Abstract
Expression of a large set of gene products required for conjugative transfer of the antibiotic resistance plasmid pCF10 is controlled by cell–cell communication between plasmid-free recipient cells and plasmid-carrying donor cells using a peptide mating pheromone cCF10. Most of the recent experimental analysis of this system has focused on the molecular events involved in initiation of the pheromone response in the donor cells, and on the mechanisms by which the donor cells control self-induction by endogenously produced pheromone. Recently, studies of the molecular machinery of conjugation encoded by the pheromone-inducible genes have been initiated. In addition, the system may serve as a useful bacterial model for addressing the evolution of biological complexity.
Keywords: quorum sensing, plasmid biology, gene regulation
1. Introduction
The antibiotic resistance plasmid pCF10 of Enterococcus faecalis is a member of a family of mobile genetic elements whose ability to transfer from a donor to a recipient bacterial cell is controlled by intercellular signalling via a peptide mating pheromone. In the case of pCF10, the heptapeptide signal molecule cCF10 (LVTLVFV) serves as a specific inducer of the pCF10 conjugation genes (Mori et al. 1988). To date, this type of plasmid has only been described in enterococci (for reviews, see Clewell & Dunny 2002; Chandler & Dunny 2004). In addition to the biological significance of pheromone-inducible conjugation as a useful paradigm for the study of cell–cell signalling, these plasmids contribute substantially to the increasing importance of the enterococci as opportunistic pathogens, because they disseminate antibiotic resistance determinants and because expression of at least one conjugation protein increases the virulence of the organisms. This form of cell–cell signalling is distinct from many of the quorum-sensing systems described elsewhere in this issue, in that the signalling occurs unidirectionally between two cell types (the donor cells use the signal to monitor recipient cell population density) and that the sensing system is encoded by a genetic element (the plasmid) that is distinct from the chromosomal determinants responsible for signal production. These features have interesting physiological and evolutionary consequences. This paper will review the general properties of the pCF10 system, summarize some recent data on the signalling mechanism and on the control of endogenous pheromone activity by the donor cells, and highlight some important questions regarding the evolution of the system in relation to the broader issue of the evolution of biological complexity.
2. Overview of system
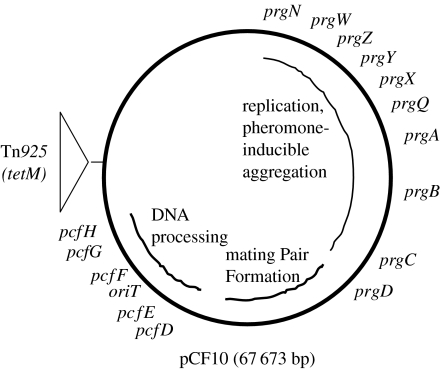
As shown in figure 1, pCF10 is slightly less than 70 kb in size and with at least half of its genetic coding capacity devoted to pheromone-inducible conjugation. A long contiguous region of the plasmid encodes regulatory loci devoted to pheromone sensing and negative control of conjugation functions in the absence of exogenous pheromone, formation of mating aggregates and the conjugative DNA processing machinery (as well as the cognate oriT target sequence). Recent DNA sequencing and transcriptional analysis of the pCF10 pheromone response (Hirt et al. 2005) suggest that the entire segment from prgQ–pcfH is coordinately induced by the addition of exogenous pheromone, while expression of the remaining pCF10 genes and the host chromosomal genes is largely unaffected. An interesting aspect of the system that will be further described below is that, in spite of the fact that all of the conjugation genes are coordinately regulated, they appear to have evolved from at least three different sources (Hirt et al. 2005). The segment of pCF10 between prgN and prgQ encodes both pheromone control and sensing, and plasmid replication and maintenance. Interestingly, pheromone production by the host cell is somehow related to stable maintenance of pCF10 and the replication initiator protein PrgW is a pheromone-binding protein (Leonard et al. 1996).
Figure 1.
Map of pCF10. The figure shows the approximate positions of a number of genes involved in replication, pheromone sensing and control, and conjugative DNA transfer as described in the text. The curved inner lines depict contiguous segments of pCF10 encoding the biological functions indicated. As described in the text, each of these segments probably originated from a different ancestral source. With the exception of prgX, all of the genes from prgW–pcfH are transcribed in the clockwise direction. The transposon Tn925 encodes the tetracycline resistance determinant tetM.
