Abstract
China has a strong background in X-ray crystallography dating back to the 1920s. Protein crystallography research in China was first developed following the successful synthesis of insulin in China in 1966. The subsequent determination of the three-dimensional structure of porcine insulin made China one of the few countries which could determine macromolecular structures by X-ray diffraction methods in the late 1960s and early 1970s. After a slow period during the 1970s and 1980s, protein crystallography in China has reached a new climax with a number of outstanding accomplishments. Here, I review the history and progress of protein crystallography in China and detail some of the recent research highlights, including the crystal structures of two membrane proteins as well as the structural genomics initiative in China.
Keywords: protein crystallography, history, structural biology, macromolecular structure
1. Introduction
There are many records about crystals, such as crystalline snowflakes, from ancient China. However, at that time the people were interested only in crystals in the aspects of art and their applications. The crystallography method was first developed in Europe in the eighteenth century and the theory of geometric crystallography was self-contained in the nineteenth century. In 1895 Roentgen discovered X-rays, which have played a dominant role over the last 100 years for the material molecular structure study. In 1820, Fraunhofer constructed the first practical diffraction gratings, which Laue later used in 1912 to demonstrate that X-ray diffraction could be used to explore the structure of crystals (Hendrickson 1995; Campbell 2002; Liang 2003). This finding was a pivotal point for protein crystallography. Unfortunately, this great scientific discovery coincided with the ‘Xin Hai’ revolution in 1911. Between the 1920s and 1940s, perhaps only three famous Chinese pioneering physicists, Gangfu Hu, Qisun Ye and Youxun Wu, who did earlier X-ray studies abroad knew the importance of X-ray crystallography (Tang 1994). In 1966, following the successful synthesis of insulin in China, several institutes from the Chinese Academy of Sciences (CAS) together with Peking University decided to develop protein crystallography research in China. The determination of the three-dimensional structure of porcine insulin made China one of the few countries which could determine macromolecular structures by X-ray diffraction methods at that time. The period from the 1970s to the mid-1980s was relatively slow while the methodologies were developing and improving. Nevertheless, in China, the determination of the crystal structure of the first inactive ribosomal protein, trichosanthin, in the early 1980s was another main achievement. During the 1990s, widespread application of technologies for DNA recombination and synchrotron radiation led to a profound revolution in quantity and quality for protein crystal structure determination. Simultaneously, progress in methodology gave birth to the field of structural biology, a scientific subject involving determination of protein structures by means of protein crystallography, nuclear magnetic resonance spectroscopy and cryo-electron microscopy. Gradually, structural biology has progressed into a leading frontier of current life sciences research (Rao 1996).
2. The key development of protein crystallography in China
Structural biology is a discipline based on the structure determination of macromolecules with the ultimate aim of understanding the fundamental principles of life at the molecular level. The study of the three-dimensional structures of macromolecules and their functions has undergone rapid development in recent years, and many subjects are now synchronous with their relative areas of biological study (Liang 2003).
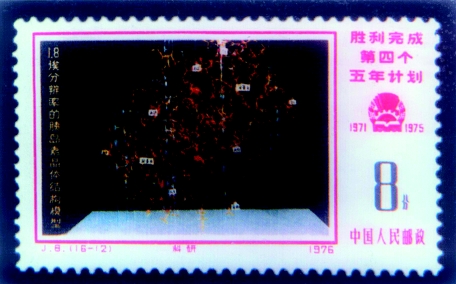
China has had a relatively strong background in protein crystallography. In 1965, for the first time in the world, Chinese scientists manually synthesized the crystalline bovine insulin. It was the first manual protein that shared the same properties with the native protein and had biological activity. Researchers from CAS and Peking University were involved in the synthesis work. Yinglai Wang, Jingyi Niu, You Wang, Chenglu Zou, Yueting Gong, Youshang Zhang et al. made great contributions towards synthesizing the first manual crystalline insulin. It is worth mentioning that many researchers at that time could not measure any activity from synthesized insulin, yet Youshang Zhang used the crystallized insulin to make successful measurements of insulin activity. From 1967, CAS and Peking University joined together to determine the structure of porcine insulin by the multiple isomorphous replacement and anomalous dispersion method. A considerable number of scientists, including Dong-Cai Liang, Zhengjiong