sativa L.) Genome Project (SRGP) is simply designed to do this and has been doing it in unprecedented speed, scale and details.
In the spring of 2000, Beijing Institute of Genomics held an open ceremony in its rented building space at the Beijing Airport Industrial Park and officially launched SRGP. What was absolutely certain in the minds of the project's leaders were three convincing arguments: (i) the agricultural crop breeding communities have been eager to explore genomic tools and hungry for basic genomic information of crop species in search of a path forward transforming their arduous and meticulous practice, as the importance of staple food crops has been emphasized in a famous Chinese phrase, ‘To the People, Food is What Under Heaven’ and the first ‘thing’ to focus on in China has to be the rice; (ii) a team of scientists, inspired by the Human Genome Project, were determined to develop tools and processes to bring a project of significant magnitude to the finishing line; and (iii) there are several major crop species of a few plant families to be studied in absolute detail and experiences on rice genomics should provide useful clues about how to proceed in this direction. Over six years into the project, we have learnt not only what rice genomics is about in unparalleled detail, but also how to focus on important issues that are practical and valuable for both the rice basic research community and the field scientists who are carrying out lifelong efforts to breed this vital food crop. In this review, we hope to evaluate our completed goals and lessons learned along the way and to emphasize what is ahead, as the project proceeds. We also hope to invite suggestions, ignite novel ideas and initiate collaborative efforts for the future and success of SRGP.
2. SRGP and its three phases
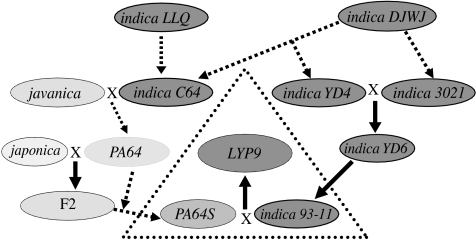
The moment when we received rice seedlings from Prof. Longping Yuan, the 2004 World Food Prize winner and Director of the China National Hybrid Rice R&D Center, SRGP was set on its charted course (figure 1). Advised by rice geneticist and molecular biologist Prof. Lihuang Zhu at the Institute of Genetics and Development of the Chinese Academy of Sciences and other experts in the rice research community, we chose a popular superhybrid rice cultivar, LYP9 or Liang-You-Pei-Jiu (it means Double Excellences Number 9; indica type; Lu & Zhou 2000), as the research object of SRGP. This ‘superhybrid’ rice cultivar produces 20–30% more grain than regular hybrid rice breeds and has a recorded breeding history (a simplified version is shown in figure 2). The project was launched with funds from the leadership of the Chinese Academy of Sciences and a research and development loan from the Hangzhou Municipal Government of Zhejiang Province. The project has never changed its direction and original promises, but has been gaining momentum towards the on-schedule completion of many deliverable goals across its three basic phases.
Figure 1.
The Chinese Superhybrid Rice Genome Project was officially launched in May 2000 with uncertainty of funding and absolute certainty of its goals. Executive Director of Beijing Institute of Genomics, Dr Jian Wang (left) is receiving rice seedlings from Mr Yongming Quan (right), Deputy Director of China National Hybrid Rice R&D Center symbolizing the beginning of SRGP.
Figure 2.
An oversimplified breeding history of LYP9. Dashed arrows depict multiple breeding processes.
In phase I of the project, we aimed at producing genome sequences of 93-11, the paternal cultivar of LYP9, an indica rice landrace and a popular choice of breeding starters (figure 3). The completion of this phase is featured by the complete genome sequence of 93-11 and comparative analyses of japonica and indica rice genomes and landmarked by two publications (Yu et al. 2002, 2005a), exactly three years apart. In phase II of the project, we have obtained the genome sequence of PA64S, the maternal cultivar of LYP9, as a working draft scheduled to be released and submitted for publication within this year. PA64S has a rather complex breeding history with a major (nuclear) genetic background of indica (estimated as greater than 55%) and minor ones of japonica and javanica (estimated as 25% and less than 20%, respectively; figure 2). Sequence analysis and acquisition of new experimental data have been tailored to molecular mechanisms of hybrid vigour at the levels of genomic sequences, transcriptomes and proteomes. Pilot studies along these directions have yielded useful data and important lessons for better experimental designs. Phase III has been planned to create a long-term effort, beginning already in the earlier phases of SRGP, and to produce a set of molecular and informatics tools for rice biologists and breeders, including high-resolution genetic maps with inter-varietal genetic markers, integrated physical/sequence maps and information on genes identified and annotated along the way of finishing the two rice genomes, and in-depth analyses and experimentation on important issues of rice biology, such as genome (gene) duplication, domestication and hybrid vigour. Our rice gene microarrays designed with 60 000 gene candidates, which include cDNA/EST-validated genes and predicted transcripts (the number is slightly greater than the current prediction), have been widely used by the rice research community within China and elsewhere, such as at the International Rice Research Institute and Yale University (Ma et al. 2005). We have been maintaining a comprehensive rice genomics database for open access (Zhao et al. 2004). We also concede that an effort to discover adequate numbers of genetic markers is of essence by an intensive sequence acquisition effort from more cultivated rice strains, as well as from wild germplasm resources. Most probably, new gene flows have to be encouraged sooner or later, and this point has also been advocated recently (McCouch 2004).
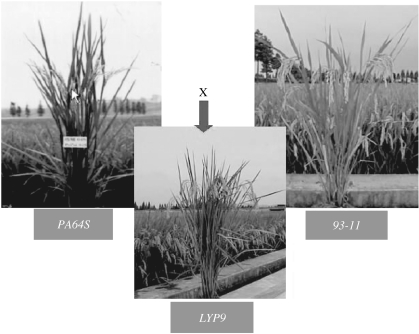
Figure 3.
A display of mature plants, which includes LYP9 and its parental cultivars, 93-11 and PA64S.
3. Lessons learnt in sequencing the rice genome
Being the second among plant genomes and the first crop genome sequenced, the rice genome has been actually sequenced independently five times. Three of the efforts targeted the same cultivar, a japonica variety, Nipponbare: one by the International Rice Genome Sequencing Consortium (2005) and the other two were from private companies, Syngenta (Goff et al. 2002) and Monsanto (their data were released to the public in a constrained way and mingled into the Consortium's rice data). We have sequenced the fourth and the fifth rice genomes: one is the indica 93-11, and the other is the PA64S that has mixed genetic backgrounds of all three cultivated rice subspecies and its chloroplast and mitochondrial genomes are japonica from its maternal origins. Most of the rice sequencing efforts have been using a whole-genome shotgun (WGS) method (reviewed by the authors recently; Yu et al. 2005b), since its genome is relatively small compared with what has been sequenced with this method, including the human genome which is six times larger than that of the rice. The International Consortium, however, had decided at the beginning of its project to use a clone-by-clone (CBC) strategy largely due to managerial difficulties, since the bulk of the work was originally allocated to different laboratories in several countries, such as the US, China and Japan. Each participating group took single or multiple chromosomes and promised to finish the sequences of targeted bacterial artificial chromosome (BAC) clones to contiguity (Sasaki & Burr 2000; Feng et al. 2002; Sasaki et al. 2002, 2005; Rice Chromosome 10 Sequencing Consortium 2003; International Rice Genome Sequencing Consortium 2005). Although it appears to be a simple promise, due to the repetitive and active nature of some repeat contents, to finish each clone to contiguity is not an easy job and certainly requires extra funding not proportional to the ordinary dollar-per-read accounting. We should all be thankful that all these efforts have served their expected purposes and data are left for the scientific community to mine and manoeuvre without restraint and charge (Wu et al. 2002, 2003; Tang et al. 2004).
