Abstract
Legume-nodulating bacteria (rhizobia) usually produce N-acyl homoserine lactones, which regulate the induction of gene expression in a quorum-sensing (or population-density)-dependent manner. There is significant diversity in the types of quorum-sensing regulatory systems that are present in different rhizobia and no two independent isolates worked on in detail have the same complement of quorum-sensing genes. The genes regulated by quorum sensing appear to be rather diverse and many are associated with adaptive aspects of physiology that are probably important in the rhizosphere. It is evident that some aspects of rhizobial physiology related to the interaction between rhizobia and legumes are influenced by quorum sensing. However, it also appears that the legumes play an active role, both in terms of interfering with the rhizobial quorum-sensing systems and responding to the signalling molecules made by the bacteria. In this article, we review the diversity of quorum-sensing regulation in rhizobia and the potential role of legumes in influencing and responding to this signalling system.
Keywords: Rhizobium, Sinorhizobium, Bradyrhizobium, homoserine lactone, nodulation, rhizosphere
1. Introduction
(a) Quorum sensing in plant growth-promoting bacteria
As described in the accompanying reviews, quorum sensing is classically a population-density-dependent signalling mechanism that allows bacteria to assess the size of their population and to behave coordinately (Dong et al. 2007; White & Winans 2007; Williams et al. 2007; Barnard et al. 2007; Bjarnsholt & Givskov 2007). Often the gene encoding the enzyme that synthesizes the signalling molecule is activated by quorum sensing, leading to the term ‘autoinducer’. In this review, we will focus mainly on quorum sensing mediated by N-acyl homoserine lactones (AHLs). Quorum-regulated genes identified to date are usually involved in adaptive changes in physiology of the bacterial population enabling them to modify aspects of their behaviour that are best undertaken when there are several other related bacteria nearby. Some of the adaptations include light production, antibiotic production and conjugation, but quorum-sensing-mediated gene expression can allow complex interactions between bacteria of different species and also with some eukaryotic organisms with which they can closely interact.
Quorum sensing is common among Gram-negative plant-associated bacteria (Pierson et al. 1998b; Parsek & Greenberg 2000; Whitehead et al. 2001b; Loh et al. 2002c; Dong et al. 2007; White & Winans 2007; Barnard et al. 2007; Bjarnsholt & Givskov 2007) and regulates several physiological traits associated with plant–bacterial interactions. AHL production by plant-associated bacteria, belonging to the genera Agrobacterium, Rhizobium, Sinorhizobium, Pantoea, Erwinia, Pseudomonas and Xanthomonas, was assessed and at least one isolate from each genera displayed AHL production (Cha et al. 1998). Most rhizobia tested were AHL producers, including isolates from Rhizobium fredii, Rhizobium leguminosarum bv. viciae, bv. phaseoli and bv. trifolii and Sinorhizobium meliloti. Different bacterial species can produce the same AHLs or AHLs with similar structures and properties suggesting that crosstalk between populations occurs and it is evident that quorum sensing via AHLs is more common among plant-associated bacteria than the general population of soil bacteria (Elasri et al. 2001; d'Angelo-Picard et al. 2005).
There are four broad classes of plant growth-promoting bacteria in soil: (i) those which enter into symbioses including the rhizobial–legume nitrogen-fixing symbioses (Wisniewski-Dye & Downie 2002; Gonzalez & Marketon 2003), (ii) the associative interactions in which bacteria such as pseudomonads suppress the growth of deleterious micro-organisms (Lugtenberg et al. 2001), (iii) the root-associated bacteria which can provide nutrients and/or growth-stimulating hormones as seen with species of Azospirillum and related bacteria (Dobbelaere et al. 2003), and (iv) bacteria which degrade environmental pollutants (Kuiper et al. 2004). Both the legume-nodulating rhizobia and the pseudomonads suppressing the growth of other micro-organisms use quorum-sensing gene regulation in relation to their stimulation of plant growth (Whitehead et al. 2001b; Wisniewski-Dye & Downie 2002; Gonzalez & Marketon 2003). In pseudomonads, one of the key biocontrol traits is the production of phenazine antibiotics, the expression of which is under quorum-sensing control via the AHL synthase encoded by phzI and the regulator encoded by phzR (Pierson & Pierson 1996; Pierson et al. 1998b; Chancey et al. 1999; Chin-A-Woeng et al. 2003). The production of these antibiotics also involves other regulators such as GacA, GacS, PsrA and RpeA (Lugtenberg et al. 2001; Whistler & Pierson 2003; Girard et al. 2006). It can also be negatively affected by the production of the fungal metabolite fusaric acid (van Rij et al. 2005) illustrating the intimate interactions between these bacteria and the fungal pathogens they suppress. In addition to influencing the growth of other micro-organisms, plant rhizosphere bacteria can activate the systemic plant defence system thereby conferring resistance to fungal pathogens. A strain of Serratia liquefaciens on the roots of tomato induced systemic resistance against the fungal leaf pathogen Alternaria alternata and this required the production of AHLs (Schuhegger et al. 2006). An important aspect of the ability of such bacteria to enhance plant growth is their ability to colonize roots and essentially form biofilms on the surface of roots, and this can be influenced by quorum-sensing regulation (Steidle et al. 2002; Ramey et al. 2004; Dubern et al. 2006).
Legume-nodulating rhizobia have several different quorum-sensing regulatory systems affecting plasmid transfer, symbiotic interactions, surface polysaccharide, growth inhibition and stationary-phase adaptation (Wisniewski-Dye & Downie 2002; Gonzalez & Marketon 2003). In this review, we will focus on the quorum-sensing regulatory systems that have been identified in a variety of different rhizobia. Most rhizobia tested were AHL producers, including isolates from R. fredii, R. leguminosarum bv. viciae, bv. phaseoli and bv. trifolii and S. meliloti. When compared with other genera, rhizobia produced the greatest diversity of AHLs, some of which were predicted to have very long acyl side chains (Cha et al. 1998; Brelles-Marino & Bedmar 2001). Where possible, we will relate the AHLs to genes and phenotypes that have been shown to be regulated by quorum sensing. We will also discuss how quorum-sensing regulation can directly or indirectly affect the interaction between rhizobia and the leguminous plants they nodulate.
(b) Acyl homoserine lactone synthases and regulators
There are two known protein families that have been shown to catalyse the synthesis of AHLs. The first group, and most widespread, is the LuxI family. These enzymes catalyse the ligation of homoserine from S-adenosylmethionine with an acylated acyl-carrier protein yielding acyl-homoserine lactone (More et al. 1996; Schaefer et al. 1996; Fuqua et al. 2001; Withers et al. 2001). A second set of AHL-biosynthetic proteins, the LuxM family, has been identified in Vibrio harveyii spp. (Bassler et al. 1994). AinS from Photobacterium (Vibrio) fisheri and VanM from Vibrio anguillarum are homologous to LuxM (Gilson et al. 1995; Milton et al. 2001), but no similar genes have been identified in rhizobia. All the AHL-response regulators identified to date in rhizobia belong to the LuxR protein family.
In many bacteria, quorum-sensing regulated genes identified to date are genes involved in bacterial interactions with their hosts (Fuqua et al. 1994, 2001; Salmond et al. 1995; Swift et al. 1996; Gray 1997; Fuqua & Greenberg 1998; Parsek & Greenberg 2000; Whitehead et al. 2001a; Withers et al. 2001; Bassler 2002; Lazdunski et al. 2004; Waters & Bassler 2005) and this is also true for rhizobia (Wisniewski-Dye & Downie 2002; Gonzalez & Marketon 2003).
(c) Acyl homoserine lactone reporters
The identification of AHL-based quorum-sensing systems in diverse bacteria and our understanding of how these systems work have been made possible through the use of AHL bioreporters. A variety of these reporter systems exist, the two most widely used being those in Chromobacterium violaceum and Agrobacterium tumefaciens, both often being used for visualization of AHLs separated by thin-layer chromatography.
In C. violaceum, the quorum-sensing genes cviI/cviR regulate a number of phenotypes including the production of the purple pigment violacein (McClean et al. 1997). A C. violaceum AHL-synthase (cviI) mutant cannot produce any AHL (and thus no violacein), but can produce only violacein in response to exogenous AHLs or to compounds that mimic AHLs, thus making a good bioreporter which is particularly sensitive to unsubstituted short-chain AHLs (C4 to C8; McClean et al. 1997).
The A. tumefaciens bioreporter is a non-AHL-producing strain carrying a traG–lacZ fusion and the LuxR homologue TraR, which induces traG in response to AHLs. This system detects 3-oxo, 3-hydroxy (3-OH) and unsubstituted AHLs with acyl side chains from C6 to C16 in length. This reporter is very sensitive, detecting AHLs at very low concentrations (Shaw et al. 1997). A reporter for detecting long-chain AHLs has been developed in S. meliloti (Llamas et al. 2004). A sinI::lacZ fusion integrated in the chromosome of S. meliloti lacking a functional sinI gene allows the detection of AHLs ranging from C12 to C18. The sensitivity can be increased, without loss of specificity, by overexpressing sinR.
Reporters based on luminescence have been constructed in Escherichia coli based on plasmids carrying the lux genes lacking the gene encoding the LuxI AHL synthase (Winson et al. 1998), but carrying genes (lasR, luxR or ahyR) encoding specific AHL receptors that activate the expression of the luxCDAB genes. An advantage is that they can be used for in situ analysis of gene expression in plants (Teplitski et al. 2000).
The use of green fluorescent protein reporter plasmids in non-AHL-producing strains of Pseudomonas putida and S. liquefaciens has made it possible to visualize AHL-mediated communication in the tomato rhizosphere (Steidle et al. 2001). Each of these strains was co-inoculated onto tomato roots with an AHL-producing bacterial strain. Visualization of fluorescence demonstrated that the bioreporter could sense the AHLs produced by the AHL-producing strain, even on some occasions when the colonies were separated by some distance. These reporter strains also made it possible to monitor AHL production by indigenous bacteria from non-sterile soil in the tomato rhizosphere. Cross-species communication has also been demonstrated between Pseudomonas and Burkholderia in mixed species biofilms and cultures (McKenney et al. 1995; Riedel et al. 2001) and between bacteria in a native wheat rhizosphere (Pierson et al. 1998a).
(d) Acyl homoserine lactone degradation
In the soil environment, micro-organisms form communities in which AHL producers interact with bacteria capable of degrading AHLs, and thus interfere with quorum sensing. AHL-degrading enzymes, classified as lactonases or acylases depending on their mode of action, have been found in different species of soil bacteria (Zhang 2003) including bacteria from the Rhizobiaceae. In A. tumefaciens, attM encodes a lactonase that can degrade AHLs (Zhang et al. 2002a) thereby affecting signal turnover and thus quorum sensing. Gamma-butyrolactones, molecules that are natural substrates of AttM, can interfere with AHL accumulation by inducing the expression of attM (Carlier et al. 2004) and related genes are found in rhizobia. This implies that rhizobia can degrade AHLs as discussed for agrobacteria in the accompanying paper by Dong et al. (2007).
2. Quorum-sensing genes in rhizobia
One of the central features of quorum sensing in rhizobia is that of diversity, because no two strains that have been analysed in detail have the same complement of quorum-sensing systems, even when such comparisons are limited to two different field isolates from within the same species. This diversity suggests that there may be no unifying paradigm of what is controlled by quorum sensing in the rhizobia. There are clearly quorum-sensing regulators and AHL synthases conserved among different species and genera, but it appears even then that different groups of genes may be controlled differently by orthologous synthases and regulators in different species. The range of genes regulated is just beginning to be identified in cultured rhizobia, but little is known about the range of rhizobial genes regulated by quorum sensing in the soil, rhizosphere or the plant environments. Most of the identified rhizobial quorum-sensing regulation systems seem to be based on AHL synthesis and perception. The LuxS-dependent biosynthetic pathway that leads to autoinducer-2 (AI-2) formation is absent in rhizobia (Winzer et al. 2002a,b). In this section, we will briefly summarize the AHL synthases, and quorum-sensing regulatory genes and the genes regulated in different rhizobia investigated so far. The various genes signalling AHLs and associated phenotypes in different rhizobia are summarized in table 1.