The genetic loci from prgZ–prgQ are involved in the initiation of the donor cell response to exogenous pheromone signal and prevention of self-induction of the donor cells by endogenously produced cCF10. These genetic functions will be addressed in detail below. The prgA, prgB and prgC genes all encode secreted cell surface proteins, with the product of prgA (Sec10 protein) likely mediating surface exclusion (Dunny et al. 1985), the Asc10 protein encoded by prgB mediating mating aggregate formation (Olmsted et al. 1991) and the function of the protein encoded by prgC being unknown. Between prgQ and prgA, there are several genetic determinants that appear to act following initiation of the pheromone response to affect the expression of downstream conjugation genes. These loci encode both positive and negative post-transcriptional regulatory factors, some of which function as small RNAs (Bensing & Dunny 1997; Bensing et al. 1997). The function of next block of genes downstream from prgC and extending through pcfD is largely unexplored, but based on ongoing analysis of amino acid sequence similarity, it might encode a Gram-positive version of a conjugative type IV secretion system involved in formation of the mating channel connecting the donor and the recipient cells. Finally, the segment including pcfE–pcfH has been shown to code for the conjugative DNA processing machinery (Staddon et al. 2004), with the non-coding region between pcfE and pcfF containing the functional origin of transfer (Staddon et al. 2006). It is noteworthy that the gene order, and the approximate order of expression of the conjugation genes following pheromone induction (Hirt et al. 2005), correlates with the order of events in conjugation, i.e. assembly of mating aggregates, followed by formation of functional mating pairs and finally DNA processing and transfer. The system has apparently evolved to produce the minimum amount of conjugation machinery required for effective transfer in response to a pheromone signal, while rapidly shutting off the expression of these genes very rapidly following induction (Bae et al. 2004).
3. The induction state of donor cells is determined by the ratio of inhibitor to pheromone peptides
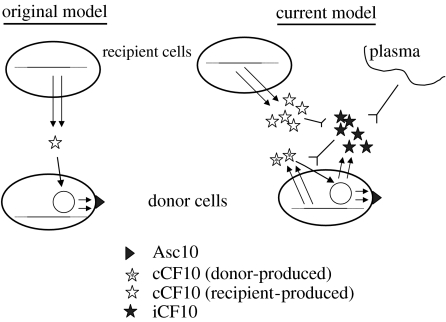
The early studies of pheromone-inducible conjugation suggested a rather simple model, where the interaction of an extracellular signal from a potential conjugative recipient cell with a donor resulted in a mating response (figure 2); it quickly became apparent that plasmid-free recipient cells produced several pheromones, each specific for a cognate plasmid, or family of related plasmids (Dunny et al. 1979). This model was supported by conditioned medium experiments where unfractionated culture filtrates of recipient strains induced mating responses by donors (Dunny et al. 1978, 1979), and by the subsequent determination of the specific peptide sequences of pheromones specific for several conjugative plasmids (Mori et al. 1984, 1986, 1988). Eventually, it was shown that seven to eight amino acid peptides that constitute the mature pheromones are generated by intramembrane processing of cleaved signal peptides from putative secreted lipoproteins (An et al. 1999; Clewell et al. 2000); cCF10 is produced from the gene product of ccfA (Antiporta & Dunny 2002). There are virtually no experimental data available to explain the functions of any of these lipoproteins, or to indicate whether the pheromones or their signal peptide precursors play any role in the physiology of the producer cell. An intramembrane protease called Eep identified by An and Clewell (An et al. 1999) is required for the production of normal levels of several pheromones. Since the mature pheromones are generally more hydrophobic than the signal peptide precursors, Eep proteolysis might be accompanied by active excretion from the cell membrane. In the case of cCF10, a considerable portion of the released pheromone remains associated with the cell wall, and some is also released into the growth medium (Buttaro(Leonard) et al. 2000).
Figure 2.
Two models for the role of signalling peptides in expression of pCF10 conjugation functions. The single arrows indicate positive control, the inverted arrows indicate negative control and the double arrows indicate polypeptide synthesis from a plasmid (iCF10, Asc10) or chromosomal (cCF10) gene. Original model: initial studies suggested a simple model whereby a single unidirectional pheromone signal from recipient cells to donors caused induction of the transfer functions, including cell aggregation mediated by Asc10. Current model: further analysis of the system has shown that both donors and recipients can produce pheromone, and that the molar ratio of chromosomally encoded pheromone (cCF10) to plasmid-encoded inhibitor (iCF10) determines the induction state of the donor cell. In a monoculture of donor cells growing in laboratory medium, the balance of these two peptides keeps the transfer system off. The balance can be shifted in favour of pheromone, either by production of pheromone by recipient cells in close proximity (upper left) or by interaction of inhibitor with plasma components (upper right) when the bacteria are growing in the bloodstream of a mammalian host.