(a) Essentials for a prolific genome project
Together with the rapid developments in genome-scale technologies, the demand for more and complete basic genomic information is only growing stronger. It is envisaged that a majority of the major agricultural crop species will be sequenced within the next decade or so, nearly 10 years after large-scale sequencing became available for the genome research community, along with hundreds of animal and plant species, relevant to health, agriculture, aquaculture and basic biology (addressing fundamental questions, such as development and evolution). Depending on the purpose of a study, the target genome for a crop species should be one of the most popular landraces. Ideally, a progenitor species should also be sequenced for comparative purpose and polymorphism discovery, since domestication is an accelerated process of genome evolution and agronomically important genes along with their functions may have been lost or altered already. An excellent example is the chicken genome projects, where the red jungle fowl was sequenced along with a comparative study on three other domesticated chickens: the layer (for egg laying); the boiler (for meat production); and the Silkie (a popular Asian delicacy for its tasty and nutrient-rich soup; International Chicken Genome Sequencing Consortium 2004; International Chicken Polymorphism Map Consortium 2004). In the study, 2.8 million single nucleotide polymorphisms (SNPs) were discovered and the chicken research community has already started an effort to genotype thousands of SNPs against thousands of collected pedigree and population samples (H. H. Cheng 2006, personal communication). A few years down the road, we will not be surprised when a community consensus is reached to sequence a wild rice variety or a probable progenitor of the cultivated rice, coupled with sampling sequences of some diverse rice cultivars. Another strategic issue is which method to use for whole-genome sequencing efforts, CBC or WGS. This is always a tough decision for a research community or a special interest group when the genome is large and its genome compositions are less known, such as GC-content, genome duplication history and repetitive sequences. What we do know at this point is that the WGS method has worked for genomes of mammalian size, approximately 3 billion base pairs. In addition, the two methods are not mutually exclusive, and a combination of the two is also valid when the project is carefully planned and adequate resources are generated. For instance, the corn genome, nearly the size of the human genome, is currently being sequenced by multiple approaches, CBC or WGS. It began with some gene-enriched portions of the genome in plasmid clones (Rabinowicz et al. 2003). There is no reason to reject a WGS approach for this agronomically important crop species if the sequencing cost becomes less of a concern as a result of some new technology advancements or even managerial improvement of the current technology. WGS may become the last choice, on the other hand, when the wheat genome is to be sequenced, which is approximately six times bigger than the human genome. Finally, when sequencing efforts are tuned to non-cultivated species or small organisms whose genomic DNA has to come from a population of individuals, it is critical to acquire preliminary data on their genetic heterogeneity and chromatin anomaly.
Aside from these strategic concerns, there are many technical issues for sequencing operators to pay adequate attention to. The most important one is the molecular resource for a project, regardless of which sequencing approaches are used. First, it includes, but is not limited to, plenty of cDNAs or expressed sequence tags (ESTs), though full-length cDNAs are often preferred, but it takes serious work, more time and adds more expense to a project. A well-planned EST discovery effort should be able to discover 50–80% of the transcripts that can serve as evidence for building gene models for the later assembled genomic sequences as well as starting material for EST-based microarrays for some focused studies. Second, large insert clone libraries are essential for long-range contiguity in physical map building. Although the currently preferred clone type is BAC, merited by its larger insert size, cosmid or fosmid is also extremely useful in closing gaps for WGS-based genome projects, since assembled contigs from a WGS are often smaller than an average BAC size that is around 150 kbp. Third, polymorphic markers are fundamental reagents for building genetic maps. When a reference genome sequence is available, SNPs can be generated by random sequencing of either clones or genomic DNA prepared from a diversity panel of related species or representative strains of targeted species. We are always grateful to the rich resources generated by the rice research community with recent heroic efforts, including physical maps, genetic markers and full-length cDNA sequences (Kikuchi et al. 2003).
(b) Whole-genome shotgun sequencing and data quality
One of the most misunderstood issues in SRGP has been related to data quality. In many ways, the situation was a replay of the controversies that dogged the Human Genome Project, which pitted a publicly funded effort led by Francis Collins, Eric Lander, John Sulston, Robert Waterston, and others (International Human Genome Sequencing Consortium 2001) against the privately funded effort led by Craig Venter of Celera (Venter et al. 2001). As participants on the public side, and one of the few genome centres to have sequenced plants (rice) and animals (human, chicken and silkworm), we offer a historical perspective.
In the earlier phases of the Human Genome Project, it was decided that the sequence must have an error rate lower than 1/10 000 bp, consented through various ad hoc committee meetings (M. V. Olson 2006, personal communication). The underlying argument was that the sequence had to be 10 times better than the variation rate for the human population (estimated to be approx. 1 part in 1000) in order to detect polymorphic differences. It was widely assumed that, notwithstanding difficult-to-clone heterochromatic regions, every base pair would be sequenced. As time passed, these quality standards were re-evaluated to reflect the needs of the intended application, particularly in the light of new technology developments and the species-specific peculiarities of the genome to be sequenced. Consider the accuracy of 1 part in 10 000. Human variation is known to be anomalously low as a result of a recent population bottleneck (Harpending & Rogers 2000). In rice, say between indica and japonica, variation for gene regions including introns is about 5 parts in 1000 and variation for intergenic regions is at least five times more than that for gene regions (Yu et al. 2005a). Hence, a more relaxed accuracy standard is justified. With the development of Phred quality scores (Ewing & Green 1998; Ewing et al. 1998), which provide accurate error probabilities for every base pair, the idea that the sequence must be 10 times better than the variation rate diminished because Phred quality scores allow one to distinguish between sequencing errors and polymorphisms, even for single-pass data (Altshuler et al. 2000) and certainly for our fourfold draft of the rice genome (Shen et al. 2004).