Table 1.
Quorum-sensing systems in the Rhizobiaceae.
| organism and strain | gene and location | signalling molecules | phenotype regulated | references |
|---|---|---|---|---|
| Agrobacterium tumefaciens | traR/traI (pTi) | 3-oxo-C8-HSL | plasmid transfer | Piper et al. (1993); Fuqua et al. (1994) and Hwang et al. (1995) |
| Rhizobium leguminosarum | ||||
| bv. viciae | cinR/cinI (chromosome) | 3-OH-C14:1-HSL | growth inhibition | Lithgow et al. (2000) |
| rhiR/rhiI (pRL1JI) | C6-HSL, C7-HSL, C8-HSL | nodulation efficiency | Cubo et al. (1992) and Rodelas et al. (1999) | |
| traR/traI (pRL1JI) | 3-oxo-C8-HSL, C8-HSL | plasmid transfer | Wilkinson et al. (2002); Danino et al. (2003) | |
| expR (chromosome) | unknown | unknown | Wisniewski-Dye & Downie (2002, unpublished) | |
| bv. phaseoli | raiR/raiI (non-symbiotic plasmid) | 3-OH-C8-HSL, C8-HSL | unknown | Wisniewski-Dye et al. (2002) |
| Rhizobium etli | ||||
| strain CNPAF512 | cinR/cinI (chromosome) | 3-OH-(slc)-HSL | nitrogen fixation, symbiosome development, growth inhibition | Daniels et al. (2002) |
| raiR/raiI (chromosome) | short-chain AHLs | nitrogen fixation, growth inhibition | Rosemeyer et al. (1998) and Daniels et al. (2002) | |
| strain CFN42 | traR/traI (p42a) unknown | 3-oxo-C8-HSL, 3-OH-C8-HSL | plasmid transfer, unknown | Tun-Garrido et al. (2003) |
| Sinorhizobium meliloti | ||||
| strain Rm1021 | sinR/sinI (chromosome) | 3-oxo-C14-HSL, C16:1-HSL, 3-oxo-C16:1-HSL, 3-oxo-C16-HSL, C18-HSL, C12-HSL | EPSII production, swarming | Marketon & Gonzalez (2002); Marketon et al. (2002, 2003); Teplitski et al. (2003) and Gao et al. (2005) |
| expR (chromosome) | C16:1-HSL | EPSII production, swarming | Pellock et al. (2002); Gao et al. (2005) | |
| mel (putative) | C8-HSL, other short-chain AHLs | unknown | Marketon et al. (2002) | |
| strain Rm41 | traR/traI (pRm41a) | 3-oxo-C8-HSL | plasmid transfer | Marketon & Gonzalez (2002) and Gonzalez & Marketon (2003) |
| RU10/406 | visN/visR (chromosome) | unknown effector | motility (flagellar regulon: fli, mot, fla and che genes) | Sourjik et al. (2000) |
| Rhizobium sp. | ||||
| strain NGR234 | traR/traI (pNGR234a) | 3-oxo-C8-HSL | plasmid transfer | He et al. (2003) |
| unknown genes (chromosome) | other AHLs | growth inhibition | ||
| Bradyrhizobium | ||||
| japonicum USDA110 | unknown | bradyoxetin | nod gene control | Brelles-Marino & Bedmar (2001); Loh et al. (2001, 2002a,b) and Pongsilp et al. (2005) |
| japonicum USDA 110/290 and B. elkanii | unknown | several AHLs detected (bioreporter) | unknown | |
(a) Rhizobium leguminosarum
There are three different biovars of R. leguminosarum: one is bv. viciae, which nodulates peas, vetch and lentils, the others are bv. trifolii, which nodulates clover, and bv. phaseoli, which nodulates Phaseolus beans. Most research has been done on R. leguminosarum bv. viciae and thus far four different LuxI-type AHL synthase genes have been identified in different isolates of R. leguminosarum bv. viciae (Wisniewski-Dye & Downie 2002). Each of these AHL synthase genes has a dedicated regulator encoded by a gene (usually) closely linked to the AHL synthase gene.
(i) cinI and cinR
Common to all the analysed strains of R. leguminosarum are the cinI and cinR genes, which are located on the chromosome (http://www.sanger.ac.uk/Projects/R_leguminosarum; Lithgow et al. 2000). CinR regulates the expression of cinI in response to CinI-made 3-OH-C14:1-HSL and there appears to be co-regulation of adjacent genes (Lithgow et al. 2000). Mutations in cinI or cinR reduce the expression of all other AHL synthase genes (Rodelas et al. 1999; Lithgow et al. 2000; Wisniewski-Dye et al. 2002; Danino et al. 2003) and it appears that the cinI/cinR system acts as an overall switch potentially influencing many aspects of rhizobial physiology (Wisniewski-Dye & Downie 2002). Nevertheless, the growth rate of cinI and cinR mutants is normal in different laboratory growth media and nodulation is relatively normal (Lithgow et al. 2000); therefore, the phenotypic changes are probably subtle, perhaps relating to survival and growth in soil, rhizosphere or in planta.
It appears that CinI-made 3-OH-C14:1-HSL plays a role in adaptation to stationary phase. Following starvation by nutrient exhaustion of C or N source, cultures entering stationary phase at high cell densities show no loss of viability over long periods (20–60 days), while cultures entering stationary phase at low cell densities lost viability rapidly. Exogenous 3-OH-C14:1-HSL promoted starvation survival in cultures of R. leguminosarum bv. phaseoli entering stationary phase at low cell density (Thorne & Williams 1999).
In R. leguminosarum, growth inhibition due to a ‘small bacteriocin’ was found to be due to 3-OH-C14:1-HSL (Gray et al. 1996; Schripsema et al. 1996), which is produced by CinI and causes growth arrest in susceptible strains (Lithgow et al. 2000; Wilkinson et al. 2002). 3-OH-C14:1-HSL growth inhibition is maximal when C8-HSL or 3-oxo-C8-HSL is added together with 3-OH-C14:1-HSL (Wilkinson et al. 2002). Resistant strains of R. leguminosarum and A. tumefaciens become susceptible when carrying cloned bisR and traR. One protein induced by BisR/TraR has been identified, this being the protein translation factor Ef-Ts (Wilkinson et al. 2002). This suggests the possibility that the observed growth inhibition may be related to the adaptation to stationary phase.
(ii) raiI and raiR
The raiI and raiR genes are located on a large (non-symbiotic) plasmid in R. leguminosarum bv. phaseoli strain 8002, but are absent from the genome of the sequenced strain of R. leguminosarum bv. viciae (http://www.sanger.ac.uk/Projects/R_leguminosarum) and appear to be absent from some other analysed strains of R. leguminosarum bv. viciae (Lithgow et al. 2000). As might be expected from their patchy distribution, mutations in these genes do not have much effect on the phenotype of R. leguminosarum either during free-living or symbiotic conditions. RaiR regulates the expression of raiI in response to the RaiI-made AHLs 3-OH-C8-HSL and C8-HSL (Wisniewski-Dye et al. 2002), but other genes regulated by RaiR are yet to be identified.
(iii) rhiI and rhiR
The rhiR gene was one of the earliest sequenced quorum-sensing regulators in the bacterial kingdom and was originally identified because it was very close to the genes (nod) required for legume nodulation and is required for the expression of the rhiA gene, which is highly expressed in the rhizosphere (Dibb et al. 1984; Economou et al. 1989; Cubo et al. 1992). RhiR regulates the expression of rhiI and rhiABC operon in response to RhiI-made C6-HSL, C7-HSL and C8-HSL (Rodelas et al. 1999). Mutations in rhiA or rhiR can cause a significant reduction in nodulation in strains already compromised for nodulation ability (Cubo et al. 1992), but although the sequence of the rhiABC genes has been known for several years, no function has been demonstrated for the gene products, which show no close similarities to proteins of known function. The observations that the rhi genes are closely linked to nodulation and nitrogen fixation genes and that the rhi genes are found only in bv. viciae but not in other biovars of R. leguminosarum suggest that they probably play a role in growth and/or survival in association with specific legume hosts.
(iv) traI and traR
TraR induces traI in response to TraI-made 3-oxo-C8-HSL. These genes are located on the symbiosis plasmid pRL1JI and together with bisR (encoding another LuxR-type regulator) are required to induce the plasmid transfer genes (see below). However, in the sequenced strain R. leguminosarum bv. viciae 3841, no equivalent genes were found on the symbiosis plasmid (which is called pRL10JI in that strain; http://www.sanger.ac.uk/Projects/R_leguminosarum). Homologues of traI and traR in strain 3841 are found on pRL7JI and pRL8JI, respectively, but their role in plasmid transfer has not been reported.
(v) Other LuxR-type regulators
In addition to the genes described above, there are three other LuxR-type regulators encoded in the genome of R. leguminosarum strain 3841. One is ExpR, which is located on the chromosome and is the orthologue of expR from S. meliloti (see below). Another gene of undefined function is located on the chromosome and yet another is on a plasmid. There are no LuxI-type (or any other candidate) AHL synthase genes associated with these regulators (http://www.sanger.ac.uk/Projects/R_leguminosarum) and their targets have not been identified.
(b) Rhizobium etli
Two different isolates of Rhizobium etli have been analysed, strains CNPAF512 and CFN42. It is clear that these are different from each other but share orthologous genes identified in different strains of R. leguminosarum.
(i) cinI and cinR
These genes in CNPAF512 seem to be very similar to those described above in R. leguminosarum, and CinI is responsible for the production of a long-chain AHL, similar to 3-OH-C14:1-HSL and referred to as 3-OH-(slc)- HSL. This AHL can inhibit the growth of a small bacteriocin-sensitive strain of R. leguminosarum, thus showing similar properties to 3-OH-C14:1-HSL from R. leguminosarum (Daniels et al. 2002). However, there is a distinct difference between R. leguminosarum bv. viciae and R. etli because mutations in cinI or cinR cause an increased lag phase and slower growth of R. etli, and this is correlated with abnormal development of nitrogen-fixing bacteria in nodules (Daniels et al. 2002). Mutations in the R. leguminosarum cinRI locus do not affect nodulation of pea or vetch (Lithgow et al. 2000), whereas mutations in the R. etli cinR or cinI genes (which are 95% identical to those of R. leguminosarum) dramatically decreased nitrogen fixation (Daniels et al. 2002). Nitrogen fixation decreased by more than 50% in both cinR and cinI mutants of R. etli and decreased even further in a cinR, cinI double mutant. The cinI mutant showed abnormal symbiosome development in nodules, decreased numbers of bacteroids packed within the symbiosome membrane and the cinI gene was shown to be expressed during the symbiosis, particularly in infection threads. (Daniels et al. 2002). The isolation of AHLs from bacteroids suggests that quorum sensing may play a role in the mature nodule (Daniels et al. 2002). At least three compounds, one of them likely to carry a long-chain AHL, were extracted from bacteroids present in nodules of bean inoculated with R. etli. Although the role, if any, played by quorum sensing has not been established, it is possible that quorum sensing may be involved in the regulation of processes that prepare bacteria for a return to a free-living state.
Mutation of cinR abolished swarming on agar plates, and this swarming could be reactivated by exogenous surfactant (Daniels et al. 2004) suggesting that genes regulated by CinR may regulate genes involved in surfactant biosynthesis as has been shown in S. liquefaciens (Lindum et al. 1998).
(ii) raiI and raiR
These genes in CNPAF512 appear to be orthologues of the R. leguminosarum bv. viciae genes described above (Rosemeyer et al. 1998). However, in R. etli, mutation of raiI caused an increase in the numbers of nodules and a parallel increase in nitrogenase activity. However, no significant net increase in symbiotic nitrogen fixation could be demonstrated based on the analysis of the growth of plants inoculated with the mutant (Rosemeyer et al. 1998). Surprisingly, while mutation of raiI increased nodulation, mutation of raiR had no effect, suggesting that RaiI-made AHLs may be involved in the suppression of nodulation (Daniels et al. 2002).