Lin, Jiahuai Wang (from Institute of Biophysics, CAS); Pengfei Li, Haifu Fan, Jinbi Dai (from Institute of Physics, CAS) and Xiaocheng Gu (from Peking University) participated in the structure determination work. In 1971 and 1972, they obtained crystal structures of porcine insulin with resolutions of 2.5 and 1.8 Å, respectively, signalling that crystal structure determination in China had reached the first-class international level. This was a particularly influential achievement (figure 1) and reached the attention of Dorothy Crowfoot Hodgkin, recipient of the Nobel Prize for Chemistry in 1964, who offered much help to early crystallographers. Hongying Liao, Youqi Tang and Dong-Cai Liang all worked in her laboratory for different periods prior to 1966. Dorothy made great efforts to promote the achievements of Chinese crystallographers to the world and, in 1975, she published a paper entitled ‘Chinese work on insulin’ in Nature. In her paper, she wrote ‘The present Peking map at 1.8 Å resolution is the most accurate map available of the insulin electron density defined by experimental, isomorphous angles—and may well remain so’ (Hodgkin 1975, p. 103). After Dorothy Hodgkin broke the deadlock in 1975 to visit China and communicate with Chinese scientists, many foreign scientists, including the Nobel Laureates Pauling, Kendrew and Lipscomb visited China in succession during the period from the end of the 1970s to the early 1980s. Simultaneously, a large number of talented researchers involved in the study of crystal structure of porcine insulin went abroad to communicate with foreign scientists. Supported by the British Royal Society, David Stuart, when he was a young up-and-coming scientist from Oxford, spent almost 2 years in the laboratory of Dong-Cai Liang (Institute of Biophysics, CAS), where he was engaged in the structure determination of an insulin analogue (despentapeptide insulin) to 1.5 Å resolution and structure refinement of insulin to 1.2 Å resolution. Henceforth, a number of famous protein crystallographers from Oxford, such as Dorothy Hodgkin, David Phillips and Louise Johnson, came to be closely affiliated with their Chinese colleagues.
Figure 1.
A commemorative stamp marking the insulin structure determined by the Peking Insulin Structure Group.
In 1978, following the structure determination of insulin, Chinese researchers embarked on the study of the crystal structure of trichosanthin, a valuable medicinal protein that was shown in clinical studies to have an effective use in abortion and against cancer. In 1973, the Shanghai Institute of Organic Chemistry (CAS) commenced protein purification and study of the primary structure of trichosanthin. In 1976, after 2 years of collaboration between the Fujian Institute of Research on the Structure of Matter (CAS) and the Shanghai Institute of Organic Chemistry (CAS), the crystal incubation and preliminary determination of unit-cell parameters of trichosanthin was achieved. Preparation of heavy-atom derivatives was also attempted at the same time. In 1978, the crystal structure of trichosanthin was determined to a resolution of 2.6 Å and a reliable molecular model of the first inactive ribosomal protein in the world was built in collaboration between the Institute of Biophysics (CAS), the Fujian Institute of Research on the Structure of Matter (CAS) and the Shanghai Institute of Organic Chemistry (CAS). The structure of trichosanthin was only the second protein structure to be independently completed by Chinese scientists and a paper entitled ‘The sequence homology of trichosanthin and ricin A chain’, written by Zhang & Wang (1986), was published in Nature. This was the first academic paper concerning protein crystallography and written by Chinese scientists to appear in the world's top-ranking scientific journal.
The development of protein crystallography in China entered into a relatively quiet stage after the completion of the crystal structures of porcine insulin and trichosanthin. The study of the three-dimensional structures of light-harvesting proteins from algae, which was established by Dong-Cai Liang in 1993, had achieved significant ground-breaking results. Other crystallographers, including Zhengjiong Liu, Yicheng Dong, Dacheng Wang, Ruchang Bi (from the Institute of Biophysics, CAS), Xiaocheng Gu (from Peking University), Liwen Niu and Maikun Teng (from the University of Science and Technology of China) and Zongxiang Xia (from the Shanghai Institute of Organic Chemistry), focused on research into three-dimensional protein structures and their relation to the biological functions of some important natural immunity-related proteins, including natural neurotoxins, anti-microbial, anti-tumour and anti-viral proteins. All of these works were indispensable in maintaining the development of Chinese protein crystallography.