Into this fray stepped the controversial proposal of a WGS for the Human Genome Project (Weber & Myers 1997). What was novel about the idea was not shotgun sequencing itself, since the idea of assembling sequences from short random reads with multiple redundant coverage of the genome was already an established method. What was audacious was the size of the genome on which it would be applied, which was three orders of magnitude larger than anything done before. Nevertheless, the immediate objections (Green 1997) were based less on the size of the genome than on the fact that if the objective is ‘finished sequence’, where almost every base pair is resolved, there was little cost advantage to the WGS idea and many uncertainties. Notwithstanding, the experiments went ahead and the uncertainties were resolved. It is now acknowledged that, if the objective is an ‘intermediate grade’ of finished sequence, this is possible using the WGS method followed only by a limited amount of computer-assisted finishing (Blakesley et al. 2004). The resultant sequence is of very high quality, with the residual gaps and errors falling mostly in repetitive sequence. Importantly, such a product is much less expensive, requiring about 40-fold less reagents and 10-fold less personnel effort than the finished product that was created for the publicly funded Human Genome Project (International Human Genome Sequencing Consortium 2004).
The history of SRGP followed much the same trajectory as the Human Genome Project, albeit with approximately a 1-year delay. In the rice effort, there was an initial publication on a fourfold draft genome, later followed by a much improved sixfold genome. Considering the limited funding for rice, our final objective was closer to an intermediate grade of finished sequence. We sought to get all the genes assembled in one piece, without fragmentation, and anchored to the maps. What happened to the intergenic sequence was less of a concern. The fact that a WGS cannot deal with repetitive sequence is less of an issue with plants than with animals, because in plants, most of these repeats are attributable to transposable elements in the intergenic regions (Bennetzen 2000), which are clearly non-functional.
In the nomenclature of shotgun sequencing, there are contigs and scaffolds. A contig is a contiguous piece of sequence where every base pair is determined. A scaffold is a series of contigs where the order and orientation between contigs is known, while the content of the gaps between contigs is not. Higher levels of organization are called super-scaffolds, ultra-scaffolds and pseudo-chromosomes (chromosome models). Baring extremes in repeat content, contig size is determined by coverage, which refers to the number of times the genome is sampled. The statistics are well understood (Lander & Waterman 1988). Assuming reads of 500 bp length and overlap detection thresholds of 26 bp, the expected contig sizes using coverages of four- and sixfold are 5.4 and 24.6 kbp, respectively. In comparison, actual N50 contig sizes (i.e. the size above which half the total length is found) were 6.7 and 24.9 kbp.
Besides the improvement in contig size, higher coverage also leads to an improvement in accuracy. Based on the Phred-derived quality scores that are computed using our RePS assembly software (Wang et al. 2002; Zhong et al. 2003), 90.8 and 83.5% of the fourfold rice sequence from 2002 had a single-base error rate of better than 10−3 and 10−4, respectively. For the sixfold sequence from 2005, the figures were 97.2 and 94.6%. If we further restrict to gene regions defined by a non-redundant set of 19 079 full-length cDNAs (Rice Full-Length cDNA Consortium et al. 2003), the figures are slightly better at 98.1 and 96.9%.
Even at the sixfold sequence, the contigs are not big enough to anchor to the maps. This objective can only be achieved at the scaffold and super-scaffold levels. In the original Celera proposal, the idea was to sequence both ends of their clone inserts (Venter et al. 1998). Eighty-five per cent of the reads would be from plasmid clones of 2 kbp insert size, 14% would be from plasmid clones of 10 kbp insert size and 1% would be from BAC clones of 150 kbp insert size. This plan was never fully implemented, since they ended up combining their assembly with the public assembly. When the mouse WGS was published (Mouse Genome Sequencing Consortium 2002), the main lesson was that insert sizes must be matched to contig sizes. BACs are actually too big. At the sixfold sequence, the ideal clones are fosmids, whose insert sizes are fixed at approximately 40 kbp.
Fortuitously, we did not have to sequence fosmid ends, because the availability of a sixfold japonica rice WGS from Syngenta (Goff et al. 2002) provided linking information at the requisite length-scale. To preserve polymorphism information from indica and japonica, we used the other subspecies only to link contigs, never to provide missing base pairs. To allow the possibility of differences in the long-range order and orientation between these two subspecies, we kept track of the assembly before and after subspecies were mixed, using the notations ‘scaffold’ and ‘super-scaffold’. For the indica assembly, the resultant mapped super-scaffolds exhibited an N50 size of 8.3 Mbp.
Many small pieces remained unmapped, owing to high repeat content, but these were from extremely gene-poor regions. As a demonstration, we aligned the non-redundant set of 19 079 full-length cDNAs against our indica and japonica assemblies, requiring not only that these genes be found among the mapped super-scaffolds, but also that they be found in one piece, without fragmentation. The outcome was that 97.7% were qualified. In other words, most of the genes were assembled in one piece, without fragmentation, and anchored to the maps. The sequence is not perfect, but we estimated that no more than 1% of these genes were misassembled, due to our inability to discriminate between recent repeats.
What is sacrificed by a WGS method are the repeats. However, in plants, the repeats are due to intergenic transposons that are extremely unstable even between rice subspecies (Ma & Bennetzen 2004). Comparing indica and japonica, at least a quarter of these two genomes could not be aligned, and where they could, SNP rates varied from as little as 3.0 SNP kbp−1 in protein-coding regions to 27.6 SNP kbp−1 in transposable elements. This vindicated our strategy to focus on genes, because such intergenic regions are highly unlikely to be functional.
All this bodes well for the future of WGS in larger cereal genomes like maize (sixfold bigger) and wheat (38-fold bigger), where the same issues arise, but on an even larger scale. A test case for plant genome sequencing with WGS will be the maize genome, which has about 75% repeat content and nearly the size of the human genome. Various gene enrichment schemes have been proposed (Palmer et al. 2003; Whitelaw et al. 2003), but it remains to be seen whether these methods will be able to recover the entire gene, without fragmentation. Cost issues aside, we now know that the WGS method works.
(c) The rice genome and its biology
Before we planned our rice sequencing project, we had realized that the rice genome is quite suitable for a WGS approach. It indeed took us only a couple of months to acquire shotgun sequence reads, equivalent to fourfold coverage of the genome. A sequence assembler had also been developed, for which an effort was initiated a couple of years ahead of the time so that it was put in use in a timely way (Wang et al. 2002). With a rather unbiased assembly in hand and the availability of genome sequences from the other rice cultivar, Nipponbare, we then set out to analyse the dynamics of the rice genome and have revealed many interesting features of the rice genome.