It soon became apparent that the ‘original’ model depicted in figure 2 was not sufficient to explain all aspects of these transfer systems. Since pheromone production is chromosomally encoded, while the response is plasmid determined, it is conceivable that transfer of a plasmid into a new host strain would give rise to constitutive expression of the conjugation genes as a result of self-induction of the newly created donor cell by endogenous pheromone. Since this has never been observed, the plasmid must encode one or more mechanism to avoid such a wasteful self-induction process. In the pCF10 system, as well as other pheromone systems that have been studied, there are two plasmid determinants devoted to preventing self-induction, and genetic disruption of either determinant leads to constitutive expression of the conjugation genes (Hedberg et al. 1996; Buttaro(Leonard) et al. 2000; Chandler et al. 2005a,b). One of these determinants, prgY in pCF10, encodes a membrane protein whose function is to reduce the level of cCF10 released from the cell membrane. The major portion of PrgY from the amino terminus and extending through nearly 60% of the amino acid sequence appears to lie outside the membrane and is anchored by several C-terminal transmembrane segments. Most mutations that retain protein stability but abolish function map in this extracellular domain. Interestingly, PrgY represents a superfamily of related proteins found in all three biological kingdoms, but not in all species (Chandler et al. 2005a). No functional analysis of any of these proteins has been completed, except for studies of PrgY and the homologues encoded by two other pheromone plasmids (Weaver & Clewell 1990; Nakayama et al. 1994a). Since the response of a donor cell to cCF10 requires pheromone import via the concerted activities of the plasmid-encoded PrgZ extracellular pheromone-binding protein and the chromosomal oligopeptide permease system (Leonard et al. 1996), one possible way that PrgY could function is to interfere with the import process. However, the cumulative data suggest a different model whereby the extracellular domain of PrgY interacts with nascent cCF10 as it is released from the membrane, and this interaction sequesters, modifies or degrades the peptide; expression of PrgY has no effect on the interaction of E. faecalis cells with exogenously added cCF10 (Chandler et al. 2005a). PrgY control of endogenous pheromone is highly specific for cCF10, and the specificity determinants for recognition by PrgY seem to reside within the mature cCF10 sequence and not in the signal peptide precursor (J. Chandler & G. Dunny 2006, unpublished data).
Although PrgY is necessary for control of endogenous pheromone activity, it is not sufficient. The donor cells produce reduced but still significant levels of cCF10 in both cell wall fractions and supernatants (Buttaro(Leonard) et al. 2000). This residual pheromone activity is neutralized by the production of a plasmid-encoded peptide iCF10 from the prgQ locus of pCF10 (Nakayama et al. 1994b). As described in more detail later, the prgQ locus is located at the extreme 5′ end of a long operon encoding many if not all of the conjugation proteins of pCF10. The effects of pheromone induction are: (i) to modestly increase the initiation of transcription from the prgQ promoter (this promoter is active at a significant level under all conditions), and (ii) to increase extension of prgQ transcripts past a putative termination region such that downstream conjugation functions are expressed (Chung & Dunny 1995; Bensing et al. 1997; Bae et al. 2004). The only polypeptide coding sequence in the mRNA produced constitutively from prgQ transcription is 66 nt Orf encoding a 22-amino acid peptide resembling a signal peptide (without an attached C-terminal secreted protein). The last codons encode the mature iCF10 sequence (AITLIFI); as is the case for cCF10, processing of the full-length precursor to the mature signal molecule occurs concomitantly with peptide export, such that iCF10 must exit the cell and be reimported to function in regulation. High performance liquid chromatography fractionation of culture supernatants of E. faecalis strains carrying pCF10 shows that these cells secrete a mixture of iCF10 and cCF10 in a molar ratio of 50–100/1 (Nakayama et al. 1994b; Hirt et al. 2002); this ratio is just sufficient for iCF10 to neutralize the cCF10 biological activity, while allowing the donor cell to remain sensitive to an extremely low (approx. 