(iii) traI and traR
The arrangement of plasmid transfer genes on plasmid p42a of R. etli CFN42 (Tun-Garrido et al. 2003) and pRL1JI from R. leguminosarum bv. viciae (Danino et al. 2003) seem to be nearly identical and are presumably regulated in the same way (see below).
(c) Sinorhizobium meliloti
Quorum-sensing regulation has been examined in two different strains of S. meliloti, but has been much more fully analysed in 1021, the genomically sequenced strain, than in the other (Rm41/AK631).
(i) sinI and sinR
The sinI gene product produces diverse long-chain AHLs, including C12-HSL, C14-HSL, 3-oxo-C14-HSL, C16-HSL, C16:1-HSL, 3-oxo-C16-HSL, 3-oxo-C16:1-HSL and C18-HSL, with the mixture varying with the culture media used (Marketon et al. 2002; Teplitski et al. 2003). The specificity and concentration dependence of responses to different SinI-made AHLs have been little explored, but numerous specific responses to particular SinI-made AHLs have been observed (Chen et al. 2003; Gao et al. 2005). It appears that SinI may be the only AHL synthase in strain 1021. A second AHL synthase that produces short-chain AHLs, including C6-HSL, 3-oxo-C6-HSL and C8-HSL, had been suggested earlier (Marketon & Gonzalez 2002), but this was not confirmed in subsequent studies (Gao et al. 2005, in press). Mutation of sinI led to changes in the accumulation of over 35 different proteins (Gao et al. 2005) and the expression of over 100 genes (Hoang et al. 2004) in laboratory cultures. Phenotypically, sinI mutants were (i) defective in mucoidy (Hoang et al. 2004), (ii) significantly reduced in the rate or efficiency of nodule initiation, and (iii) defective for swarming on agar plates (Gao et al. 2005). The swarming could be restored by the addition of 5 nM C16:1-HSL (Gao et al. 2005).
sinR is adjacent to sinI on the chromosome, and SinR induces sinI expression in response to SinI-made AHLs, thereby enhancing AHL production 3–10-fold in vitro (Marketon et al. 2002). Slightly delayed nodulation has been reported for the sinR mutant. In microarray studies, three sinI-dependent genes had altered expression in the sinR mutant, consistent with SinI–AHL-dependent regulation via SinR (Hoang et al. 2004). Curiously, the expression of another 23 genes, including symbiotically relevant fixS1, was dependent on sinR, but independent of sinI, raising the question of what signal molecules besides SinI-made AHLs might be responsible for regulating the expression of these genes via SinR (Hoang et al. 2004).
(ii) expR
In strain 1021, the expR gene encoding a LuxR-type regulator is interrupted by a native insertion sequence (Pellock et al. 2002). Spontaneous excision of the insertion sequence generates a functional ExpR that regulates the expression of many genes, including genes in the exp operon required for the synthesis of EPSII (Pellock et al. 2002; Marketon et al. 2003; Hoang et al. 2004; Gao et al. 2005). EPSII is one of the three S. meliloti exopolysaccharides capable of eliciting responses in the host that permit infection (Gonzalez et al. 1996). Most wild-type strains of S. meliloti tested (Pellock et al. 2002) have an uninterrupted expR gene. Although the expR mutant has no obvious nodulation phenotype, the mutant is significantly altered in the levels of over 50 proteins and the expression of over 80 genes in the laboratory cultures (Gao et al. 2005; Hoang et al. 2004). These genes and proteins encompass a broad range of functions including central metabolism, regulation, transport, transposases, motility and symbiotically related behaviours. Remarkably, there was virtually no overlap of the 80 expR-dependent genes identified by microarray and the 50 expR-dependent proteins identified by proteome analysis (Gao et al. 2005). The observed lack of overlap may be due to specific culture conditions. However, if culture conditions so strongly influence which sets of genes are significantly affected by quorum sensing, then the really important focus in the future will be to identify the set of genes in rhizobia that are quorum-sensing regulated in natural environments—the soil, rhizosphere and host plant.
(iii) Other LuxR-type regulators
In addition to SinR and ExpR, the genome sequence for strain 1021 indicates the presence of six other proteins with good homology to LuxR-like regulators (Smc0658, 0877, 0878, 3015, 3016 and 4032), all chromosomally located (http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/). None of these six has been carefully tested, but if they prove to be functional quorum-sensing receptors, then quorum-sensing regulation in strain 1021 could be potentially much more complex than outlined above, especially in view of evidence for the formation of heterodimers as well as homodimers of the three AHL receptors in P. aeruginosa (Ledgham et al. 2003; Ventre et al. 2003). There is good evidence that many genes in strain 1021 may be regulated via LuxR-type regulators other than ExpR. Two earlier proteome studies (Chen et al. 2003; Teplitski et al. 2004) identified over 75 proteins in strain 1021 that were responsive to added SinI-made AHLs. In addition, another 60 proteins in strain 1021 that showed AHL-dependent accumulation have been identified (Gao et al. in press). Since strain 1021 lacks a functional ExpR, these results suggest that either SinR and/or one or more of the six putative AHL receptors may be either directly or indirectly responsible for the AHL responsiveness of this diverse and symbiotically relevant group of approximately 135 proteins. In this regard, it is worth noting that Smc3015 and 3016 in strain 1021 correspond to visN and visR in S. meliloti strain SU10/406. The visNR genes in SU10/406 encode LuxR-like regulators that are active as global regulators of flagellar motility and taxis, probably functioning as a heterodimer (Sourjik et al. 2000). The motility genes regulated by visNR in SU10/406 were not affected by added culture filtrate from stationary-phase cells, but the effects of added SinI AHLs were not tested. Thus, the role of visNR as AHL receptors remains uncertain.
(iv) traI, traR and melI
While strain 1021 lacks traI and traR, strain Rm41 (and its derivative AK631) has traI and traR next to plasmid transfer genes (Marketon & Gonzalez 2002). In addition, strain Rm41 has an AHL synthase encoded by melI (Marketon & Gonzalez 2002), which produces a range of AHLs with short acyl side chains in addition to the sinI-determined long-chain AHLs, including several in common with the SinI-made AHLs of strain 1021 (Teplitski et al. 2003).
(d) Rhizobium sp. NGR234
It appears that the transfer of the symbiotic plasmid of this strain has the potential to be under quorum-sensing regulation because traI and traR genes are on the plasmid pNGR234a. TraI synthesizes 3-oxo-C8-HSL, and two more AHLs have been detected in a traI mutant, indicating that the corresponding synthase(s) is encoded elsewhere in the genome (He et al. 2003). Quorum-sensing-regulated growth inhibition has also been reported in Rhizobium sp. NGR234. The inhibitory effect appears to be a result of TraR-dependent inhibition of growth in response to traI-made 3-oxo-C8-HSL. Addition of 3-oxo-C8-HSL to this strain leads to a significantly reduced growth rate. The inhibition requires traR and traI genes from pNGR234a and undefined genes elsewhere in the NGR234 genome (He et al. 2003).
(e) Mesorhizobium
Two LuxI-type AHL synthase genes are present in the genome of Mesorhizobium loti, one adjacent to a LuxR-type regulator gene and other genes that might be associated with conjugal transfer (http://www.kazusa.or.jp/rhizobase/); however, the effects of mutations have not yet been reported. A strain of Mesorhizobium huakuii has been shown to produce AHLs (Zhu et al. 2003) and reduced AHL production in this strain was correlated with the formation of a thinner biofilm (Wang et al. 2004).
(f) Bradyrhizobium
Approximately 20% of Bradyrhizobium japonicum and Bradyrhizobium elkanii strains tested using an AHL-detection bioassay made detectable amounts of AHLs (Pongsilp et al. 2005); but only a little has been done with regard to the identification of the AHL synthesis genes or the genes regulated by AHLs (Brelles-Marino & Bedmar 2001). However, in B. japonicum strain USDA110, the nodulation genes are expressed in a population-density-dependent manner and this is regulated by a factor in the growth medium called bradyoxetin. This compound was shown to be an iron chelator and so is probably not among the classic types of quorum-sensing regulation signals (Loh & Stacey 2001; Loh et al. 2002a).
Bradyoxetin is similar in structure to the siderophore mugineic acid (Loh et al. 2002a). The production of bradyoxetin is iron regulated, being maximal in iron-depleted conditions (Loh et al. 2002a; Loh & Stacey 2003). At high cell densities, the nod genes are repressed by a cascade initiated by the regulator NswB in response to high concentrations of bradyoxetin. NswB functions by inducing NolA that activates NodD2, which then represses the nod genes (Loh et al. 2001, 2002b). This repression of nod genes has been demonstrated in vitro and in planta (Loh et al. 2002b). Bradyoxetin has also been found in other Alphaproteobacteria (Loh et al. 2002a; Loh & Stacey 2003), but its role in these species is yet to be established.
3. Plasmid transfer
The paradigm for quorum-sensing control of plasmid transfer in the Rhizobiaceae was established in A. tumefaciens, which contain plasmids that carry genes for pathogenesis and transformation of plants (White & Winans 2007). Agrobacterium tumefaciens strains carrying the appropriate plasmids can induce crown gall disease on susceptible plant hosts. This disease is caused by the transfer of a part of the pathogenesis (Ti) plasmid into plant cells, where it recombines with the chromosome and initiates the production of a gall and metabolites (opines) that can be selectively metabolized by agrobacteria in the rhizosphere. The spread of this tumour-inducing (Ti) plasmid can allow horizontal transfer of the pathogenicity traits to other Agrobacterium strains lacking such characteristics (Zhu et al. 2000). The plasmid transfer genes on the Ti plasmid are strongly induced only in the presence of opines and only when the bacteria enter late exponential phase and are at an appropriate population density. This is achieved due to the action of the regulator TraR, which is transcriptionally induced by opines and activated by the accumulation of AHLs (principally 3-oxo-C8-HSL) produced by TraI (Piper et al. 1993; Zhang et al. 1993). TraR bound to the AHL has been crystallized and it is clear that the AHL is almost completely encased within the dimers of TraR (Zhang et al. 2002b). This TraR–AHL complex induces plasmid transfer operons and is much more stable to proteolytic degradation than TraR lacking the AHL (Zhu & Winans 2001). One of the induced operons contains traI as the first gene and so there is positive feedback that enhances the production of 3-oxo-C8-HSL and TraR-mediated gene induction (Zhu et al. 2000). Clearly, any such positive feedback system requires some kind of governor to prevent it from being induced prematurely. The traM gene product plays such a role: TraM forms a dimer (Chen et al. 2004; Qin et al. 2004) and binds to TraR–AHL preventing the induction of plasmid transfer operons. However, as the levels of AHL-activated TraR accumulate, these titrate out the available TraM and so can then initiate gene induction and the positive feedback effect on TraI-made AHLs.
It is clear that a similar quorum-sensing-based control of plasmid transfer is at the heart of the mechanisms of induction of transfer of several different plasmids in rhizobia. The systems best understood include various symbiotic plasmids such as pRL1JI from R. leguminosarum bv. viciae (Wilkinson et al. 2002; Danino et al. 2003), pNGR234 from Rhizobium sp. NGR234 (Freiberg et al. 1997; He et al. 2003) and pRme41a from S. meliloti (Marketon & Gonzalez 2002; Gonzalez & Marketon 2003), and non-symbiotic plasmids such as p42a from R. etli strain CFN42 can also use such a plasmid transfer induction system (Tun-Garrido et al. 2003). All of these systems share traI, traR and traM genes and use TraI-made 3-oxo-C8-HSL to activate TraR, although other AHLs such as C8-HSL and 3-OH-C8-HSL can also activate TraR (Wilkinson et al. 2002; Danino et al. 2003; Gonzalez & Marketon 2003). It is important to note that this is not the only mechanism of regulation of plasmid transfer in rhizobia; for example, R. etli strain CFN42 contains another plasmid whose regulation is mediated via two genes, rctA and rctB, and whose control appears to be independent of quorum-sensing regulation (Perez-Mendoza et al. 2005).