(i) A dramatic GC-content increase unique to genomes of the Gramineae family among plants
The GC-content difference between rice and Arabidopsis genomes was first noticed when we compared the two sequences shredded in a 500 bp window (Wong et al. 2002; Yu et al. 2002). The GC-content increase has two important characteristics: directionality (GC-content at 5′ end of the sequence is higher than that at the 3′ end) and transcript-specificity (GC-content increase is not limited to mRNA or protein-coding sequences, i.e. exons, but also observed in flanking introns). It became clear that this strong GC-content gradient might be unique to the Gramineae family (the order Poales) of plants and certainly was not found in all monocotyledon plants, since it was absent in Lycoris longituba, a perennial herbaceous plant of the Amaryllidaceae family (the order Asparagales; Cui et al. 2004) and onion (the order Asparagales; Kuhl et al. 2004). Furthermore, we believe that it has to be related to transcription-coupled DNA repair (TCR), where repair errors have left their signatures in nucleotide composition as a GC-content gradient with more in the upstream and less in the downstream sequences along actively transcribed genes. The gradient does not seem to be related to replication errors because replication does not discriminate genes and their transcription process. However, it is difficult to demonstrate the precise mechanism that leads to such a gradient, since TCR is somewhat universal and found in almost all organisms from E. coli to yeast and to humans. In other words, this GC-content gradient is not a direct result of TCR but related to TCR-associated mechanisms that are unique to the organisms that manifest it. Furthermore, most of the bioinformatics analyses are not really transcript-centric or gene-centric, where the sequence compositional effect of genes is easily masked by fluctuations of the genomic background, let alone DNA repair mechanisms which are also complex in plants and remain to be elucidated (Kimura et al. 2004). Nevertheless, the GC-content gradient is an excellent indicator for the fast-evolving rice genes after a single round of whole-genome duplication (WGD), and the spare set of genes can undergo subfunctionalization or neofunctionalization for developing phenotypic novelties (Yu et al. 2005b).
(ii) Organizationally, the genomes of higher plants are different from higher animals
It was noted before we started SRGP that the plant genomes are organized differently compared with higher animals (Wong et al. 2000, 2001), based on limited information from several sequenced organisms including the only one plant genome, i.e. Arabidopsis. On the one hand, in animals, an overwhelming majority of their genomes are gene coding and most of the transposon insertions are in the introns so that a very limited portion of the genome space is left for intergenic regions. On the other hand, the plant genes are clustered, and most of the plant-specific transposable elements are inserted into intergenic spaces, albeit some fall near the genes (such as miniature inverted-repeat transposable elements, or MITEs) but seldom within genes (exons and introns). This notion not only has been held true for most, if not all, of the sequenced genomes of multi-cellular organisms, but has also strongly supported the idea of carrying out a WGS sequencing endeavour for the rice genome. When the genome sequences are compared between the two rice subspecies, 16% of them are not alignable, attributable mostly to intergenic retrotransposons (Yu et al. 2002). Moreover, this concept is rather important for plotting out sequencing strategies for large plant genomes, together with the ample literatures on genome duplications and polyploidy (Adams & Wendel 2005). Based on the history of genome duplication, it is feasible to estimate the gene content of a plant genome, since the gene sizes of higher plants are fairly constant in a range of 4–5 kbp. For instance, the gene content of a rice genome is from 200 to 250 Mbp, capable of harbouring 40 000–50 000 genes in an average size of 5 kbp. If we count maize genome as 2.5 billion base pairs in size and it has duplicated after it diverged from its Gramineae ancestor (Devos 2005), the gene content of the genome may be only 16–20% and the rest are intergenic sequences.
(iii) The rice genome has duplicated once after divergence from a common angiosperm ancestor but small-scale duplications are ongoing
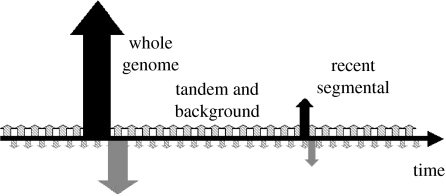
There are several types of sequence duplications, including WGD, segmental duplication, tandem duplication and background duplication (single gene-sized duplicated copies inserted into genomes without regularity). A schematic summary of the different duplication scenarios in the rice genome is shown in figure 4. When we applied a cDNA homologue pairing scheme to the discovery of duplicated chromosome segments, there were 18 pairs of duplicated segments that covered nearly 66% of the length of all the mapped rice sequences. The mean (median) number of homologue pairs per segment was 74 (53). The segment sizes were 6.9 Mbp (5.4 Mbp), and they differ by 43% (42%) within a segment pair. It revealed an ancient WGD, a recent segmental duplication on chromosomes 11 and 12 and massive ongoing individual gene duplications. Depending on the method used to calculate Ks, the fraction of synonymous site, the WGD event was dated to 53 and 94 million years ago (Myr ago), assuming a neutral evolutionary rate of 6.53×109 substitutions per silent site per year (Gaut et al. 1996). This result is consistent with the literature as reviewed recently (Blanc & Wolfe 2004; Adams & Wendel 2005; Devos 2005). The recent segmental duplication was dated ca 21 Myr ago, after rice split from corn and wheat within the grass family. Tandem duplication was also very significant and these duplicated genes were identified to be 16.5% of the sequenced rice genome. All these duplication scenarios seemed independent of each other in terms of the underlying mechanisms but the driving mutation forces may be similar, creating a new class of genes that have very limited homology from the original copies. The GC-content gradient in rice transcripts is just one of such sequence signatures observable and concrete—it is transcription-related. In other words, only genes that are actively expressed after duplication would maintain a GC-content gradient or accumulate mutation events over time, resulting in a GC-content gradient, and the inactive copies may fade away in their composition specificity.
Figure 4.
Duplications and gene losses of the rice genome over evolutionary time-scale. Horizontal arrow depicts time of genome duplication events and upward or downward vertical arrows denote gene duplication and gene loss events over time, respectively. Genome and segmental duplications are rare events (larger arrows), but tandem and background duplications (smaller arrows) are ongoing.
(iv) Two classes of genes defined according to the degree of similarities between rice and Arabidopsis genomes
When compared with Arabidopsis genes in a genome-wide fashion, rice genes can be theoretically classified into two categories according to a homology threshold: those that have homologues in the Arabidopsis genome and the rest that do not have homologues. Since plant genes are generally redundant and true orthologues are hard to determine, we did not use the term orthologue here. These two classes of genes are tentatively named as HH (with high homology) and LH (with low homology) genes for those of the rice, defined with a threshold: an E-value of 1×10−7 covering 50% of the aligned sequences or 100 amino acids in length. Although we started our analysis with computer-predicted genes (or gene models), there were 34.3% LH genes found in a full-length cDNA collection (Kikuchi et al. 2003). Furthermore, the reasons why we have classified the gene content into different categories are twofold; one is to simply stratify the genes at the genome level in addition to function-based categories that are largely incomplete, and the other is to see whether there are lineage-specific genes in the grass family of plants. We can stratify genes based on sequence homology, GC-content bias, duplication history, polymorphism rate, chromosomal location, expression level and structural elements (such as introns, exons, repeats and regulatory sequences) and relate them to functions.