10−11 M) level of exogenous cCF10 as would be produced by the recipient cells in close proximity (Mori et al. 1988). Thus, in the ‘real world’, the induction state of donor cells is dependent on the relative concentrations of the two peptides rather than the absolute cCF10 concentration. Production of iCF10 by the donor cells auto-represses the prgQ promoter through an interaction with PrgX (Kozlowicz et al. 2004), and the first direct result of induction by exogenous pheromone seems to increase synthesis of iCF10. This probably serves to shut the system back off after a transient period of expression. Consistent with this regulatory model, we recently found that genetic disruption of prgY causes an increase in prgQ transcription and presumably yielding an increased production of iCF10 to compensate for the elevated levels of cCF10 production by prgY mutants (Chandler et al. 2005b). At first blush, the involvement of two different extracellular peptide signals in controlling pCF10 conjugation functions might seem unnecessarily complicated. However, we recently found that this system is also used by pCF10-containing cells to activate expression of aggregation substance (Asc10) when growing in the mammalian bloodstream (Hirt et al. 2002; Chandler et al. 2005b). A host factor, probably an albumin/lipid complex, selectively sequesters or degrades iCF10, leading to induction (by endogenous cCF10) of conjugation proteins including Asc10. Expression of the latter protein increases virulence of E. faecalis in several model systems (McCormick et al. 2000), and genetic disruption of either the pheromone-sensing machinery of pCF10 or the ability of the plasmid-containing cells to produce cCF10 abolishes in vivo induction. Therefore, it is apparent that the more complex system actually allows for detecting two different kinds of environmental signals (shown in figure 2) that are ultimately of great importance to both the host bacterium and the plasmid.
4. The molecular switch controlling the initiation of the pheromone response is a shift in the structure of the C-terminus of prgx as a result of pheromone binding
The transcription of the prgX/prgQ region of pCF10 has been studied extensively (Chung et al. 1995; Bensing et al. 1996, 1997; Bae et al. 2000, 2004). The prgQ promoter is active in uninduced cells, but the resulting transcripts (termed Qs) are only about 400 nt in length with a 3′ end in the vicinity of an inverted repeat structure IRS1 that probably functions in transcription termination, mRNA processing or both (Chung & Dunny 1995; Bensing et al. 1997; Bae et al. 2004). Pheromone induction results in a modest increase (2–5X) in prgQ promoter activity, and northern blot analysis reveals the presence of extended forms of mRNAs, the most abundant being QL, which is about 130 nt longer than Qs. Reverse transcriptase polymerase chain reaction analyses show that there is extensive processing of Q transcripts under all conditions, but in induced cells they can extend through prgB and beyond (Chung & Dunny 1992, 1995; Bensing et al. 1996, 1997). Thus, the most significant result of pheromone induction is to extend transcription from the prgQ promoter through the IRS1 region.
The prgX gene is adjacent to prgQ, but in the opposite orientation. Contrary to initial expectations, prgX transcription does not initiate from the region between prgX and prgQ. Instead, this transcript is produced from a promoter within the prgQ locus, but on the non-template strand (Bae et al. 2000). Thus, an unprocessed prgX mRNA contains several hundred nucleotides of non-coding sequence at its 5′ end. As is the case for prgQ transcripts, the prgX message is subject to extensive processing. The most abundant and stable form of this transcript, Qa (Q antisense), consists of the 5′ 102 nt (Bae et al. 2004). Qa, which is complementary to a segment of Qs, also plays a critical role in regulation of conjugation. Genetic evidence shows that Qa functions to block production of prgQ transcripts that extend past IRS1; this activity of Qa was not affected by pheromone. Both Qa and Qs have potential for extensive secondary structure, and it was predicted that pairing of these two RNAs would shift the structure of the 3′ terminus of Qs into a functional terminator, whereas unpaired Qs would fold in an alternative non-terminating structure (Bae et al. 2004). In uninduced cells, the synthesis of Qa and Qs is balanced such that none of the Qs transcripts extend past IRS1. The increase in prgQ transcription initiation resulting from pheromone abolition of PrgX repression (next paragraph) essentially titrates all of the Qa and leaves unpaired Qs transcripts that extend past IRS1, ultimately leading to expression of downstream conjugation proteins. This model is currently being examined experimentally.