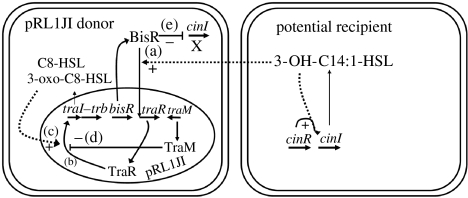
A critical part of the induction of plasmid transfer is the control of traR expression, because unless traR is induced, the quorum-sensing induction of plasmid transfer does not occur. In Agrobacterium strains, the traR genes are co-induced with the opine catabolism genes induced in response to the opines secreted from transformed plant cells (Kim & Farrand 1998; Zhu et al. 2000). The mechanism of regulation of traR expression in the rhizobial strains analysed is different. As shown in figure 1, on the R. leguminosarum bv. viciae plasmid pRL1JI, the expression of traR is under the control of a second LuxR-type regulator called BisR (bifunctional signalling regulator). The bisR gene is on pRL1JI upstream of traR (Wilkinson et al. 2002), and BisR specifically induces traR expression in response to 3-OH-C14:1-HSL (there is no induction of traR by TraI-made 3-oxo-C8-HSL or C8-HSL; Danino et al. 2003). The 3-OH-C14:1-HSL is made by another AHL synthase encoded by the chromosomal gene cinI, which is strongly expressed in most strains of R. leguminosarum and is under the control of CinR, which induces cinI expression in response to CinI-made 3-OH-C14:1-HSL (Lithgow et al. 2000). However, in those strains carrying pRL1JI, little or no 3-OH-C14:1-HSL is made because BisR represses cinI expression. Consequently, there is little induction of traR by BisR (because the appropriate AHL is not present), even though BisR is present in the cells. Any low-level induction of traR is inhibited by the production of TraM, which by analogy with A. tumefaciens TraM (Chen et al. 2004; Qin et al. 2004) titres out TraR (Danino et al. 2003). When bacteria carrying pRL1JI come close to other strains of R. leguminosarum that produce 3-OH-C14:1-HSL, BisR detects this AHL and induces traR resulting in enough TraR to titre out TraM. The result is strong induction of plasmid transfer genes under TraI–TraR quorum-sensing control. This system can result in very high rates of plasmid transfer (up to 1 transconjugant per 100 recipients) under optimal conditions (Danino et al. 2003). This recipient-induced mode of plasmid transfer relies on the bifunctional (inducer and repressor) nature of BisR and can promote the spread of pea and vetch nodulation characteristics into Rhizobium strains that do not have a plasmid that carries bisR and so does not repress cinI expression. Based on the identified genes (Tun-Garrido et al. 2003), it is probable that the non-symbiotic plasmid p42a in R. etli CFN42 has a similar mechanism of plasmid transfer, but this is yet to be demonstrated.
Figure 1.
Model for the regulation of the plasmid transfer traI–trb operon on pRL1JI in R. leguminosarum bv. viciae. The effects of AHLs on activation or inhibition of regulators are shown as dashed lines, and the inducing or repressing effects of regulators are shown as solid lines. (a) In cells that act as donors of pRL1JI, BisR induces traR in response to 3-OH-C14:1-HSL. This occurs predominantly in response to 3-OH-C14:1-HSL from potential recipients of pRL1JI. (b) Induction of traR produces TraR, which induces the traI–trb operon in response to 3-oxo-C8-HSL and C8-HSL. (c) There is autoinduction of the traI–trb operon by the TraI-made AHLs, 3-oxo-C8-HSL and C8-HSL. (d) Induction of the traI–trb operon by low levels of TraR is inhibited by TraM. The TraM-mediated repression of traI expression is overcome following induction of traR by BisR (a), in response to 3-OH-C14:1-HSL from potential recipients. (e) Endogenous production of 3-OH-C14:1-HSL is greatly reduced because BisR represses the expression of the chromosomal gene cinI, whose product produces 3-OH-C14:1-HSL. This repression enables the pRL1JI donors to respond primarily to potential recipient strains lacking bisR (and hence pRL1JI) but not to strains carrying pRL1JI. This figure was modified and reproduced (with permission) from a previous publication (Danino et al. 2003).
In contrast to the high rates of plasmid transfer observed with pRL1JI and p42a, only low rates of plasmid transfer (around 1 transconjugant per 107–109 recipients) have been observed with pNGR234 from Rhizobium sp. NGR234 (He et al. 2003) or pRme41a from S. meliloti strain Rm1021 (Gonzalez & Marketon 2003). The mechanism of regulation of transfer of these plasmids is different from pRL1JI because these plasmids lack a bisR gene (Freiberg et al. 1997). Possibly these low frequencies occur because optimal induction of traR has not been achieved, or possibly because some other component of the plasmid transfer system is not fully functional (He et al. 2003).
4. Responses of eukaryotic hosts to bacterial quorum sensing
Quorum-sensing mutants of bacterial pathogens such as P. aeruginosa and Erwinia carotovora have much reduced virulence in their plant and animal hosts (Winzer & Williams 2001; Von Bodman et al. 2003), while quorum-sensing mutants of Vibrio fisheri, the bioluminescent symbiont of squid (Lupp & Ruby 2004; Nyholm & McFall-Ngai 2004) and some rhizobia (see above) are impaired in establishing normal symbiotic interactions with their hosts. Thus, both bacterial pathogens and symbionts have come to rely on quorum sensing during interactions with their hosts. It seems reasonable to expect that host organisms, in turn, have evolved mechanisms for neutralizing, disrupting or manipulating this regulation in the bacteria they encounter. Recent studies provide evidence that eukaryotic hosts can indeed suppress or manipulate quorum sensing in a diversity of bacteria by synthesizing compounds that mimic the signals used by these bacteria. In addition, there is evidence that eukaryotes can detect bacterial quorum-sensing signals and make sophisticated responses to them, including altered regulatory, metabolic and defence responses. Both plant and animal hosts have also been shown to possess enzymes that rapidly and rather specifically inactivate bacterial AHL quorum-sensing signals. An examination of what has been learned about these capabilities of eukaryotic hosts may be valuable in appreciating the potential role of legume hosts in affecting AHL-mediated regulation in rhizobia.
(a) Eukaryotic agonists and antagonists of bacterial quorum sensing
Eukaryotic hosts that have no immune system often rely on the production of diverse secondary metabolites that help protect them from pests and pathogens. As described more fully in the accompanying review by Bjarnsholt & Givskov (2007), some very potent antifouling substances were recently found in a marine red alga, Delisea pulchra. This alga was remarkably free of the thick layer of bacteria and other organisms that colonize most biotic and abiotic surfaces in marine waters. The antifouling substances were identified as a set of approximately 30 different halogenated furanones. These furanones have structural similarity to AHLs and are potent and specific inhibitors of quorum sensing in many bacteria (Givskov et al. 1996; Manefield et al. 2000; Hentzer et al. 2002, 2003a,b; Hentzer & Givskov 2003; Martinelli et al. 2004). The furanones appear to interact directly with LuxR-type regulators and promote their proteolysis (Manefield et al. 1999, 2002; Koch et al. 2005), effectively preventing AHLs from activating gene expression. Mutations affecting AHL binding by LuxR had relatively little effect on inhibition by the furanones (Koch et al. 2005) suggesting that the furanones and AHLs may not compete for the same binding site.
Higher plants, including various legumes, rice, garlic and tomato, also secrete compounds that affect bacterial quorum sensing (Teplitski et al. 2000; Daniels et al. 2002; Gao et al. 2003; Keshavan et al. 2005; Rasmussen et al. 2005a; Bjarnsholt & Givskov 2007). Plant roots appear to secrete sufficient amounts of the active compounds to elicit changes in quorum-sensing-regulated gene expression in bacteria on the root surface (Teplitski et al. 2000). The set of plant-secreted effectors appears to change during seedling development, and bacteria may be exposed to a different set of AHL ‘mimic’ compounds once they get inside the root (Gao et al. 2003).
The active compounds from plants have not yet been chemically identified, so their mode of action and possible structural similarity to known quorum-sensing signals are unknown. However, the plant compounds were detected by their ability to activate or inhibit AHL-mediated gene induction in reporter bacteria that produce no AHLs. Thus, the active plant compounds appear to be substances that interact directly with AHL receptors in the reporters. Most of the active compounds secreted by pea and Medicago truncatula were soluble in methanol extracts of freeze-dried root exudates, but were less soluble or insoluble in ethyl acetate (Teplitski et al. 2000; Gao et al. 2003, in press) suggesting that these plant compounds are probably not AHLs. l-Canavanine, present in seed exudates of alfalfa, was shown to inhibit quorum-sensing-regulated gene expression in Chromobacterium and Sinorhizobium (Keshavan et al. 2005). However, canavanine is an analogue of arginine that can be incorporated into proteins, thereby causing polypeptide misfolding and inhibiting bacterial growth. The effects of canavanine on quorum sensing in bacteria may therefore be an indirect and general result of protein misfolding on various transcriptional regulators, including AHL regulators.
In conventional screens, quorum-sensing antagonists are difficult to distinguish from compounds that inhibit growth or are toxic to reporter bacteria. A novel screening method that distinguishes between quorum-sensing inhibitors and toxic substances has been developed (Persson et al. 2005). Several plant extracts and off-the-shelf chemicals were found to inhibit quorum sensing (Rasmussen et al. 2005a), with garlic extract and 4-nitro-pyridine-N-oxide being the most potent. Transcriptome analysis indicated that both of these rather specifically affected quorum-sensing-regulated gene expression in P. aeruginosa (Rasmussen et al. 2005a). The same kind of screen for antagonists revealed that most of the 50 Penicillium species surveyed produced compounds that inhibited AHL-mediated quorum sensing (Rasmussen et al. 2005b). Penicillic acid and patulin were among the active compounds and both of these have structural similarity to the Delisea furanones. Thus, it appears that fungi, as well as plants and algae, produce compounds that can alter quorum sensing in the bacteria they encounter.
Similar to higher plants, the green alga Chlamydomonas secretes agonists and antagonists of both AHL- and AI-2-mediated quorum-sensing gene expression in bacteria (Teplitski et al. 2004). A purified Chlamydomonas compound rather specifically targeted quorum sensing-regulated protein accumulation in S. meliloti. This agonist, like S. meliioti's own AHLs, stimulated the accumulation of some proteins; however, it also prevented AHL-induced accumulation of other proteins (Teplitski et al. 2004). This suggests that individual agonist or antagonist compounds may have markedly different effects on different AHL-regulated functions.
Another AHL-receptor agonist from Chlamydomonas has been chemically identified as lumichrome, which is a normal degradation product of the vitamin riboflavin (W. D. Bauer et al. 2007, unpublished data). Interestingly, lumichrome had previously been identified as a compound secreted by S. meliloti that was able to stimulate root respiration and shoot growth in alfalfa (Phillips et al. 1999). Thus, lumichrome may contribute to signalling and regulation in both S. meliloti and its legume hosts during symbiotic interactions.
Most of the active compounds detected after HPLC fractionation of pea, M. truncatula and Chlamydomonas exudates affected only one or two of the half-dozen reporter strains used. Both the plant and algal compounds therefore appear to interact rather specifically with certain AHL receptors and not others. This is in contrast with the ability of the Delisea furanones to inhibit regulation mediated by most (but not all) of the AHL regulators tested. Many of the quorum-sensing active compounds produced by plants and algae may act by a different mechanism than the furanones.