The majority of the LH genes seems to be real and functional genes but evolving with a fast pace. Several lines of evidence support this notion. First, about 66% of the LH genes are confirmed with RNA level evidence, including full-length cDNAs, ESTs and SAGE tags, as opposed to 85% confirmation rate of the HH genes. Second, LH genes are also confirmed at the protein level, albeit with a lower confirmation rate; approximately 11% in a dataset containing over 3276 are confirmed in proteomics, including LH and HH genes. On the other hand, the HH genes have a much higher confirmation rate than the LH genes, nearly 21%. Third, the LH genes have smaller open reading frames (ORFs) than the HH genes, on average, half the size of HH genes. Fourth, LH genes have higher overall GC-contents than HH genes. We believe that shorter ORFs and higher GC-contents are both signatures of fast evolving genes, in which the GC-content gradient is continuously increasing to the extent that the first one or two exons of the LH genes may have lost their homology to the ancestors and thus may have lost their original functions or, to a lesser extent, gained new functions. Finally, we used a popular test for selecting the ratio of Ka/Ks, where Ka and Ks are the fraction of non-synonymous and synonymous sites, respectively, that are polymorphic. The expectation is that this ratio is 1 for neutrally evolving genes and less than 1 for any genes under purifying selection. Considering the small number of polymorphisms between indica and japonica, we would get a lot of zeros and infinities if we computed Ka/Ks for individual genes. We therefore computed Ka/Ks for HH and LH genes as a group. On average, the Ka/Ks ratios were 0.59 and 0.30 for the HH and LH genes, respectively. The result indicated that LH genes are under weaker purifying selection compared with HH genes.
Taken together, we have seen a dynamic nature of novel gene creation process in rice, perhaps extendable to other grass genomes: new copies of existing genes are generated by duplication mechanisms (birth of the LH genes), and, at the mean time, some of the duplicated genes are fast mutating, driven by novel mutation mechanisms that relate to gene expression (making of the LH genes), so that these newborn genes either evolve to have new functions or are quickly eliminated from the gene pool (death of the LH genes). We are currently carrying out detailed analyses and experiments to characterize genes in the two classes and to relate them to functional classifications. This hypothesis has been consistent with our observations and theories on duplicated genes proposed over time (Moore & Purugganan 2005). It will be tested vigorously when new grass family plants are sequenced in the year to come, which may include corn, sorghum, sugar cane, barley and wheat.
(v) Sequence signatures are found related to rice domestication
Like other major domestic crops and animals, rice was also domesticated ca 10 000 years ago, when Earth was gradually recovering from the last ice age and reached its lowest temperature point ca 18 000 years ago ending ca 10 000 years ago. Agriculture and human population increased together with the long warming process after the last ice age: agricultural practice started in major continents and human population was growing to 4 000 000 strong. Therefore, domestication as an interesting scientific subject studied since Charles Darwin should be more generalized and brought to the depth of genomics.
In searching for obvious sequence signatures of rice domestication, we aligned the sequence of 93-11, PA64S and Nipponbare. The pairwise alignments yielded clear bimodalities in SNP rate distributions, where two distinct modes were formed: a major mode at approximately 7 SNP kbp−1 and a minor mode at near zero. When we further divided the two modes in the 93-11 versus Nipponbare alignment, approximately 13% of the aligned sequences were found inside the minor mode at an average SNP rate per kbp. In other words, nearly 13% of the sequences in these two rice cultivars are almost identical. Similar bimodalities were also discovered in other genome-wide pairwise comparisons. The size range of these SNP-poor or ‘SNP desert’ regions is also striking, from 20 kbp to even 1 Mbp, which is constituted by both genic and intergenic sequences when annotated with a set of gene models. Some important genes have been found in the rice SNP desert, such as MADS-box genes and the Waxy gene. Dating based on neutral rate assumptions suggested that the minor mode are roughly formed around 10 000 years ago and the major mode cannot be easily calculated due to the polyphyletic nature of the domesticated rice species (Cheng et al. 2003). Detailed statistical analyses revealed positive selections on functional genes (some of the mutations may have been deleterious and loss-of-function) and degenerative effects on gene expression in the SNP deserts.
It is appealing to speculate that phenotypic (human artificial) selection may have encouraged selective sweeps and/or hitchhiking, which have been conserving the variation-poor SNP desert and building the minor mode since agriculture began. Climate changes, geographical isolations and human activities may have produced the three major rice ecotypes or subspecies, as well as polyphyletic signatures on their genomes. The human role in this process may have been just to bring the progenitors of a few rice subspecies together along their migrating routes and kept the genes flowing from one cultivar to another. Further studies along this line should focus on the molecular details of phenotype–genotype relationship to identify genes and their functional networks, which are related to certain domestication traits, and thus agronomically important.
4. Studying gene expression in hybrid rice and its parental cultivars
Our gene expression studies have been focused on comparative analysis among the triad, the parental cultivars and their F1 progeny at two basic levels. We have begun studies at the transcriptome level by using EST sampling, SAGE (serial analysis of gene expression; Bao et al. 2005) and GST (gene sequence tag)-based microarrays (Ma et al. 2005). Some of our EST data have been published (Zhou et al. 2003) to facilitate gene identifications and others are still in the pipeline to be analysed and published. At the second level, we have initiated analysis on the rice proteome (Zhao et al. 2005). Although the data acquisition effort is still underway, we have learnt significant lessons as to how to design better experiments and to acquire comparable data for functional studies.
There are two essential goals in studying gene expression: one is to discover genes expressed in targeted tissues (space) and at a particular developmental stage (time) and the other is to correlate the gene expression to particular cellular functions or molecular mechanisms in a context of regulated gene networks or systems (relationship). The gene discovery part is relatively straightforward, in which gene models (full sequences of genes) or tags (partial sequences of genes) can be verified, quantified and documented with some exhaustive efforts and sample resources. Since plant anatomy is not as complicated as that of the animal, the great majority of plant genes can be discovered in the above-mentioned ways and some of the genome-scale gene discovery efforts have been attempted in Arabidopsis and humans (Bertone et al. 2004; Hilson et al. 2004). The difficult part comes with designing experiments for acquiring expression data used to infer relationship among functional networks. It is relatively feasible when varied parameters (such as normal and challenged growth conditions) do not exceed the numbers of designed experiments (duplicated samples and controls should not be counted) and by strictly using the same cultivar as gene sources. However, when different cultivars are to be compared, such as in the case of our hybrid rice studies, true parallel comparisons are not easily achieved since the parental lines are to some extent only distantly related to each other and their growth and development behaviour (morphology and ontogeny) are certainly diverse. For instance, PA64S is male-infertile, therefore its reproductive organs and underlying genes are surely developed differently from the paternal 93-11. In addition, there are more than enough subtleties of phenotypic plasticity in the face of changing environments, genetic heterogeneities and newly created allelic differences in the hybrid progeny, which are all making functional interpretation of experimental results a nightmare, especially when some of the observed differences are not statistically significant enough and limited by the technology itself. Let us take our SAGE experiment as an example. We made nine SAGE libraries, three each from the triad: the leaf, the root and the panicles. We acquired nearly half million SAGE tags in comparable amounts from each library, which assembled nearly 70 000 unique tags. We then did an initial annotation of some unique SAGE tags based on a collection of full-length cDNAs, which allowed us to identify 595 upregulated and 25 downregulated genes in LYP9, the hybrid, through comparing its gene expression profiles with those of the parental cultivars. Most of the tag-identified, upregulated genes in LYP9 are related to enhancing carbon and nitrogen assimilation, including photosynthesis in leaves, nitrogen uptake in roots and rapid growth in both roots and panicles. However, it is difficult to evaluate downregulated genes since they are 24 times less than upregulated genes. Even with hundreds of upregulated genes to hand, we have a hard time to draw convincing conclusions because pictures of regulatory networks that involve the identified genes are still incomplete (Bao et al. 2005).