3. A new stage of protein crystallography in China
Recent advances in molecular biology, such as the widespread use of DNA recombination technology, coupled with the progress of technology and methods centred on the third generation of synchrotron radiation, have ushered in a new and rich stage of protein crystallography. Internationally, the field of protein crystallography developed rapidly following a quiet period during the 1970s and 1980s. From the late 1980s to 1990s, huge achievements were made in basic research, which they had a simultaneous impact on applied research in areas such as medical science and pharmaceutical design. Protein crystallography entered into a new phase with substantial growth in the numbers of relatively difficult macromolecular structures determined by X-ray diffraction during this period. In summer 1996, Dazhong Wang, the then president of Tsinghua University, was visiting England and approved a report entitled ‘To develop structural biology in China and to build a laboratory of structural biology at Tsinghua University’ proposed by Rao (2003). It was decided to grant a $500 000 outlay from the ‘211 Project’ to Zihe Rao towards the construction of a new structural biology laboratory. The Laboratory of Structural Biology in Tsinghua University was established to focus on systematic and thorough research of the structure–function relationship based on the techniques of molecular biology, protein chemistry and X-ray crystallography. In recent years, the Laboratory of Structural Biology in Tsinghua University has developed quickly through the efforts of its laboratory members, and has become an international base for protein science research largely due to the support and funding from the Ministry of Science and Technology ‘863 Project’ and ‘973 Project’, and the National Natural Science Foundation of China (NSFC; Rao 2003). Since 1996, a collaborative consortium of Chinese protein crystallographers has developed smoothly. The Institute of Biophysics, CAS, Tsinghua University, University of Science and Technology of China, Peking University, Institute of Physics, CAS, Institute of High Energy Physics, CAS, Shanghai Institute for Biological Sciences, CAS, Fujian Institute of Research on the Structure of Matter, CAS and the Shanghai Fudan University are now major research bases in China for structural biology and have made great contributions towards its growth and maturity as a discipline.
Rapid developments in structure determination and the results of human genome sequencing projects have consequently led to the emergence of structural genomics as a relatively new field. Structural genomics is a worldwide initiative to determine the three-dimensional structures of all proteins encoded by a genome and infer their molecular functions (Kim 1998). This formal definition of structural genomics was initially put forward at the first international structural genomics meeting in Hinxton, UK, in April 2000 (Burley 2000).
Chinese researchers became aware of this new field in the pilot phase of the international structural genomics initiatives. In response, China officially announced in 2001 its intention to launch structural genomics projects based both on the worldwide momentum and on the mobilization of Chinese crystallographers by Dong-Cai Liang, the pioneering protein crystallographer in China (from the Institute of Biophysics CAS). Leading research bodies in China, including the Ministry of Education, the Ministry of Science and Technology, CAS and NSFC separately pledged to support the basic research work of structural genomics and construction of the related technology platforms. Two independent and mostly collaborative projects were initiated, one involving leading Chinese universities and the other led by the CAS. After sufficient consideration of the specialty of national structural genomics and the Chinese level of structural biology research, it was decided that the aims of the China structural genomics initiatives should be to complete 100–200 protein structures in the following 5 years. Proteins were selected for their relevance to significant human diseases and for their potential as targets for the design of new drugs. All projects involved high-throughput protein selection, expression and purification of the target proteins, and research into the three-dimensional structures and functions of such proteins using current technology for structural biology. At the same time, efforts were made to establish high-throughput technology platforms for gene cloning, protein expression and purification, crystallization, structure determination and drug screening. China did not have crystallization robots at this stage and instead relied on ‘human robotics’ to achieve its goals (table 1).
Table 1.
Major structural genomics initiatives in China.
| date | group leaders | related institutions | target |
|---|---|---|---|
| 2001–2003 | Zihe Rao | Chinese Universities | proteins related to virus and tumour |
| 2001–2003 | Yunyu Shi | Chinese Academy of Sciences | proteins related to haematopoietic stem/progenitor cells and blood system disease |
| 2004 | Fuchu He & Zihe Rao | whole of China | China National Human Liver Proteomics Project (CNHLPP), including structural proteomics |
Phase I of the China structural genomics initiative was from 2001 to 2003, when research was interrupted by the outbreak of SARS. At that stage, it was decided to review and revise the goals of the China structural genomics initiative before embarking on phase II. Since the structural genomics initiatives in China were started, the most important returns have been as follows: broadening the consortium of Chinese structural biologists; constructing the technology platforms for protein science; and cultivating a number of excellent protein crystallographers.