Pea, M. truncatula and Chlamydomonas make several chromatographically separable compounds that act on specific AHL or AI-2 reporters. For example, Chlamydomonas produces about half a dozen substances that stimulate the LasR AHL receptor and produces a different half-dozen compounds that stimulate the CepR AHL receptor from Burkholderia (Teplitski et al. 2004). Why should eukaryotic hosts produce several compounds that affect the same receptor? The answer may lie in natural variation in binding specificity among the AHL receptors. Since different bacteria are likely to have receptors that differ somewhat in amino acid sequences and binding specificity, their hosts may need to produce several structural variants of their active compounds to ensure that at least one will act strongly in each of the different bacteria they encounter. If host organisms do employ a ‘shotgun’ approach like this to disrupt quorum sensing, how do highly co-evolved symbionts such as rhizobia avoid being hit? One possibility is that rhizobia may use signals such as the long-chain AHLs so that receptors for these signals are not much affected by ‘signal-mimic’ compounds that target other bacteria. Plant-made metabolites that influence quorum-sensing gene expression are described in the accompanying article by Bjarnsholt & Givskov (2007).
Most of the plant compounds so far detected in root exudates by reporter strains are agonists rather than antagonists of AHL signalling (Teplitski et al. 2000; Daniels et al. 2002; Gao et al. 2003, in press). Biologically, the ability of plant hosts to specifically stimulate gene expression in the bacteria they encounter greatly increases the potential for sophisticated host manipulation of bacterial behaviour. Instead of simply preventing the activation of gene expression by bacterial quorum-sensing signals, the stimulatory plant compounds could enable the host directly to induce bacterial genes, inducing changes that are beneficial to the host. The host may have a limited window of opportunity to influence gene expression when it makes first contact with low numbers of bacteria that are not yet quorate. One of the central questions regarding the role of quorum sensing in the Rhizobium–legume symbiosis is whether compounds produced by the plant host serve to stimulate events such as infection initiation and bacteroid formation and, perhaps, block others such as unrestrained bacterial multiplication within the host.
(b) Responses of hosts to bacterial quorum-sensing signals
Exposure of M. truncatula roots to physiological (nanomolar) concentrations of bacterial AHLs significantly altered the accumulation of over 7% of the root proteins resolved by two-dimensional gels (Mathesius et al. 2003). These results suggest that plants ‘listen’ very actively to bacterial conversations. The global responsiveness of the plant to bacterial AHL signals was unexpected and the biological consequences of the responses are not at all clear yet. Various proteins related to host defences, hormones, regulation, metabolism, protein processing and cytoskeleton were reduced in level, while others increased. Certain proteins changed levels in response to one AHL but not another, and many changes were time dependent (Mathesius et al. 2003). In addition, some of the responses to bacterial signals are tissue specific while others are systemic (Hartmann et al. 2003; Mathesius et al. 2003; U. Mathesius 2006, unpublished data).
If both the bacteria and the hosts can respond to the same signals, then the role of quorum sensing in modulating the outcome of interactions becomes much more complex. Animals respond in a variety of ways to AHL signals including modulation of immune responses, blood-vessel relaxation, selective apoptosis and chemotaxis (Lawrence et al. 1999; Joint et al. 2002; Smith et al. 2002; Williams 2002). Thus, eukaryotic responses to quorum-sensing signals are likely to have been established for a considerable time in host–bacterium interactions. As yet, no ‘receptors’ for the signalling molecules have been identified in any eukaryote and there have been no studies to establish the consequences of defective host perception of bacterial quorum-sensing signals on the outcome of host–bacterial interactions.
One can imagine that natural selection might favour the development of additional feedback layers of related interactions between specific partners. For example, perception of certain quorum-sensing signals might induce a host to produce a different set of agonistic or antagonistic signals (Mathesius et al. 2003). Alternatively, compounds from the host may stimulate or inhibit the synthesis of quorum-sensing signals by the bacterium. The antagonism between Candida albicans and P. aeruginosa provides an interesting example of such feedback-layered responses. Both of these microbes are opportunistic pathogens frequently co-cultured from cystic fibrosis patients. P. aeruginosa 3-oxo-C12-HSL inhibits the dimorphic shift of C. albicans from yeast-to-mycelial forms (Hogan et al. 2004), thus mimicking the action of farnesol, the yeast's own signal. In turn, farnesol is able to strongly suppress AHL synthesis in P. aeruginosa (J. Robinson 2006, unpublished data).
Another potentially important facet of host responses to quorum sensing is the ability of enzymes in both plants and animals to inactivate AHL signals (Chun et al. 2004; Delalande et al. 2005; Yang et al. 2005; Dong et al. 2007). If the primary function of these enzymes proves to be the disruption of quorum sensing in bacteria or modulation of host responses to related bacterial signals, then the production, location and specificity of the signal-degrading enzymes become rather important factors in the pattern of reciprocal action and reaction between hosts and bacteria.
5. Perspectives, questions and conclusions
Various rhizobia establish fairly effective symbioses even when genes for AHL synthases and certain AHL receptors are mutated. Thus, rhizobia do not appear to be as dependent as many bacterial pathogens on quorum sensing for successful interactions with their hosts. At present, it appears that quorum-sensing-regulated functions in the bacterium and host serve primarily to optimize various interactions between the partners; for example, the enhancement of infection initiation by R. leguminosarum (Cubo et al. 1992) and S. meliloti (Gao et al. 2005) and the enhancement of the number of R. etli bacteroids in symbiosomes (Daniels et al. 2002). There are many opportunities for both the host and the bacterium to modify the levels of many symbiotically relevant genes and proteins in both partners. This has been artificially done in relation to plant pathogenesis using transgenic plants expressing AHL-degrading enzymes; as reviewed in the accompanying article by Dong et al. (2007), such plants have increased resistance to some bacterial pathogens. The induction of systemic resistance dependent on quorum sensing in a bacterial pathogen illustrates the types of changes that can occur naturally in plant–bacterial interactions (Schuhegger et al. 2006). Furthermore, it has been shown that a bacterial AHL synthase targeted to potato and tobacco plastids can result in transgenic plants that produce AHLs, which can be released into the rhizosphere and can influence interactions with pathogens (Fray et al. 1999; Toth et al. 2004; Scott et al. 2006). In principle, similar experiments with transgenic legumes carrying genes to produce or degrade AHLs could be used to analyse the role of quorum sensing in their interactions with rhizobia.
In addition to a role in optimizing interactions with a host, quorum sensing in rhizobia may yet prove to be one of the required facets of successful symbiosis. All the rhizobia tested so far have additional LuxR-type regulators that have not been mutated and so the roles of quorum sensing in regulating some crucial aspects of the symbiosis are yet to be fully explored. For example, while AHLs inhibit cell division in R. leguminosarum and affect expression of cell division related genes in S. meliloti, the regulation of rhizobial cell division in planta is still unclear either when bacterial replication stops in mature nodules or reinitiates in senescing nodules. It will be interesting to learn from future studies whether all the diverse strategies for quorum-sensing regulation seen among different isolates and strains of rhizobia contain some elements in common that are required to establish and maintain mutually beneficial relations with their legume partners.
Acknowledgments
M.S.-C and J.A.D. are supported by a grant-in-aid and a grant (208/P19980) from the BBSRC. We thank Paul Williams for his comments on the manuscript. Unpublished studies described in this review by W.D.B., J.B.R. and M.G. were supported by USDA grants NRI 2003-01177 and 2005-35319-15318.
Footnotes
One contribution of 12 to a Theme Issue ‘Bacterial conversations: talking, listening and eavesdropping’.
References
- Barnard A.M.L, Bowden S.D, Burr T, Coulthurst S.J, Monson R.E, Salmond G.P.C. Quorum sensing, virulence and secondary metabolite production in plant soft-rotting bacteria. Phil. Trans. R. Soc. B. 2007;362:1165–1183. doi: 10.1098/rstb.2007.2042. doi:10.1098/rstb.2007.2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler B.L. Small talk. Cell-to-cell communication in bacteria. Cell. 2002;109:421–424. doi: 10.1016/s0092-8674(02)00749-3. doi:10.1016/S0092-8674(02)00749-3 [DOI] [PubMed] [Google Scholar]
- Bassler B.L, Wright M, Silverman M.R. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. doi:10.1111/j.1365-2958.1994.tb00422.x [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Givskov M. Quorum-sensing blockade as a strategy for enhancing host defences against bacterial pathogens. Phil. Trans. R. Soc. B. 2007;362:1213–1222. doi: 10.1098/rstb.2007.2046. doi:10.1098/rstb.2007.2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelles-Marino G, Bedmar E.J. Detection, purification and characterisation of quorum-sensing signal molecules in plant-associated bacteria. J. Biotechnol. 2001;91:197–209. doi: 10.1016/s0168-1656(01)00330-3. [DOI] [PubMed] [Google Scholar]
- Carlier A, Chevrot R, Dessaux Y, Faure D. The assimilation of gamma-butyrolactone in Agrobacterium tumefaciens C58 interferes with the accumulation of the N-acyl-homoserine lactone signal. Mol. Plant Microbe Interact. 2004;17:951–957. doi: 10.1094/MPMI.2004.17.9.951. [DOI] [PubMed] [Google Scholar]
- Cha C, Gao P, Chen Y.C, Shaw P.D, Farrand S.K. Production of acyl-homoserine lactone quorum-sensing signals by Gram-negative plant-associated bacteria. Mol. Plant Microbe Interact. 1998;11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- Chancey S.T, Wood D.W, Pierson L.S. Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 1999;65:2294–2299. doi: 10.1128/aem.65.6.2294-2299.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.C, Teplitski M, Robinson J.B, Rolfe B.G, Bauer W.D. Proteomic analysis of wild-type Sinorhizobium meliloti responses to N-acyl homoserine lactone quorum-sensing signals and the transition to stationary phase. J. Bacteriol. 2003;185:5029–5036. doi: 10.1128/JB.185.17.5029-5036.2003. doi:10.1128/JB.185.17.5029-5036.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Malenkos J.W, Cha M.R, Fuqua C, Chen L. Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR. Mol. Microbiol. 2004;52:1641–1651. doi: 10.1111/j.1365-2958.2004.04110.x. doi:10.1111/j.1365-2958.2004.04110.x [DOI] [PubMed] [Google Scholar]