What should we do to study hybrid vigour at the level of gene expression and regulation? After initial attempts in comparing samples from the triad in parallel at a certain developmental stage, we realized that the simple-minded approach we used would not work at all, because the triad develops quite differently and their comparability cannot be plainly measured by the date of seeding or the harvesting time judged upon similarity in growth status. The experiments have to be designed to integrate information of gene expression and regulation both vertically within cultivars and horizontally between cultivars. SAGE and GST-based microarray are probably better methods to start with. First, each cultivar should be studied in a parallel fashion and in a series of experiments that sample different tissues at different developmental stages. Results from similar or comparable stages should be compared among the triad and the better matched stages should be defined. Second, data should be collected with higher density of the sampling points focused on a particular interested stage of development. The goal here is to identify genes or regulatory networks of interest and to narrow the number of targeted genes down to the extent that other low throughput methods, such as northern and RT-PCR, can be used for validation of results since both SAGE and microarray methods have their own drawbacks. Finally, quantitative methods and allelic information should be used to trace the origin and regulation of the genes with major effects on hybrid growth and development, and to determine mechanisms of hybrid vigour. Based on our preliminary results from EST, SAGE and microarray experiments, we envisage that the rice hybrid we are studying may obtain its supreme vigour from a collective effect of massive genetic complementation, where lowly expressed genes become prosperous, deleterious genes are replaced by effective ones and the regulatory networks are balanced by phenotypic plasticity and ontogeny, artificially selected by breeding practice over decades of time. We should remind ourselves that neither all expressed genes are functional nor all functionally expressed genes contribute to hybrid vigour. Only when systematically studied could the true facets of heterosis be unveiled.
5. Revealing the molecular details of hybrid vigour
We have anticipated that studying hybrid rice is not an easy endeavour, and we understand that eventually many of the important tasks will have to be passed on to the rice research community and not be kept for genomicists. Before it all happens, the trick here is to dig in; we have two parental genome sequences and a solid start in gene expression studies with all available technical platforms. With the genome sequence of PA64S to hand, we will identify allele-specific markers and subtle differences in gene expression between the two parental cultivars. However, we still need to decipher molecular mechanisms of hybrid vigour and not just identify some gene expression differences among the triad.
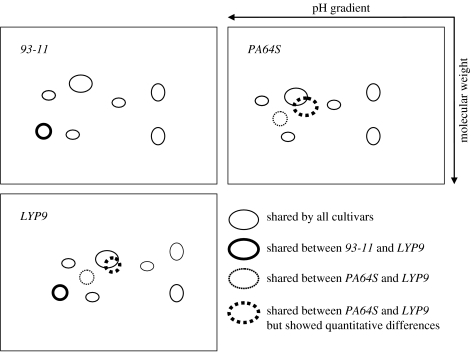
Some recent preliminary data from rice proteome studies gave us excellent hints for where to start. These experiments were originally designed to survey protein and RNA abundance in rice seeds (endosperm and embryo), with a hope to find enough protein markers to evaluate hybrid rice and other rice cultivars. It is also noteworthy that seeds from many cultivars and wild rice strains can be surveyed broadly and massively once proven useful. In this experimental design, we started from the seeds of the triad; they were sowed and the plants grew in conditions as identical as possible. Rice seeds have two major compartments: the endosperm and the embryo. The latter has more protein species to be identified; nearly 400 protein spots are identifiable on a regular Coomassie blue-stained two-dimensional polyacrylamide gel. Among the identifiable protein spots on the gel, nearly 10% of them showed differentially expressed patterns in three categories: unique to each parental embryo, absent in the hybrid and inherited from the parental embryos. Furthermore, almost all spots identified in LYP9 are traceable from either 93-11 or PA64S. Variations in isoelectric points, secondary modifications and other electrophoretic features were all detected in the type of experiments described here (Xie et al. 2006). A schematic chart showing this phenomenon is illustrated in figure 5. With better resolution and fractionation methods, it is conceivable that nearly a thousand protein spots could be identified on two-dimensional gels for such comparative protein analyses in the embryo alone. The other dimension of the proteomic experiments is to focus protein identification and expression studies on different organs or tissues during development. A recently published paper from our group has demonstrated the feasibility of such a focused study (Zhao et al. 2005). In the study, we investigated protein expression among six different developmental phases and identified 49 differentially expressed proteins (or spots on two-dimensional gels). Of these, 89.8% were confirmed to be rice proteins. After the confirmation of some of the interesting rice proteins with immunoblotting, we have drawn three major conclusions from the experiments. First, protein expression in rice leaves, at least for high or middle abundance proteins, is attenuated during growth (especially some chloroplast proteins) and the expression profiles are relatively stable during rice development, despite the fact that the change was not very dramatic. Second, ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), a major protein in rice leaves, is expressed at constant levels at different growth stages. Interestingly, a high ratio of degradation of the RuBisCO large subunit was found in all samples. The degraded fragments were similar to other digested products of RuBisCO mediated by free radicals. Third, the expression of antioxidant proteins such as superoxide dismutase and peroxidase declines at the early ripening stage. Such detailed investigations at the protein level are of importance to paint a complete picture of gene regulation networks and to relate them to molecular mechanisms governing growth and development.
Figure 5.
Schematic chart shows the inheritance pattern of proteins identified on two-dimensional gels for analysing urea-soluble proteins isolated from embryos of hybrid rice LYP9 and its parental cultivars.