4. Recent progress
The last five years have seen a new climax in protein crystallography in China with a number of outstanding accomplishments. Since the beginning of structural genomics in early 2001, and accelerating up to the present day, Chinese crystallographers have already produced nearly 1000 clones and more than 100 protein and protein complex structures, including those of proteins associated with significant human diseases and important physiological functions. In addition, two structures of membrane proteins have been determined. The most notable achievements are listed as follows:
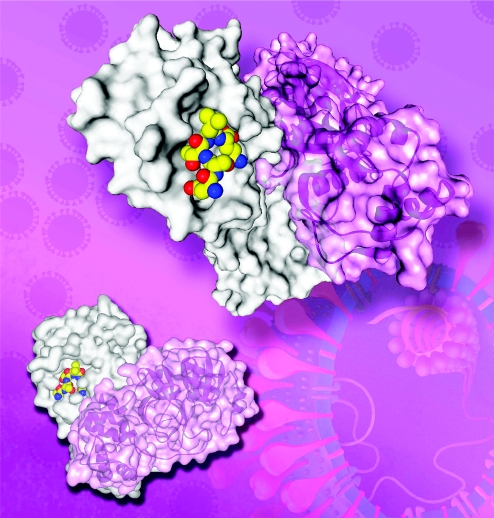
A collaborative research group between the Laboratory of Structural Biology (Tsinghua University) and the National Laboratory of Biomacromolecules (Institute of Biophysics, CAS), led by Zihe Rao, commenced studies on the structure and function of virus proteins, and drug design, for SARS (severe acute respiratory syndrome, an atypical highly contagious pneumonia) during the outbreak in China in May 2003. Since then, a number of significant results have been achieved. These results have been published in nine papers in internationally renowned journals, such as Proceedings of the National Academy of Sciences USA, Journal of Biological Chemistry and Biochemistry. Furthermore, three patents related to this work have been applied for. The results of these basic researches can be divided into four main aspects: (a) The structure of the SARS coronavirus main proteinase (Mpro or 3CLpro) and its complex with an inhibitor was determined (figure 2). This was the first structure of any protein from the SARS coronavirus to be determined in the world. The analysis of the Mpro structure has vital significance for basic research of the SARS coronavirus and for anti-SARS drug design (Yang et al. 2003). This research group has also produced a follow-up study in which they designed a wide-spectrum inhibitor against coronavirus Mpro, including SARS, using the SARS-CoV Mpro crystal structure as a basis (Yang et al. 2005). (b) During the SARS outbreak, the BJ01 strain of SARS was used to clone the genes of all structural and non-structural proteins (nsp), and to perform the expression and purification of 48 proteins and important functional fragments as the basis for a structural proteomics study of the SARS coronavirus. (c) The crystal structure of the SARS-CoV membrane fusion protein was determined with the aim of providing a new target for the design of anti-SARS therapeutics. This was the third crystal structure of a SARS coronavirus protein to be determined in the world, and the second structure from China. Taking the SARS-CoV spike (S) protein fusion core as a basis, together with the crystal structure of the mouse hepatitis virus S protein fusion core, a molecular mechanism by which the S protein mediates coronavirus membrane fusion and subsequent viral entry was proposed (Xu et al. 2004a–c). (d) The crystal structure of the hexadecameric nsp7–nsp8 super-complex from the SARS coronavirus has been determined successfully (Zhai et al. 2005). Coronavirus replication/transcription machinery involves multiple virus-encoded nsp. The mechanism by which it mediates genome replication and mRNA transcription remains unclear. This structure provides the first insight into the sophisticated architecture of this machinery. nsp8 demonstrates a novel ‘golf-club’ fold and exists in two conformations. The framework of the super-complex is a unique hollow cylinder-like structure assembled from eight copies of nsp8 and held tightly together by eight copies of nsp7. With an internal diameter of approximately 30 Å, the dimensions and the electrostatic properties of the central channel are favourable for duplex RNA binding, as confirmed by electrophoretic mobility shift assay. The results imply that the hexadecameric super-complex may confer processivity on RNA-dependent RNA polymerase. A model system for the SARS-CoV replication/transcription machinery was proposed on the basis of the nsp7–nsp8 super-complex and provides potential explanations for coronavirus recombination and the putative proofreading ability.
One group in the National Laboratory of Biomacromolecules (Institute of Biophysics, CAS), led by Wenrui Chang, determined the crystal structure of the major light-harvesting complex of photosystem II (LHC-II; figure 3). The LHC-II is an important membrane protein and a complicated molecular system. Besides its light-harvesting function, LHC-II has also been shown to function in the non-radiative dissipation of excess excitation energy formed under high light conditions. Moreover, LHC-II also takes part in regulating the distribution of excitation energy to photosystems I and II. It is a highly challenging task to solve the crystal structure of a membrane protein, and it is also an important indication of the level of protein crystallography in a particular country. The researchers have obtained innovative achievements both in the field of membrane crystallography and mechanism of light-harvesting and light-protection: (a) they reveal an elegant arrangement of membrane proteins in the icosahedral proteoliposome assembly, and show that membrane proteins can be crystallized in a way that differs from those that have been proposed before and (b) they show the pigment arrangement pattern in the LHC-II trimer (Liu et al. 2004). This is the first high-resolution structure of a light-harvesting protein to be determined by X-ray crystallography.