The other functional product produced from the Qa promoter is PrgX. Genetic analysis of PrgX function showed that this protein represses (albeit modestly) transcription initiation from the prgQ promoter, and that repression is sensitive to pheromone (Bae et al. 2004); interestingly, PrgX protein also positively regulates its own expression and that of Qa at a post-transcriptional level (Bae et al. 2000; Kozlowicz et al. 2004). These cumulative results show that the gene products produced by transcription from the Qa promoter affect prgQ transcription by two independent mechanisms; once produced, each gene product can function independently of the other, but their mode of synthesis makes them interdependent in wild-type donor cells. Extensive genetic and molecular analyses of PrgX indicate that it is multifunctional. In addition to the functions described previously, PrgX specifically binds pCF10 DNA at two sites in the region between prgX and prgQ; it exists in E. faecalis cells in an oligomeric state, and pheromone addition causes a reduction in the oligomerization state of PrgX (Bae & Dunny 2001). A compilation of the important features of both PrgX and Qa is presented in table 1.
Table 1.
Important properties of PrgX and Qa.
| property | PrgX | Qa |
|---|---|---|
| molecular composition | 317 amino acid protein | 102 nt RNA |
| source | translated from 3′ end of PQa transcript | processed from 5′ end of PQa transcript |
| molecular target | two binding sites in pCF10 DNA | Qs RNAa |
| functions | reduce activity of prgQ promoter; positively regulate Qa mRNA | inhibit prgQ transcription |
| processing | elongation | |
| pheromone sensitivity | ++ | −− |
predicted.
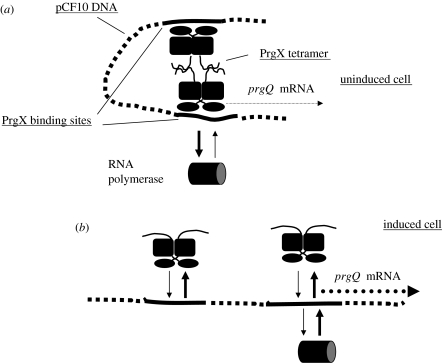
The most recent analysis of PrgX focused on mutants that were deficient in prgQ repression, but were not dominant negative and that retained sufficient positive auto-regulatory function to produce normal levels of protein in E. faecalis (Kozlowicz et al. 2004). Although it was not clear at the outset whether such mutants could be obtained, we were successful in this endeavour. Three phenotypic classes of mutants were identified, with most containing substitutions in the C-terminal region of the protein. In the same study, we also confirmed directly that the N-terminus contained the functional DNA-binding domain and we provided biochemical evidence for binding of cCF10 to PrgX. Further analysis showed that all of these mutants were partially defective in DNA binding and/or oligomerization such that prgQ repression was impacted more severely than auto-regulation. These results, along with those from previous studies were used to formulate a DNA looping model (figure 3) that accounts for both the regulatory functions of PrgX via a single mechanism. Previous biochemical analyses indicated that the affinity of PrgX binding to pCF10 DNA was not extremely high, with the secondary binding site (which overlaps the prgQ promoter and is therefore most important for repression) being particularly weak (Bae et al. 2002). We proposed that protein–protein interactions between PrgX molecules attached to the two sites could result in formation of a DNA loop that yields a much more stable complex than those involving only a single binding site in the DNA. We further suggested that the primary effect of pheromone on PrgX could be on the regions involved in protein–protein interactions rather than on the DNA-binding domain; when this model was initially proposed, there was not sufficient evidence to indicate whether each pCF10 DNA-binding site was occupied by a PrgX monomer or a dimer. Structural data described below has demonstrated that each site is bound by a dimer, such that the looped complex contains a PrgX tetramer. Pheromone binding causes disruption of tetramers, destabilizing the loop and ultimately reducing the PrgX occupancy of each DNA target site. While it is easy to envision how formation of this complex could repress the prgQ promoter, the mechanism of positive auto-regulation is less obvious. We believe that the looped complex could affect the dynamics of elongation, processing and decay of mRNAs synthesized from the Qa promoter by acting as a roadblock to RNA polymerase molecules. One way that this could work is to block or delay extension of transcription past the secondary PrgX-binding site to allow for folding of the 5′ end of the message into a stable structure, i.e. mature Qa, prior to the synthesis of a distal sequence that functions to accelerate 3′→5′ decay of the entire message. Obviously, this is the most speculative aspect of the model, but there are compelling data that force us to reject simpler alternatives that might appear to be more obvious (Bae et al. 2000); further experimental testing of this model is a high priority for current and future work.
Figure 3.