It is quite clear that systematic investigations with a combination of RNA/protein qualification and allelic discrimination techniques will allow us to identify an adequate amount of genes and their products, which are differentially expressed and/or regulated among a triad. The study can take a solid start around a quiescent stage (the embryo) of the plant, extend initially to embryogenesis and germination, and later to rooting, tillering and flowering stages. Ultimately, these experiments will lead to a basic construct with connections and modules in the context of regulatory networks and metabolic pathways, and which, in turn, leads to a comprehensive understanding of how hybrid vigour comes into play for plant physiology in satisfactory molecular detail. We are certainly at the dawn of this new era.
Acknowledgments
We would like to thank Ms Wei Gong for her excellent assistance in editing this manuscript. SRGP is supported by the grants from Chinese Academy of Sciences (KSCX1-SW-03; KSCX2-SW-223; KSCX2-SW-306), Commission for Economy Planning, Ministry of Science and Technology (2001AA225041; 2002AA229021; 2002AA2Z1001; 2002AA104250; 2002AA234011; 2001AA231061; 2001AA231011; 2001AA231101; 2004AA231050; 2003AA207160), National Natural Science Foundation of China (30399120; 30200159; 30370330; 30370872; 30200163; 90208019), Beijing Municipal Government, Zhejiang Provincial Government, Hangzhou Municipal Government.
Footnotes
One contribution of 14 to a Theme Issue ‘Biological science in China’.
References
- Adams K.L, Wendel J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. doi:10.1016/j.pbi.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Adams M.D, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. doi:10.1126/science.287.5461.2185 [DOI] [PubMed] [Google Scholar]
- Altshuler D, Pollara V.J, Cowles C.R, Van Etten W.J, Baldwin J, Linton L, Lander E.S. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. doi: 10.1038/35035083. doi:10.1038/35035083 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. doi:10.1038/35048692 [DOI] [PubMed] [Google Scholar]
- Bao J, et al. Serial analysis of gene expression study of a hybrid rice strain (LYP9) and its parental cultivars. Plant Physiol. 2005;138:1216–1231. doi: 10.1104/pp.105.060988. doi:10.1104/pp.105.060988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J.L. Comparative sequence analysis of plant nuclear genomes: microcolinearity and its many exceptions. Plant Cell. 2000;12:1021–1029. doi: 10.1105/tpc.12.7.1021. doi:10.1105/tpc.12.7.1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. doi:10.1126/science.1103388 [DOI] [PubMed] [Google Scholar]
- Blakesley R.W, et al. An intermediate grade of finished genomic sequence suitable for comparative analyses. Genome Res. 2004;14:2235–2244. doi: 10.1101/gr.2648404. doi:10.1101/gr.2648404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. doi:10.1105/tpc.021345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Motohashi R, Tsuchimoto S, Fukuta Y, Ohtsubo H, Ohtsubo E. Polyphyletic origin of cultivated rice: based on the interspersion pattern of SINEs. Mol. Biol. Evol. 2003;20:67–75. doi: 10.1093/molbev/msg004. doi:10.1093/molbev/msg004 [DOI] [PubMed] [Google Scholar]
- Cui Y, et al. Identification and expression analysis of EST-based genes in the bud of Lycoris longituba. Genomics Proteomics Bioinformatics. 2004;2:43–46. doi: 10.1016/S1672-0229(04)02006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos K.M. Updating the ‘Crop Circle’. Curr. Opin. Plant Biol. 2005;8:155–162. doi: 10.1016/j.pbi.2005.01.005. doi:10.1016/j.pbi.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl M.C, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Feng Q, et al. Sequence and analysis of rice chromosome 4. Nature. 2002;420:316–320. doi: 10.1038/nature01183. doi:10.1038/nature01183 [DOI] [PubMed] [Google Scholar]
- Gaut B.S, Morton B.R, McCaig B.C, Clegg M.T. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl Acad. Sci. USA. 1996;93:10 274–10 279. doi: 10.1073/pnas.93.19.10274. doi:10.1073/pnas.93.19.10274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A, et al. A draft sequence of the rice genome (Oryza sativa L. ssp, japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. doi:10.1126/science.1068275 [DOI] [PubMed] [Google Scholar]
- Green P. Against a whole-genome shotgun. Genome Res. 1997;7:410–417. doi: 10.1101/gr.7.5.410. [DOI] [PubMed] [Google Scholar]
- Harpending H, Rogers A. Genetic perspectives on human origins and differentiation. Annu. Rev. Genomics Hum. Genet. 2000;1:361–385. doi: 10.1146/annurev.genom.1.1.361. doi:10.1146/annurev.genom.1.1.361 [DOI] [PubMed] [Google Scholar]
- Hilson P, et al. Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res. 2004;14:2176–2189. doi: 10.1101/gr.2544504. doi:10.1101/gr.2544504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. doi:10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- International Chicken Polymorphism Map Consortium. A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature. 2004;432:717–722. doi: 10.1038/nature03156. doi:10.1038/nature03156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. doi:10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. doi:10.1038/nature03001 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. doi:10.1038/nature03895 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. doi:10.1126/science.1081288 [DOI] [PubMed] [Google Scholar]
- Kimura S, Tahira Y, Ishibashi T, Mori Y, Mori1 T, Hashimoto J, Sakaguchi K. DNA repair in higher plants; photoreactivation is the major DNA repair pathway in non-proliferating cells while excision repair (nucleotide excision repair and base excision repair) is active in proliferating cells. Nucleic Acids Res. 2004;32:2760–2767. doi: 10.1093/nar/gkh591. doi:10.1093/nar/gkh591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl J.C, et al. A unique set of 11,008 onion expressed sequence tags reveals expressed sequence and genomic differences between the monocot orders Asparagales and Poales. Plant Cell. 2004;16:114–125. doi: 10.1105/tpc.017202. doi:10.1105/tpc.017202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S, Waterman M.S. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. doi:10.1016/0888-7543(88)90007-9 [DOI] [PubMed] [Google Scholar]
- Lu C, Zhou J. Breeding and utilization of two-line interspecific hybrid rice LYP9. Hybrid Rice. 2000;15:4–5. (in Chinese). [Google Scholar]
- Ma J, Bennetzen J.L. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl Acad. Sci. USA. 2004;101:12 404–12 410. doi: 10.1073/pnas.0403715101. doi:10.1073/pnas.0403715101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, et al. A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res. 2005;15:1274–1283. doi: 10.1101/gr.3657405. doi:10.1101/gr.3657405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K.A, Kaufman T.C, Gelbart W.M. Research resources for Drosophila: the expanding universe. Nat. Rev. Genet. 2005;6:179–193. doi: 10.1038/nrg1554. doi:10.1038/nrg1554 [DOI] [PubMed] [Google Scholar]