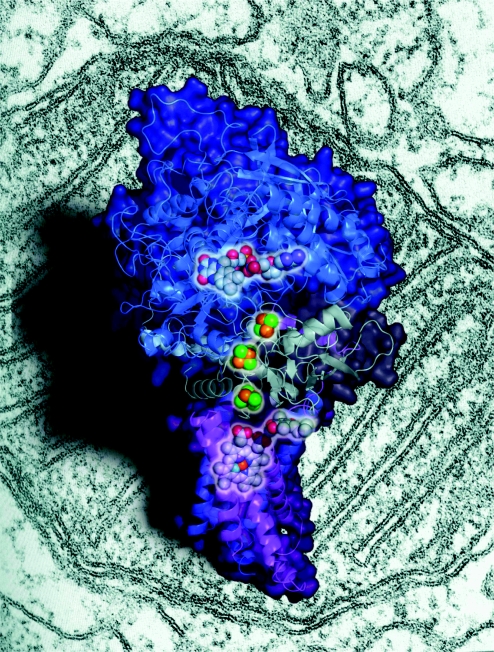
Recently, a collaborative research group between the Laboratory of Structural Biology (Tsinghua University) and the National Laboratory of Biomacromolecules (Institute of Biophysics, CAS), led by Zihe Rao, determined the crystal structural of the mitochondrial respiratory membrane protein complex II (figure 4). The results of this work were published in Cell (Sun et al. 2005). This represents the first purely ‘Chinese’ Cell paper from the native country to be published during the last 25 years, having a great impact on the international academic group. The mitochondrial respiratory system, consisting of five membrane protein complexes (I–V), produces most of the energy in eukaryotic cells. To 2005, the structures of complexes III, IV and V had been determined. However, no breakthroughs had been made on the structures of complex I or II. This research group solved the crystal structure of complex II from porcine heart at 2.4 Å resolution, together with its complex structure with inhibitors 3-nitropropionate and 2-thenoyltrifluoroacetone (TTFA) at 3.5 Å resolution. Elucidation of the complex II structure is a milestone and fills a blank in the field mitochondrial electron transfer research. Complex II contains four proteins: the FAD-binding protein or flavoprotein, the iron–sulphur protein and two membrane-anchor proteins with a total of six transmembrane helices. The overall structure is shaped like the letter ‘q’, with a hydrophilic head and a hydrophobic multipass transmembrane-anchor tail. Based on the above, complex II is assigned as a transmembrane protein, correcting a common mistake in biology textbooks in which complex II is regarded as a peripheral membrane protein. From the structure complexed with inhibitors, two binding sites were found. One was located near the FMN group of FAD, while the other was found at the distal side of the transmembrane anchor. Inactivation of mitochondrial complex II either by premature termination or by mutation has manifested a range of diseases, such as familial pheochromoctoma, familial head and neck paraganglioma and Leigh syndrome. The complex II structure provides a bona fide model for the study of human mitochondrial diseases related to mutations in this complex. Twenty years ago, the determination of the first membrane protein (a photosynthetic reaction centre) structure countered the dogma that membrane proteins cannot be crystallized. Henceforth, purification and crystallization of membrane proteins has become a hotspot in protein crystallography. The crystal structure of the mitochondrial respiratory membrane protein complex II is one of the few membrane structures to have been solved to date. It is the second membrane structure from China after the LHC-II structure was solved by the National Laboratory of Biomacromolecules (Institute of Biophysics, CAS). This research result has brought great international attention to our work, and indicates that the study of protein crystallography in China has reached the international level.