- Chin-A-Woeng T.F.C, Bloemberg G.V, Lugtenberg B.J.J. Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol. 2003;157:503–523. doi: 10.1046/j.1469-8137.2003.00686.x. doi:10.1046/j.1469-8137.2003.00686.x [DOI] [PubMed] [Google Scholar]
- Chun C.K, Ozer E.A, Welsh M.J, Zabner J, Greenberg E.P. Inactivation of a Pseudomonas aeruginosa quorum-sensing signal by human airway epithelia. Proc. Natl Acad. Sci. USA. 2004;101:3587–3590. doi: 10.1073/pnas.0308750101. doi:10.1073/pnas.0308750101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubo M.T, Economou A, Murphy G, Johnston A.W, Downie J.A. Molecular characterization and regulation of the rhizosphere-expressed genes rhiABCR that can influence nodulation by Rhizobium leguminosarum biovar viciae. J. Bacteriol. 1992;174:4026–4035. doi: 10.1128/jb.174.12.4026-4035.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Angelo-Picard C, Faure D, Penot I, Dessaux Y. Diversity of N-acyl homoserine lactone-producing and -degrading bacteria in soil and tobacco rhizosphere. Environ. Microbiol. 2005;7:1796–1808. doi: 10.1111/j.1462-2920.2005.00886.x. doi:10.1111/j.1462-2920.2005.00886.x [DOI] [PubMed] [Google Scholar]
- Daniels R, et al. The cin quorum sensing locus of Rhizobium etli CNPAF512 affects growth and symbiotic nitrogen fixation. J. Biol. Chem. 2002;277:462–468. doi: 10.1074/jbc.M106655200. doi:10.1074/jbc.M106655200 [DOI] [PubMed] [Google Scholar]
- Daniels R, Vanderleyden J, Michiels J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 2004;28:261–289. doi: 10.1016/j.femsre.2003.09.004. doi:10.1016/j.femsre.2003.09.004 [DOI] [PubMed] [Google Scholar]
- Danino V.E, Wilkinson A, Edwards A, Downie J.A. Recipient-induced transfer of the symbiotic plasmid pRL1JI in Rhizobium leguminosarum bv. viciae is regulated by a quorum-sensing relay. Mol. Microbiol. 2003;50:511–525. doi: 10.1046/j.1365-2958.2003.03699.x. doi:10.1046/j.1365-2958.2003.03699.x [DOI] [PubMed] [Google Scholar]
- Delalande L, et al. N-hexanoyl-l-homoserine lactone, a mediator of bacterial quorum-sensing regulation, exhibits plant-dependent stability and may be inactivated by germinating Lotus corniculatus seedlings. FEMS Microbiol. Ecol. 2005;52:13–20. doi: 10.1016/j.femsec.2004.10.005. doi:10.1016/j.femsec.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Dibb N.J, Downie J.A, Brewin N.J. Identification of a rhizosphere protein encoded by the symbiotic plasmid of Rhizobium leguminosarum. J. Bacteriol. 1984;158:621–627. doi: 10.1128/jb.158.2.621-627.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere S, Vanderleyden J, Okon Y. Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit. Rev. Plant Sci. 2003;22:107–149. [Google Scholar]
- Dong Y.H, Wang L.H, Zhang L.H. Quorum-quenching microbial infections: mechanisms and implications. Phil. Trans. R. Soc. B. 2007;362:1201–1211. doi: 10.1098/rstb.2007.2045. doi:10.1098/rstb.2007.2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubern J.F, Lugtenberg B.J.J, Bloemberg G.V. The ppuI–rsaL–ppuR quorum-sensing system regulates biofilm formation of Pseudomonas putida PCL1445 by controlling biosynthesis of the cyclic lipopeptides putisolvins I and II. J. Bacteriol. 2006;188:2898–2906. doi: 10.1128/JB.188.8.2898-2906.2006. doi:10.1128/JB.188.8.2898-2906.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou A, Hawkins F.K, Downie J.A, Johnston A.W. Transcription of rhiA, a gene on a Rhizobium leguminosarum bv. viciae Sym plasmid, requires rhiR and is repressed by flavanoids that induce nod genes. Mol. Microbiol. 1989;3:87–93. doi: 10.1111/j.1365-2958.1989.tb00107.x. doi:10.1111/j.1365-2958.1989.tb00107.x [DOI] [PubMed] [Google Scholar]
- Elasri M, Delorme S, Lemanceau P, Stewart G, Laue B, Glickmann E, Oger P.M, Dessaux Y. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl. Environ. Microbiol. 2001;67:1198–1209. doi: 10.1128/AEM.67.3.1198-1209.2001. doi:10.1128/AEM.67.3.1198-1209.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fray R.G, Throup J.P, Daykin M, Wallace A, Williams P, Stewart G, Grierson D. Plants genetically modified to produce N-acylhomoserine lactones communicate with bacteria. Nat. Biotechnol. 1999;17:1017–1020. doi: 10.1038/13717. doi:10.1038/13717 [DOI] [PubMed] [Google Scholar]
- Freiberg C, Fellay R, Bairoch A, Broughton W.J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. doi:10.1038/387394a0 [DOI] [PubMed] [Google Scholar]
- Fuqua C, Greenberg E.P. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr. Opin. Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. doi:10.1016/S1369-5274(98)80009-X [DOI] [PubMed] [Google Scholar]
- Fuqua W.C, Winans S.C, Greenberg E.P. Quorum sensing in bacteria: the LuxR–LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C, Parsek M.R, Greenberg E.P. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. doi:10.1146/annurev.genet.35.102401.090913 [DOI] [PubMed] [Google Scholar]
- Gao M, Teplitski M, Robinson J.B, Bauer W.D. Production of substances by Medicago truncatula that affect bacterial quorum sensing. Mol. Plant Microbe Interact. 2003;16:827–834. doi: 10.1094/MPMI.2003.16.9.827. [DOI] [PubMed] [Google Scholar]
- Gao M, Chen H, Eberhard A, Gronquist M.R, Robinson J.B, Rolfe B.G, Bauer W.D. sinI- and expR-dependent quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 2005;187:7931–7944. doi: 10.1128/JB.187.23.7931-7944.2005. doi:10.1128/JB.187.23.7931-7944.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., Chen, H., Eberhard, A., Gronquist, M. R., Robinson, J. B., Connolly, M., Teplitski, M., Rolfe, B. G. & Bauer, W. D. In press. Effects of AiiA-mediated quorum quenching in Sinorhizobium meliloti on quorum sensing signals, proteome patterns and symbiotic interactions. Mol. Plant Microbe Interact [DOI] [PubMed]
- Gilson L, Kuo A, Dunlap P.V. AinS and a new family of autoinducer synthesis proteins. J. Bacteriol. 1995;177:6946–6951. doi: 10.1128/jb.177.23.6946-6951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard G, van Rij E.T, Lugtenberg B.J.J, Bloemberg G.V. Regulatory roles of psrA and rpoS in phenazine-1-carboxamide synthesis by Pseudomonas chlororaphis PCL1391. Microbiology. 2006;152:43–58. doi: 10.1099/mic.0.28284-0. doi:10.1099/mic.0.28284-0 [DOI] [PubMed] [Google Scholar]
- Givskov M, de Nys R, Manefield M, Gram L, Maximilien R, Eberl L, Molin S, Steinberg P.D, Kjelleberg S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol. 1996;178:6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J.E, Marketon M.M. Quorum sensing in nitrogen-fixing rhizobia. Microbiol. Mol. Biol. Rev. 2003;67:574–592. doi: 10.1128/MMBR.67.4.574-592.2003. doi:10.1128/MMBR.67.4.574-592.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J.E, Reuhs B.L, Walker G.C. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc. Natl Acad. Sci. USA. 1996;93:8636–8641. doi: 10.1073/pnas.93.16.8636. doi:10.1073/pnas.93.16.8636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray K.M. Intercellular communication and group behavior in bacteria. Trends Microbiol. 1997;5:184–188. doi: 10.1016/S0966-842X(97)01002-0. doi:10.1016/S0966-842X(97)01002-0 [DOI] [PubMed] [Google Scholar]
- Gray K.M, Pearson J.P, Downie J.A, Boboye B.E, Greenberg E.P. Cell-to-cell signaling in the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum: autoinduction of a stationary phase and rhizosphere-expressed genes. J. Bacteriol. 1996;178:372–376. doi: 10.1128/jb.178.2.372-376.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, A., Gantner, S., Schuhegger, R., Steidle, A., Durrl, C., Schmid, M., Langebartels, C., Dazzo, F. B. & Eberl, L. 2003 Presented at the Molecular Plant-Microbe Interactions Symposium, St. Petersburg, Russia.
- He X, Chang W, Pierce D.L, Seib L.O, Wagner J, Fuqua C. Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J. Bacteriol. 2003;185:809–822. doi: 10.1128/JB.185.3.809-822.2003. doi:10.1128/JB.185.3.809-822.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M, Givskov M. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Invest. 2003;112:1300–1307. doi: 10.1172/JCI20074. doi:10.1172/JCI200320074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer M, et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology. 2002;148:87–102. doi: 10.1099/00221287-148-1-87. [DOI] [PubMed] [Google Scholar]
- Hentzer M, Eberl L, Nielsen J, Givskov M. Quorum sensing: a novel target for the treatment of biofilm infections. BioDrugs. 2003a;17:241–250. doi: 10.2165/00063030-200317040-00003. doi:10.2165/00063030-200317040-00003 [DOI] [PubMed] [Google Scholar]
- Hentzer M, et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003b;22:3803–3815. doi: 10.1093/emboj/cdg366. doi:10.1093/emboj/cdg366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang H.H, Becker A, Gonzalez J.E. The LuxR homolog ExpR, in combination with the Sin quorum sensing system, plays a central role in Sinorhizobium meliloti gene expression. J. Bacteriol. 2004;186:5460–5472. doi: 10.1128/JB.186.16.5460-5472.2004. doi:10.1128/JB.186.16.5460-5472.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D.A, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 2004;54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. doi:10.1111/j.1365-2958.2004.04349.x [DOI] [PubMed] [Google Scholar]
- Hwang I, Cook D.M, Farrand S.K. A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J. Bacteriol. 1995;177:449–458. doi: 10.1128/jb.177.2.449-458.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joint I, Tait K, Callow M.E, Callow J.A, Milton D, Williams P, Camara M. Cell-to-cell communication across the prokaryote–eukaryote boundary. Science. 2002;298:1207. doi: 10.1126/science.1077075. doi:10.1126/science.1077075 [DOI] [PubMed] [Google Scholar]
- Keshavan N.D, Chowdhary P.K, Haines D.C, Gonzalez J.E. l-Canavanine made by Medicago sativa interferes with quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 2005;187:8427–8436. doi: 10.1128/JB.187.24.8427-8436.2005. doi:10.1128/JB.187.24.8427-8436.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Farrand S.K. Opine catabolic loci from Agrobacterium plasmids confer chemotaxis to their cognate substrates. Mol. Plant Microbe Interact. 1998;11:131–143. doi: 10.1094/MPMI.1998.11.2.131. [DOI] [PubMed] [Google Scholar]
- Koch B, Liljefors T, Persson T, Nielsen J, Kjelleberg S, Givskov M. The LuxR receptor: the sites of interaction with quorum-sensing signals and inhibitors. Microbiology. 2005;151:3589–3602. doi: 10.1099/mic.0.27954-0. doi:10.1099/mic.0.27954-0 [DOI] [PubMed] [Google Scholar]
- Kuiper I, Lagendijk E.L, Bloemberg G.V, Lugtenberg B.J.J. Rhizoremediation: a beneficial plant-microbe interaction. Mol. Plant Microbe Interact. 2004;17:6–15. doi: 10.1094/MPMI.2004.17.1.6. [DOI] [PubMed] [Google Scholar]
- Lawrence R.N, Dunn W.R, Bycroft B, Camara M, Chhabra S.R, Williams P, Wilson V.G. The Pseudomonas aeruginosa quorum-sensing signal molecule, N-(3-oxododecanoyl)-l-homoserine lactone, inhibits porcine arterial smooth muscle contraction. Br. J. Pharmacol. 1999;128:845–848. doi: 10.1038/sj.bjp.0702870. doi:10.1038/sj.bjp.0702870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazdunski A.M, Ventre I, Sturgis J.N. Regulatory circuits and communication in Gram-negative bacteria. Nat. Rev. Microbiol. 2004;2:581–592. doi: 10.1038/nrmicro924. doi:10.1038/nrmicro924 [DOI] [PubMed] [Google Scholar]