- McCouch S. Diversifying selection in plant breeding. PLoS Biol. 2004;2:e347. doi: 10.1371/journal.pbio.0020347. doi:10.1371/journal.pbio.0020347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R.C, Purugganan M.D. The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 2005;8:122–128. doi: 10.1016/j.pbi.2004.12.001. doi:10.1016/j.pbi.2004.12.001 [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. doi:10.1038/nature01262 [DOI] [PubMed] [Google Scholar]
- Palmer L.E, Rabinowicz P.D, O'Shaughnessy A.L, Balija V.S, Nascimento L.U, Dike S, de la Bastide M, Martienssen R.A, McCombie W.R. Maize genome sequencing by methylation filtration. Science. 2003;302:2115–2117. doi: 10.1126/science.1091265. doi:10.1126/science.1091265 [DOI] [PubMed] [Google Scholar]
- Rabinowicz P.D, McCombie W.R, Martienssen R.A. Gene enrichment in plant genomic shotgun libraries. Curr. Opin. Plant Biol. 2003;6:150–156. doi: 10.1016/s1369-5266(03)00008-6. doi:10.1016/S1369-5266(03)00008-6 [DOI] [PubMed] [Google Scholar]
- Rice Chromosome 10 Sequencing Consortium. In-depth view of structure, activity, and evolution of rice chromosome 10. Science. 2003;300:1566–1569. doi: 10.1126/science.1083523. doi:10.1126/science.1083523 [DOI] [PubMed] [Google Scholar]
- Rice Full-Length cDNA Consortium; National Institute of Agrobiological Sciences Rice Full-Length cDNA Project Team; Foundation of Advancement of International Science Genome Sequencing & Analysis Group; RIKEN. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. doi:10.1126/science.1081288 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Burr B. International rice genome sequencing project: the effort to completely sequence the rice genome. Curr. Opin. Plant Biol. 2000;3:138–141. doi: 10.1016/s1369-5266(99)00047-3. doi:10.1016/S1369-5266(99)00047-3 [DOI] [PubMed] [Google Scholar]
- Sasaki T, et al. The genome sequence and structure of rice chromosome 1. Nature. 2002;420:312–316. doi: 10.1038/nature01184. doi:10.1038/nature01184 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Matsumoto T, Antonio B.A, Nagamura Y. From mapping to sequencing, post-sequencing and beyond. Plant Cell Physiol. 2005;46:3–13. doi: 10.1093/pcp/pci503. doi:10.1093/pcp/pci503 [DOI] [PubMed] [Google Scholar]
- Shen Y.J, et al. Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol. 2004;135:1198–1205. doi: 10.1104/pp.103.038463. doi:10.1104/pp.103.038463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, et al. A comparison of rice chloroplast genomes. Plant Physiol. 2004;135:412–420. doi: 10.1104/pp.103.031245. doi:10.1104/pp.103.031245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J.C, Adams M.D, Sutton G.G, Kerlavage A.R, Smith H.O, Hunkapiller M. Shotgun sequencing of the human genome. Science. 1998;280:1540–1542. doi: 10.1126/science.280.5369.1540. doi:10.1126/science.280.5369.1540 [DOI] [PubMed] [Google Scholar]
- Venter J.C, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. doi:10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]
- Wang J, et al. RePS: a sequence assembler that masks exact repeats identified from the shotgun data. Genome Res. 2002;12:824–831. doi: 10.1101/gr.165102. doi:10.1101/gr.165102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.L, Myers E.W. Human whole-genome shotgun sequencing. Genome Res. 1997;7:401–409. doi: 10.1101/gr.7.5.401. [DOI] [PubMed] [Google Scholar]
- Wernersson R, et al. Pigs in sequence space: a 0.66X coverage pig genome survey based on shotgun sequencing. BMC Genomics. 2005;6:70. doi: 10.1186/1471-2164-6-70. doi:10.1186/1471-2164-6-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw C.A, et al. Enrichment of gene-coding sequences in maize by genome filtration. Science. 2003;302:2118–2120. doi: 10.1126/science.1090047. doi:10.1126/science.1090047 [DOI] [PubMed] [Google Scholar]
- Wong G.K, Passey D.A, Huang Y, Yang Z, Yu J. Is “junk” DNA mostly intron DNA? Genome Res. 2000;10:1672–1678. doi: 10.1101/gr.148900. doi:10.1101/gr.148900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G.K, Passey D.A, Yu J. Most of the human genome is transcribed. Genome Res. 2001;11:1975–1977. doi: 10.1101/gr.202401. doi:10.1101/gr.202401 [DOI] [PubMed] [Google Scholar]
- Wong G.K, Wang J, Tao L, Tan J, Zhang J.G, Passey D.A, Yu J. Compositional gradients in Gramineae genes. Genome Res. 2002;12:851–856. doi: 10.1101/gr.189102. doi:10.1101/gr.189102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, et al. A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell. 2002;14:525–535. doi: 10.1105/tpc.010274. doi:10.1105/tpc.010274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, et al. Physical maps and recombination frequency of six rice chromosomes. Plant J. 2003;36:720–730. doi: 10.1046/j.1365-313x.2003.01903.x. doi:10.1046/j.1365-313X.2003.01903.x [DOI] [PubMed] [Google Scholar]
- Xie Z, Wang J, Cao M, Zhao C, Zhao K, Shao J, Lei T, Xu N, Liu S. Pedigree analysis of an elite rice hybrid using proteomic approach. Proteomics. 2006;6:474–486. doi: 10.1002/pmic.200500227. doi:10.1002/pmic.200500227 [DOI] [PubMed] [Google Scholar]
- Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. doi:10.1126/science.1068037 [DOI] [PubMed] [Google Scholar]
- Yu J, et al. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005a;3:e38. doi: 10.1371/journal.pbio.0030038. doi:10.1371/journal.pbio.0030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Wong, G. K.-S., Wang, J. & Yang, H., 2005b Shotgun sequencing (SGS). In Encyclopedia of molecular cell biology and molecular medicine, vol. 13, pp. 71–114, 2nd edn. Weinheim, Germany: Wiley-VCH.
- Zhao W, et al. BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res. 2004;32:D377–D382. doi: 10.1093/nar/gkh085. doi:10.1093/nar/gkh085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, et al. Proteomic changes in rice leaves during development of field-grown rice plants. Proteomics. 2005;5:961–972. doi: 10.1002/pmic.200401131. doi:10.1002/pmic.200401131 [DOI] [PubMed] [Google Scholar]
- Zhong L, Zhang K, Huang X, Ni P, Han Y, Wang K, Wang J, Li S. A statistical approach designed for finding mathematically defined repeats in shotgun data and determining the length distribution of clone-inserts. Genomics Proteomics Bioinformatics. 2003;1:43–51. doi: 10.1016/S1672-0229(03)01006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, et al. Gene identification and expression analysis of 86,136 expressed sequence tags (EST) from the rice genome. Genomics Proteomics Bioinformatics. 2003;1:26–42. doi: 10.1016/S1672-0229(03)01005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]