In 2005, a group from the Key Laboratory of Proteomics, Shanghai Institutes for Biological Sciences, CAS, led by Jianping Ding, determined the three-dimensional structures of human Rheb in complexes with GDP, GTP and GppNHp (5′-(β,γ-imide)triphosphate), which reveal novel structural features of Rheb and provide a molecular basis for its distinct properties (Yu et al. 2005). The small GTPase Rheb displays unique biological and biochemical properties different from other small GTPases and functions as an important mediator between the tumour suppressor proteins TSC1 and TSC2 and the mammalian target of rapamycin to stimulate cell growth. During GTP/GDP cycling, switch I of Rheb undergoes conformational change while switch II maintains a stable, unusually extended conformation, which is substantially different from the α-helical conformation seen in other small GTPases. In 2004, the same group also published the crystal structure of T2-hTrpRS at 2.5 Å resolution (Yu et al. 2004). Human tryptophanyl-tRNA synthetase (hTrpRS) produces a full-length and three N-terminus-truncated forms through alternative splicing and proteolysis. The shortest fragment that contains the aminoacylation catalytic fragment (T2-hTrpRS) exhibits the most potent angiostatic activity. The results of their structural analysis suggested that mammalian and bacterial TrpRSs might use different mechanisms to recognize the substrate, and that angiostatic activity is probably located at the α-helical domain, which resembles the short chain cytokines.
In 2005, a group from the Key Laboratory of Structural Biology, University of Science and Technology of China led by Liwen Niu deduced that Agkistrodon acutus venom serine proteinases I and II, previously isolated from the venom of A. acutus, are encoded by two almost identical genes, with only the single substitution of Asp for Asn at residue 62. Amidolytic assays indicated that they possess slightly different enzymatic properties. Crystal structures of A. acutus venom serine proteinases I and II were determined at resolution of 2.0 and 2.1 Å with the identification of trisaccharide (NAG(301)–FUC(302)–NAG(303)) and monosaccharide (NAG(301)) residues in them, respectively (Zhu et al. 2005). The substrate-binding sites S3 of the two proteinases appear much shallower than that of Trimeresurus stejnegeri venom plasminogen activator despite the overall structural similarity. Based on structural analysis, they showed that these Asn(35)-linked oligosaccharides collide spatially with some inhibitors, such as soybean trypsin inhibitor, and would therefore hinder their inhibitory binding. Difference of the carbohydrates in both the proteinases might also lead to their altered catalytic activities.
In early 2005, a group from the Department of Biochemistry and Molecular Biology, College of Life Sciences, Peking University, led by Xiao-dong Su, determined the structure of human AK 6 to 2.0 Å resolution (Ren et al. 2005). Adenylate kinases (AKs) play important roles in nucleotide metabolism in all organisms and in cellular energetics by means of phosphotransfer networks in eukaryotes. Sequence analyses revealed that human AK6 belongs to a distinct subfamily of AKs present in all eukaryotic organisms sequenced so far. Enzymatic assays show that human AK6 has properties similar with other AKs, particularly with AK5. Fluorescence microscopy showed that human AK6 is localized predominantly to the nucleus of HeLa cells. The identification of a nuclear-localized AK sheds light on nucleotide metabolism in the nucleus and the energetic communication between mitochondria and nucleus by means of phosphotransfer networks. The authors revealed that human AK6, although having much in common with other AK isoforms in terms of general structure, owing to its sequence, nuclear localization and enzymatic properties, is a unique AK and offers exciting avenues for future research.
Figure 2.
The SARS-CoV Mpro structure complexed with a substrate-analogue inhibitor.
Figure 3.
The structure of an icosahedral particle packed by 20 trimers of LHC-II on the cover of Nature, 18 March 2004. Reprinted by permission from Macmillan Publishers Ltd: Nature Physics Volume 428, Number 6980, © 2004.
Figure 4.
Schematic figure of three-dimensional structure of mitochondrial respiratory membrane protein complex II (the background shows the electron microscopic figure of mitochondria).
Furthermore, in 2005, a consortium of Chinese structural biologists began the ‘Human Liver Structural Proteomics’ project led by Weimin Gong (Institute of Biophysics, CAS). The ‘Human Liver Structural Proteomics’ project forms part of a larger national initiative, the ‘China National Human Liver Proteomics Project’, under the leadership of Fuchu He (China National Centre of Biomedical Analysis) and Zihe Rao (Tsinghua University/Institute of Biophysics, CAS). The research groups chose normal and pathological liver cells as chief experimental materials, with the objective to select and determine the structures of new proteins and liver proteins with the potential for drug design. The ultimate aims of the project are to elucidate the molecular mechanisms of liver cells and to provide a basis for research into new drugs against significant liver diseases.