- Ledgham F, Ventre I, Soscia C, Foglino M, Sturgis J.N, Lazdunski A. Interactions of the quorum sensing regulator QscR: interaction with itself and the other regulators of Pseudomonas aeruginosa LasR and RhlR. Mol. Microbiol. 2003;48:199–210. doi: 10.1046/j.1365-2958.2003.03423.x. doi:10.1046/j.1365-2958.2003.03423.x [DOI] [PubMed] [Google Scholar]
- Lindum P.W, Anthoni U, Christophersen C, Eberl L, Molin S, Givskov M. N-acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 1998;180:6384–6388. doi: 10.1128/jb.180.23.6384-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow J.K, Wilkinson A, Hardman A, Rodelas B, Wisniewski-Dye F, Williams P, Downie J.A. The regulatory locus cinRI in Rhizobium leguminosarum controls a network of quorum-sensing loci. Mol. Microbiol. 2000;37:81–97. doi: 10.1046/j.1365-2958.2000.01960.x. doi:10.1046/j.1365-2958.2000.01960.x [DOI] [PubMed] [Google Scholar]
- Llamas I, Keshavan N, Gonzalez J.E. Use of Sinorhizobium meliloti as an indicator for specific detection of long-chain N-acyl homoserine lactones. Appl. Environ. Microbiol. 2004;70:3715–3723. doi: 10.1128/AEM.70.6.3715-3723.2004. doi:10.1128/AEM.70.6.3715-3723.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh J.T, Stacey G. Feedback regulation of the Bradyrhizobium japonicum nodulation genes. Mol. Microbiol. 2001;41:1357–1364. doi: 10.1046/j.1365-2958.2001.02603.x. doi:10.1046/j.1365-2958.2001.02603.x [DOI] [PubMed] [Google Scholar]
- Loh J, Stacey G. Nodulation gene regulation in Bradyrhizobium japonicum: a unique integration of global regulatory circuits. Appl. Environ. Microbiol. 2003;69:10–17. doi: 10.1128/AEM.69.1.10-17.2003. doi:10.1128/AEM.69.1.10-17.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh J.T, Yuen-Tsai J.P, Stacey M.G, Lohar D, Welborn A, Stacey G. Population density-dependent regulation of the Bradyrhizobium japonicum nodulation genes. Mol. Microbiol. 2001;42:37–46. doi: 10.1046/j.1365-2958.2001.02625.x. doi:10.1046/j.1365-2958.2001.02625.x [DOI] [PubMed] [Google Scholar]
- Loh J, Carlson R.W, York W.S, Stacey G. Bradyoxetin, a unique chemical signal involved in symbiotic gene regulation. Proc. Natl Acad. Sci. USA. 2002a;99:14 446–14 451. doi: 10.1073/pnas.222336799. doi:10.1073/pnas.222336799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh J, Lohar D.P, Andersen B, Stacey G. A two-component regulator mediates population-density-dependent expression of the Bradyrhizobium japonicum nodulation genes. J. Bacteriol. 2002b;184:1759–1766. doi: 10.1128/JB.184.6.1759-1766.2002. doi:10.1128/JB.184.6.1759-1766.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh J, Pierson E.A, Pierson L.S, 3rd, Stacey G, Chatterjee A. Quorum sensing in plant-associated bacteria. Curr. Opin. Plant Biol. 2002c;5:285–290. doi: 10.1016/s1369-5266(02)00274-1. doi:10.1016/S1369-5266(02)00274-1 [DOI] [PubMed] [Google Scholar]
- Lugtenberg B.J.J, Dekkers L, Bloemberg G.V. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 2001;39:461–490. doi: 10.1146/annurev.phyto.39.1.461. doi:10.1146/annurev.phyto.39.1.461 [DOI] [PubMed] [Google Scholar]
- Lupp C, Ruby E.G. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 2004;186:3873–3881. doi: 10.1128/JB.186.12.3873-3881.2004. doi:10.1128/JB.186.12.3873-3881.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield M, de Nys R, Kumar N, Read R, Givskov M, Steinberg P, Kjelleberg S. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology. 1999;145(Pt 2):283–291. doi: 10.1099/13500872-145-2-283. [DOI] [PubMed] [Google Scholar]
- Manefield M, Harris L, Rice S.A, de Nys R, Kjelleberg S. Inhibition of luminescence and virulence in the black tiger prawn (Penaeus monodon) pathogen Vibrio harveyi by intercellular signal antagonists. Appl. Environ. Microbiol. 2000;66:2079–2084. doi: 10.1128/aem.66.5.2079-2084.2000. doi:10.1128/AEM.66.5.2079-2084.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield M, Rasmussen T.B, Henzter M, Andersen J.B, Steinberg P, Kjelleberg S, Givskov M. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology. 2002;148:1119–1127. doi: 10.1099/00221287-148-4-1119. [DOI] [PubMed] [Google Scholar]
- Marketon M.M, Gonzalez J.E. Identification of two quorum-sensing systems in Sinorhizobium meliloti. J. Bacteriol. 2002;184:3466–3475. doi: 10.1128/JB.184.13.3466-3475.2002. doi:10.1128/JB.184.13.3466-3475.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marketon M.M, Gronquist M.R, Eberhard A, Gonzalez J.E. Characterization of the Sinorhizobium meliloti sinR/sinI locus and the production of novel N-acyl homoserine lactones. J. Bacteriol. 2002;184:5686–5695. doi: 10.1128/JB.184.20.5686-5695.2002. doi:10.1128/JB.184.20.5686-5695.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marketon M.M, Glenn S.A, Eberhard A, Gonzalez J.E. Quorum sensing controls exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 2003;185:325–331. doi: 10.1128/JB.185.1.325-331.2003. doi:10.1128/JB.185.1.325-331.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli D, Grossmann G, Sequin U, Brandl H, Bachofen R. Effects of natural and chemically synthesized furanones on quorum sensing in Chromobacterium violaceum. BMC Microbiol. 2004;4:25. doi: 10.1186/1471-2180-4-25. doi:10.1186/1471-2180-4-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U, Mulders S, Gao M, Teplitski M, Caetano-Anolles G, Rolfe B.G, Bauer W.D. Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc. Natl Acad. Sci. USA. 2003;100:1444–1449. doi: 10.1073/pnas.262672599. doi:10.1073/pnas.262672599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean K.H, et al. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143(Pt 12):3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- McKenney D, Brown K.E, Allison D.G. Influence of Pseudomonas aeruginosa exoproducts on virulence factor production in Burkholderia cepacia: evidence of interspecies communication. J. Bacteriol. 1995;177:6989–6992. doi: 10.1128/jb.177.23.6989-6992.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton D.L, Chalker V.J, Kirke D, Hardman A, Camara M, Williams P. The luxM homologue vanM from Vibrio anguillanrum directs the synthesis of N-(3-hydroxyhexanoyl)homoserine lactone and N-hexanoylhomoserine lactone. J. Bacteriol. 2001;183:3537–3547. doi: 10.1128/JB.183.12.3537-3547.2001. doi:10.1128/JB.183.12.3537-3547.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- More M.I, Finger L.D, Stryker J.L, Fuqua C, Eberhard A, Winans S.C. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. doi:10.1126/science.272.5268.1655 [DOI] [PubMed] [Google Scholar]
- Nyholm S.V, McFall-Ngai M.J. The winnowing: establishing the squid–vibrio symbiosis. Nat. Rev. Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. doi:10.1038/nrmicro957 [DOI] [PubMed] [Google Scholar]
- Parsek M.R, Greenberg E.P. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl Acad. Sci. USA. 2000;97:8789–8793. doi: 10.1073/pnas.97.16.8789. doi:10.1073/pnas.97.16.8789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellock B.J, Teplitski M, Boinay R.P, Bauer W.D, Walker G.C. A LuxR homolog controls production of symbiotically active extracellular polysaccharide II by Sinorhizobium meliloti. J. Bacteriol. 2002;184:5067–5076. doi: 10.1128/JB.184.18.5067-5076.2002. doi:10.1128/JB.184.18.5067-5076.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Mendoza D, et al. Identification of the rctA gene, which is required for repression of conjugative transfer of rhizobial symbiotic megaplasmids. J. Bacteriol. 2005;187:7341–7350. doi: 10.1128/JB.187.21.7341-7350.2005. doi:10.1128/JB.187.21.7341-7350.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson T, Hansen T.H, Rasmussen T.B, Skinderso M.E, Givskov M, Nielsen J. Rational design and synthesis of new quorum-sensing inhibitors derived from acylated homoserine lactones and natural products from garlic. Org. Biomol. Chem. 2005;3:253–262. doi: 10.1039/b415761c. doi:10.1039/b415761c [DOI] [PubMed] [Google Scholar]
- Phillips D.A, Joseph C.M, Yang G.P, Martinez-Romero E, Sanborn J.R, Volpin H. Identification of lumichrome as a sinorhizobium enhancer of alfalfa root respiration and shoot growth. Proc. Natl Acad. Sci. USA. 1999;96:12 275–12 280. doi: 10.1073/pnas.96.22.12275. doi:10.1073/pnas.96.22.12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson L.S, Pierson E.A. Phenazine antibiotic production in Pseudomonas aureofaciens: role in rhizosphere ecology and pathogen suppression. FEMS Microbiol. Lett. 1996;136:101–108. doi:10.1111/j.1574-6968.1996.tb08034.x [Google Scholar]
- Pierson E.A, Wood D.W, Cannon J.A, Blachere F.M, Pierson L.S. Interpopulation signaling via N-acyl-homoserine lactones among bacteria in the wheat rhizosphere. Mol. Plant Microbe Interact. 1998a;11:1078–1084. [Google Scholar]
- Pierson L.S, Wood D.W, Pierson E.A. Homoserine lactone-mediated gene regulation in plant-associated bacteria. Annu. Rev. Phytopathol. 1998b;36:207–225. doi: 10.1146/annurev.phyto.36.1.207. doi:10.1146/annurev.phyto.36.1.207 [DOI] [PubMed] [Google Scholar]
- Piper K.R, Beck von Bodman S, Farrand S.K. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature. 1993;362:448–450. doi: 10.1038/362448a0. doi:10.1038/362448a0 [DOI] [PubMed] [Google Scholar]
- Pongsilp N, Triplett E.W, Sadowsky M.J. Detection of homoserine lactone-like quorum sensing molecules in bradyrhizobium strains. Curr. Microbiol. 2005;51:250–254. doi: 10.1007/s00284-005-4550-5. doi:10.1007/s00284-005-4550-5 [DOI] [PubMed] [Google Scholar]
- Qin Y, Smyth A.J, Su S, Farrand S.K. Dimerization properties of TraM, the antiactivator that modulates TraR-mediated quorum-dependent expression of the Ti plasmid tra genes. Mol. Microbiol. 2004;53:1471–1485. doi: 10.1111/j.1365-2958.2004.04216.x. doi:10.1111/j.1365-2958.2004.04216.x [DOI] [PubMed] [Google Scholar]
- Ramey B.E, Koutsoudis M, von Bodman S.B, Fuqua C. Biofilm formation in plant-microbe associations. Curr. Opin. Microbiol. 2004;7:602–609. doi: 10.1016/j.mib.2004.10.014. doi:10.1016/j.mib.2004.10.014 [DOI] [PubMed] [Google Scholar]
- Rasmussen T.B, Bjarnsholt T, Skindersoe M.E, Hentzer M, Kristoffersen P, Kote M, Nielsen J, Eberl L, Givskov M. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 2005a;187:1799–1814. doi: 10.1128/JB.187.5.1799-1814.2005. doi:10.1128/JB.187.5.1799-1814.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T.B, et al. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology. 2005b;151:1325–1340. doi: 10.1099/mic.0.27715-0. doi:10.1099/mic.0.27715-0 [DOI] [PubMed] [Google Scholar]
- Riedel K, et al. N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology. 2001;147:3249–3262. doi: 10.1099/00221287-147-12-3249. [DOI] [PubMed] [Google Scholar]
- Rodelas B, Lithgow J.K, Wisniewski-Dye F, Hardman A, Wilkinson A, Economou A, Williams P, Downie J.A. Analysis of quorum-sensing-dependent control of rhizosphere-expressed (rhi) genes in Rhizobium leguminosarum bv. viciae. J. Bacteriol. 1999;181:3816–3823. doi: 10.1128/jb.181.12.3816-3823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosemeyer V, Michiels J, Verreth C, Vanderleyden J. luxI- and luxR-homologous genes of Rhizobium etli CNPAF512 contribute to synthesis of autoinducer molecules and nodulation of Phaseolus vulgaris. J. Bacteriol. 1998;180:815–821. doi: 10.1128/jb.180.4.815-821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmond G.P, Bycroft B.W, Stewart G.S, Williams P. The bacterial ‘enigma’: cracking the code of cell–cell communication. Mol. Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. doi:10.1111/j.1365-2958.1995.tb02424.x [DOI] [PubMed] [Google Scholar]