5. Impacts on life sciences
Chinese protein crystallography has made tremendous progress in recent years, and the development of structural genomics in particular has accelerated the growth of life sciences research as a whole. More information concerning protein structures will provide more direct and useful resources for the study of life sciences. First, the determination of three-dimensional protein structures provides a basis for elucidating their molecular mechanisms and for designing and screening potential lead compounds for drug development (Hol 2000). Second, structural genomics is likely to facilitate the engineering of industrial enzymes by providing a wealth of structures of thermostable proteins (Hol 2000). Finally, the development of protein crystallography should lead to new growth in other areas, such as bioinformatics, molecular and cellular biology, as well as many other fields. Bioinformatics plays a key role in target selection, homology modelling, fold recognition and functional annotation of proteins. It will have broader applications as the number of available protein structures increases and the protein fold space is completed. Similarly, molecular and cellular biology will benefit from greater advances in gene cloning, protein expression, protein purification and especially the automation of purification procedures (Sun & Rao 2001).
The growth of Chinese protein crystallography has also facilitated the development of related engineering projects, such as the Beijing Synchrotron Radiation Facility (BSRF) and the Shanghai Synchrotron Radiation Facility (SSRF), to meet increasing demand for synchrotron radiation from Chinese crystallographers. Despite its limitations, the BSRF experimental station completed in 2002 has provided an excellent technological platform for structural genomics research. The largest scientific installation, the SSRF, is under construction and is expected to be open to users in 2009.
As a sign of its growing status in international X-ray crystallography, China has hosted a number of important conferences in protein crystallography and related areas. In June 2004, the 10th International Conference on the Crystallization of Biomacromolecules (ICCBM10) was held in Beijing under the organization of Zihe Rao and attracted nearly 500 participants, including many renowned experts in the field of protein crystallization. The Tsinghua International Conference of Protein Sciences (TICPS), held annually since 2001, has become an important fixture and is now in its fifth year. Also in 2005, Peking University will organize the International Workshop on Recent Advances in Phasing Methods for High-Throughput Protein Structure Determination chaired by Xiao-Dong Su and Tom Terwilliger (USA). In 2006, Beijing will play host to the prestigious Fourth International Conference of Structural Genomics (ICSG2006), the most important meeting for structural genomics researchers. The decision to award ICSG2006 to Beijing recognizes the emergence of China as a key player in the global structural genomics initiative. ICSG2006 will be chaired by Zihe Rao and Shigeyuki Yokoyama (Japan), and follows on from previous meetings held in Yokohama, Japan (2000), Berlin, Germany (2002) and Washington DC, USA (2004).
China is also attracting wider interest and collaborations with crystallographers from overseas. For example, Robert Huber, the 1998 Nobel Laureate in Chemistry for the determination of the three-dimensional structure of a photosynthetic reaction centre, is the chairman of the international advisory board in the Institute of Biophysics, CAS and a visiting professor in the Laboratory of Structural Biology, Tsinghua University.
6. Future plans
In the face of strong international competition and cooperation, Chinese researchers are participating in international structural genomics and proteomics projects. Recently, the protein science research platform constructed by the Institute of Biophysics, CAS has entered into its phase I stage. There are five central components to the operational platform, namely the high-throughput expression and purification system; the proteomics and bioinformatics system; the protein function analysis system; the structural genomics system; and the antibody R&D system. The construction of the protein science research platform is an innovative work and should have a great impact on the expansion of Chinese life sciences, technological advances and cultivation of talent. Operation of this platform will not only provide a consummate protein science research system and an excellent research facility, but also stimulates key technology innovation, first-class research achievements and the industrialization of these research products. The future growth of Chinese protein crystallography is focused on four main points. First, to consolidate and strengthen the basic important research bases including the Institute of Biophysics, CAS, Tsinghua University, the University of Science and Technology of China, Peking University, the Shanghai Institute for Biological Sciences (CAS), and the Fujian Institute of Research on the Structure of Matter (CAS), and to attempt to introduce protein crystallography to every University and pharmaceutical research institution which has the ability to do such research work. Second, to take membrane proteins, protein–protein complexes and molecular machinery as the main research objectives; to integrate more closely with problems in biology and medical science; and to combine with other research fields to study the relationship between protein structure and function. Third, to design and build technology platforms for gene cloning, protein expression and purification, and structure determination of biomacromolecules. Fourth and finally, to continue to strengthen international communication and cooperation in protein crystallography.
Acknowledgments
I am grateful to Yuna Sun and Mark Bartlam for their kind help in preparation of this manuscript.
Footnotes
One contribution of 14 to a Theme Issue ‘Biological science in China’.
References
- Burley S.K. An overview of structural genomics. Nat. Struct. Biol. 2000;7(Suppl.):932–934. doi: 10.1038/80697. doi:10.1038/80697 [DOI] [PubMed] [Google Scholar]
- Campbell I.D. Timeline: the march of structural biology. Nat. Rev. Mol. Cell. Biol. 2002;3:377–381. doi: 10.1038/nrm800. doi:10.1038/nrm800 [DOI] [PubMed] [Google Scholar]
- Hendrickson W.A. X-rays in molecular biophysics. Phys. Today. 1995;48:11. [Google Scholar]
- Hodgkin D. Chinese work on insulin. Nature. 1975;255:103. [Google Scholar]
- Hol W.G. Structural genomics for science and society. Nat. Struct. Biol. 2000;7(Suppl.):964–966. doi: 10.1038/80744. doi:10.1038/80744 [DOI] [PubMed] [Google Scholar]
- Kim S.H. Shining a light on structural genomics. Nat. Struct. Biol. 1998;5(Suppl.):643–645. doi: 10.1038/1334. doi:10.1038/1334 [DOI] [PubMed] [Google Scholar]
- Liang, D.-C. 2003 Protein crystallography. Institute of Biophysics, CAS
- Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W. Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature. 2004;428:287–292. doi: 10.1038/nature02373. doi:10.1038/nature02373 [DOI] [PubMed] [Google Scholar]
- Rao Z. The proposal to develop Chinese structural biology. Mag. Chin. Acad. Sci. 1996;3:158–160. [Google Scholar]
- Rao Z. Basic research report. Chin. Scientist. 2003;3:24–25. [Google Scholar]
- Ren H, et al. The crystal structure of human adenylate kinase 6: an adenylate kinase localized to the cell nucleus. Proc. Natl Acad. Sci. USA. 2005;102:303–308. doi: 10.1073/pnas.0407459102. doi:10.1073/pnas.0407459102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Rao Z. The current research conditions and prospects of structural genomics. Res. Global Technol. Dev. 2001;3:7–10. [Google Scholar]
- Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, Rao Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. doi:10.1016/j.cell.2005.05.025 [DOI] [PubMed] [Google Scholar]
- Tang Y. Crystallography of China. Physics. 1994;2:65–67. [Google Scholar]
- Xu Y, et al. Structural basis for coronavirus-mediated membrane fusion. Crystal structure of mouse hepatitis virus spike protein fusion core. J. Biol. Chem. 2004a;279:30 514–30 522. doi: 10.1074/jbc.M403760200. doi:10.1074/jbc.M403760200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Lou Z, Liu Y, Pang H, Tien P, Gao G.F, Rao Z. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 2004b;279:49 414–49 419. doi: 10.1074/jbc.M408782200. doi:10.1074/jbc.M408782200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, et al. Characterization of the heptad repeat regions, HR1 and HR2, and design of a fusion core structure model of the spike protein from severe acute respiratory syndrome (SARS) coronavirus. Biochemistry. 2004c;43:14 064–14 071. doi: 10.1021/bi049101q. doi:10.1021/bi049101q [DOI] [PubMed] [Google Scholar]
- Yang H, et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl Acad. Sci. USA. 2003;100:13 190–13 195. doi: 10.1073/pnas.1835675100. doi:10.1073/pnas.1835675100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. doi:10.1371/journal.pbio.0030324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Liu Y, Shen N, Xu X, Xu F, Jia J, Jin Y, Arnold E, Ding J. Crystal structure of human tryptophanyl-tRNA synthetase catalytic fragment: insights into substrate recognition, tRNA binding, and angiogenesis activity. J. Biol. Chem. 2004;279:8738–8788. doi: 10.1074/jbc.M311284200. [DOI] [PubMed] [Google Scholar]
- Yu Y, Li S, Xu X, Li Y, Guan K, Arnold E, Ding J. Structural basis for the unique biological function of small GTPase RHEB. J. Biol. Chem. 2005;280:17 093–17 100. doi: 10.1074/jbc.M501253200. doi:10.1074/jbc.M501253200 [DOI] [PubMed] [Google Scholar]
- Zhai Y, Sun F, Li X, Pang H, Xu X, Bartlam M, Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.J, Wang J.H. Homology of trichosanthin and ricin A chain. Nature. 1986;321:477–478. doi: 10.1038/321477b0. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Liang Z, Zhang T, Zhu Z, Xu W, Teng M, Niu L. Crystal structures and amidolytic activities of two glycosylated snake venom serine proteinases. J. Biol. Chem. 2005;280:10 524–10 529. doi: 10.1074/jbc.M412900200. doi:10.1074/jbc.M412900200 [DOI] [PubMed] [Google Scholar]