- Schaefer A.L, Hanzelka B.L, Eberhard A, Greenberg E.P. Quorum sensing in Vibrio fischeri: probing autoinducer–LuxR interactions with autoinducer analogs. J. Bacteriol. 1996;178:2897–2901. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schripsema J, deRudder K.E.E, vanVliet T.B, Lankhorst P.P, deVroom E, Kijne J.W, vanBrussel A.A.N. Bacteriocin small of Rhizobium leguminosarium belongs to the class of N-acyl-l-homoserine lactone molecules, known as autoinducers and as quorum sensing co-transcription factors. J. Bacteriol. 1996;178:366–371. doi: 10.1128/jb.178.2.366-371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhegger R, et al. Induction of systemic resistance in tomato by N-acyl-l-homoserine lactone-producing bacteria. Plant Cell Environ. 2006;29:909–918. doi: 10.1111/j.1365-3040.2005.01471.x. doi:10.1111/j.1365-3040.2005.01471.x [DOI] [PubMed] [Google Scholar]
- Scott R.A, Weil J, Le P.T, Williams P, Fray R.G, von Bodman S.B, Savka M.A. Long- and short-chain plant-produced bacterial N-acyl-homoserine lactones become components of phyllosphere, rhizosphere, and soil. Mol. Plant Microbe Interact. 2006;19:227–239. doi: 10.1094/MPMI-19-0227. [DOI] [PubMed] [Google Scholar]
- Shaw P.D, Ping G, Daly S.L, Cha C, Cronan J.E, Jr, Rinehart K.L, Farrand S.K. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl Acad. Sci. USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. doi:10.1073/pnas.94.12.6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.S, Harris S.G, Phipps R, Iglewski B. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J. Bacteriol. 2002;184:1132–1139. doi: 10.1128/jb.184.4.1132-1139.2002. doi:10.1128/jb.184.4.1132-1139.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Muschler P, Scharf B, Schmitt R. VisN and VisR are global regulators of chemotaxis, flagellar, and motility genes in Sinorhizobium (Rhizobium) meliloti. J. Bacteriol. 2000;182:782–788. doi: 10.1128/jb.182.3.782-788.2000. doi:10.1128/JB.182.3.782-788.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidle A, et al. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 2001;67:5761–5770. doi: 10.1128/AEM.67.12.5761-5770.2001. doi:10.1128/AEM.67.12.5761-5770.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidle A, Allesen-Holm M, Riedel K, Berg G, Givskov M, Molin S, Eberl L. Identification and characterization of an N-acylhomoserine lactone-dependent quorum-sensing system in Pseudomonas putida strain IsoF. Appl. Environ. Microbiol. 2002;68:6371–6382. doi: 10.1128/AEM.68.12.6371-6382.2002. doi:10.1128/AEM.68.12.6371-6382.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S, Throup J.P, Williams P, Salmond G.P, Stewart G.S. Quorum sensing: a population-density component in the determination of bacterial phenotype. Trends Biochem. Sci. 1996;21:214–219. doi:10.1016/0968-0004(96)10027-X [PubMed] [Google Scholar]
- Teplitski M, Robinson J.B, Bauer W.D. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant Microbe Interact. 2000;13:637–648. doi: 10.1094/MPMI.2000.13.6.637. [DOI] [PubMed] [Google Scholar]
- Teplitski M, Eberhard A, Gronquist M.R, Gao M, Robinson J.B, Bauer W.D. Chemical identification of N-acyl homoserine lactone quorum-sensing signals produced by Sinorhizobium meliloti strains in defined medium. Arch. Microbiol. 2003;180:494–497. doi: 10.1007/s00203-003-0612-x. doi:10.1007/s00203-003-0612-x [DOI] [PubMed] [Google Scholar]
- Teplitski M, Chen H, Rajamani S, Gao M, Merighi M, Sayre R.T, Robinson J.B, Rolfe B.G, Bauer W.D. Chlamydomonas reinhardtii secretes compounds that mimic bacterial signals and interfere with quorum sensing regulation in bacteria. Plant Physiol. 2004;134:137–146. doi: 10.1104/pp.103.029918. doi:10.1104/pp.103.029918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne S.H, Williams H.D. Cell density-dependent starvation survival of Rhizobium leguminosarum bv. phaseoli: identification of the role of an N-acyl homoserine lactone in adaptation to stationary-phase survival. J. Bacteriol. 1999;181:981–990. doi: 10.1128/jb.181.3.981-990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth I.K, Newton J.A, Hyman L.J, Lees A.K, Daykin M, Ortori C, Williams P, Fray R.G. Potato plants genetically modified to produce N-acylhomoserine lactones increase susceptibility to soft rot Erwiniae. Mol. Plant Microbe Interact. 2004;17:880–887. doi: 10.1094/MPMI.2004.17.8.880. [DOI] [PubMed] [Google Scholar]
- Tun-Garrido C, Bustos P, Gonzalez V, Brom S. Conjugative transfer of p42a from Rhizobium etli CFN42, which is required for mobilization of the symbiotic plasmid, is regulated by quorum sensing. J. Bacteriol. 2003;185:1681–1692. doi: 10.1128/JB.185.5.1681-1692.2003. doi:10.1128/JB.185.5.1681-1692.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij E.T, Girard G, Lugtenberg B.J.J, Bloemberg G.V. Influence of fusaric acid on phenazine-1-carboxamide synthesis and gene expression of Pseudomonas chlororaphis strain PCL1391. Microbiology. 2005;151:2805–2814. doi: 10.1099/mic.0.28063-0. doi:10.1099/mic.0.28063-0 [DOI] [PubMed] [Google Scholar]
- Ventre I, Ledgham F, Prima V, Lazdunski A, Foglino M, Sturgis J.N. Dimerization of the quorum sensing regulator RhlR: development of a method using EGFP fluorescence anisotropy. Mol. Microbiol. 2003;48:187–198. doi: 10.1046/j.1365-2958.2003.03422.x. doi:10.1046/j.1365-2958.2003.03422.x [DOI] [PubMed] [Google Scholar]
- Von Bodman S.B, Bauer W.D, Coplin D.L. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2003;41:455–482. doi: 10.1146/annurev.phyto.41.052002.095652. doi:10.1146/annurev.phyto.41.052002.095652 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhong Z, Cai T, Li S, Zhu J. Heterologous overexpression of quorum-sensing regulators to study cell-density-dependent phenotypes in a symbiotic plant bacterium Mesorhizobium huakuii. Arch. Microbiol. 2004;182:520–525. doi: 10.1007/s00203-004-0735-8. doi:10.1007/s00203-004-0735-8 [DOI] [PubMed] [Google Scholar]
- Waters C.M, Bassler B.L. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. doi:10.1146/annurev.cellbio.21.012704.131001 [DOI] [PubMed] [Google Scholar]
- Whistler C.A, Pierson L.S. Repression of phenazine antibiotic production in Pseudomonas aureofaciens strain 30–84 by RpeA. J. Bacteriol. 2003;185:3718–3725. doi: 10.1128/JB.185.13.3718-3725.2003. doi:10.1128/JB.185.13.3718-3725.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E.W, Winans S.C. Cell–cell communication in the plant pathogen Agrobacterium tumefaciens. Phil. Trans. R. Soc. B. 2007;362:1135–1148. doi: 10.1098/rstb.2007.2040. doi:10.1098/rstb.2007.2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead N.A, Barnard A.M, Slater H, Simpson N.J, Salmond G.P. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 2001a;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. doi:10.1111/j.1574-6976.2001.tb00583.x [DOI] [PubMed] [Google Scholar]
- Whitehead N.A, Barnard A.M.L, Slater H, Simpson N.J.L, Salmond G.P.C. Quorum-sensing in gram-negative bacteria. FEMS Microbiol. Rev. 2001b;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. doi:10.1111/j.1574-6976.2001.tb00583.x [DOI] [PubMed] [Google Scholar]
- Wilkinson A, Danino V, Wisniewski-Dye F, Lithgow J.K, Downie J.A. N-acyl-homoserine lactone inhibition of rhizobial growth is mediated by two quorum-sensing genes that regulate plasmid transfer. J. Bacteriol. 2002;184:4510–4519. doi: 10.1128/JB.184.16.4510-4519.2002. doi:10.1128/JB.184.16.4510-4519.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. Quorum sensing: an emerging target for antibacterial chemotherapy? Expert Opin. Ther. Targets. 2002;6:257–274. doi: 10.1517/14728222.6.3.257. doi:10.1517/14728222.6.3.257 [DOI] [PubMed] [Google Scholar]
- Williams P, Winzer K, Chan W, Cámara M. Look who's talking: communication and quorum sensing in the bacterial world. Phil. Trans. R. Soc. B. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. doi:10.1098/rstb.2007.2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winson M.K, Swift S, Fish L, Throup J.P, Jorgensen F, Chhabra S.R, Bycroft B.W, Williams P, Stewart G.S.A.B. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. doi:10.1111/j.1574-6968.1998.tb13044.x [DOI] [PubMed] [Google Scholar]
- Winzer K, Williams P. Quorum sensing and the regulation of virulence gene expression in pathogenic bacteria. Int. J. Med. Microbiol. 2001;291:131–143. doi: 10.1078/1438-4221-00110. doi:10.1078/1438-4221-00110 [DOI] [PubMed] [Google Scholar]
- Winzer K, et al. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology. 2002a;148:909–922. doi: 10.1099/00221287-148-4-909. [DOI] [PubMed] [Google Scholar]
- Winzer K, Hardie K.R, Williams P. Bacterial cell-to-cell communication: sorry, can't talk now—gone to lunch! Curr. Opin. Microbiol. 2002b;5:216–222. doi: 10.1016/s1369-5274(02)00304-1. doi:10.1016/S1369-5274(02)00304-1 [DOI] [PubMed] [Google Scholar]
- Wisniewski-Dye F, Downie J.A. Quorum-sensing in Rhizobium. Antonie Van Leeuwenhoek. 2002;81:397–407. doi: 10.1023/a:1020501104051. [DOI] [PubMed] [Google Scholar]
- Wisniewski-Dye F, Jones J, Chhabra S.R, Downie J.A. raiIR Genes are part of a quorum-sensing network controlled by cinI and cinR in Rhizobium leguminosarum. J. Bacteriol. 2002;184:1597–1606. doi: 10.1128/JB.184.6.1597-1606.2002. doi:10.1128/JB.184.6.1597-1606.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers H, Swift S, Williams P. Quorum sensing as an integral component of gene regulatory networks in Gram-negative bacteria. Curr. Opin. Microbiol. 2001;4:186–193. doi: 10.1016/s1369-5274(00)00187-9. doi:10.1016/S1369-5274(00)00187-9 [DOI] [PubMed] [Google Scholar]
- Yang F, Wang L.H, Wang J, Dong Y.H, Hu J.Y, Zhang L.H. Quorum quenching enzyme activity is widely conserved in the sera of mammalian species. FEBS Lett. 2005;579:3713–3717. doi: 10.1016/j.febslet.2005.05.060. doi:10.1016/j.febslet.2005.05.060 [DOI] [PubMed] [Google Scholar]
- Zhang L.H. Quorum quenching and proactive host defense. Trends Plant Sci. 2003;8:238–244. doi: 10.1016/S1360-1385(03)00063-3. doi:10.1016/S1360-1385(03)00063-3 [DOI] [PubMed] [Google Scholar]
- Zhang L, Murphy P.J, Kerr A, Tate M.E. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature. 1993;362:446–448. doi: 10.1038/362446a0. doi:10.1038/362446a0 [DOI] [PubMed] [Google Scholar]
- Zhang H.B, Wang L.H, Zhang L.H. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl Acad. Sci. USA. 2002a;99:4638–4643. doi: 10.1073/pnas.022056699. doi:10.1073/pnas.022056699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R.G, et al. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 2002b;417:971–974. doi: 10.1038/nature00833. doi:10.1038/nature00833 [DOI] [PubMed] [Google Scholar]
- Zhu J, Winans S.C. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl Acad. Sci. USA. 2001;98:1507–1512. doi: 10.1073/pnas.98.4.1507. doi:10.1073/pnas.98.4.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Oger P.M, Schrammeijer B, Hooykaas P.J, Farrand S.K, Winans S.C. The bases of crown gall tumorigenesis. J. Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. doi:10.1128/JB.182.14.3885-3895.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chai Y, Zhong Z, Li S, Winans S.C. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl. Environ. Microbiol. 2003;69:6949–6953. doi: 10.1128/AEM.69.11.6949-6953.2003. doi:10.1128/AEM.69.11.6949-6953.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]