DNA looping model for PrgX. (a) Uninduced donor cell. PrgX molecules bound to each DNA target site interact (the model shown here has each site occupied by a PrgX dimer; the two dimers interact to form a tetramer) to form a DNA loop that stabilizes DNA/protein interactions and increases occupancy of both binding sites. RNA polymerase access to the prgQ promoter, which overlaps the lower binding site, is restricted. This reduces prgQ mRNA synthesis. (b) Pheromone-induced donor cell. Pheromone interaction with PrgX breaks up tetramers, opening the loop, decreasing occupancy of the binding sites and allowing for increased polymerase access to the prgQ promoter. This increases production of prgQ mRNA.
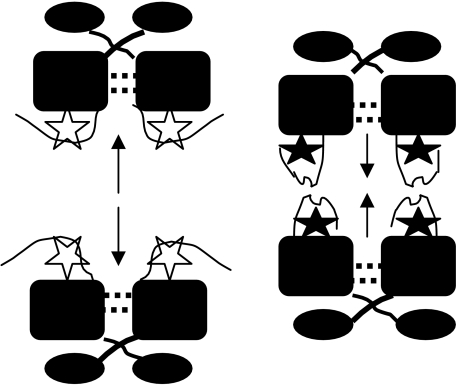
A substantial body of published genetic and biochemical evidence (Bae et al. 2000, 2002, 2004; Bae & Dunny 2001; Kozlowicz et al. 2004) is consistent with the model shown in figure 3, and recent genetic analysis of constructs where the spacing between the two binding sites has been changed to affect their position on the face of the DNA helix (B. Kozlowicz & G. Dunny 2005, unpublished data) also supports the model. In addition, recent determination of high-resolution structures of PrgX and PrgX/cCF10 complexes (Shi et al. 2005) has provided physical evidence for crucial features of this model, including the direct interaction between PrgX and cCF10, and the effects of cCF10 binding on the PrgX structure and oligomerization state. The structural analysis indicates that a very large portion of each PrgX molecule is α-helical and, consistent with previous biochemical data (Kozlowicz et al. 2004), there is a DNA-binding motif near the N-terminus. Extensive atomic interactions between numerous residues in a large central segment of the protein, as well as domain swapping between N-terminal domains of adjacent monomers, promote stabilization of dimers. In PrgX crystals, the molecules are actually in the form of tetramers, where pairs of dimers are held together by a rather small loop region near the C-terminus (figure 4). Pheromone binds to PrgX in a pocket formed by several parallel and anti-parallel helices, and the overall structure of PrgX bound to cCF10 is quite similar to that of the unbound protein. The major difference is that upon occupancy of the binding pocket by cCF10, a major rotation of the C-terminus (approx. 20 amino acid residues) occurs such that a region including residues 296–298 covers the bound pheromone in the pocket, shifts its conformation from a short α-helix to a β-sheet and moves the C-terminal loop involved in stabilization of tetramers such that this interaction is disrupted. It is highly probable that, in vivo, these structural changes are sufficient to disrupt the looped structure of the PrgX–pCF10 DNA complex, reducing the occupancy of the operator sites on the DNA and leading to increased activity of the prgQ promoter. Recent genetic, biochemical and structural data (Kozlowicz et al. 2006) suggest that the iCF10 inhibitor peptide binds PrgX in the same pocket as cCF10, but that the bound peptide interacts with a different set of amino acid residues near the C-terminus of PrgX, such that the amino acid segment mediating tetramer formation is actually stabilized in a conformation that favours the tetramer-promoting interaction. A significant focus of future investigations will be to examine the interactions of PrgX with itself and with its target DNA in solution, and to develop an in vitro transcription system to confirm the molecular mechanisms of regulation suggested by the structural data. In addition, the isolation and characterization of additional mutations in the C-terminal region of PrgX, such as those that give a pheromone-insensitive ‘super-repressor’ phenotype, should also help to confirm and refine the looping model.
Figure 4.
Structural consequences of pheromone and inhibitor binding to PrgX. Extensive intermolecular interactions between amino acid residues in the N-terminal and central domains of PrgX allow the protein to form a stable dimer in all structures examined. In the absence of exogenous peptides, as well as in the presence of iCF10, the PrgX C-terminus assumes a conformation that promotes interaction between pairs of dimers to form stable tetramers; this structure favours formation of DNA loops as shown in figure 3. When complexed with cCF10, the C-terminal arm changes the structure and rotates such that the protein–protein interactions favouring tetramer formation are weakened significantly. Both peptides occupy the same binding cleft formed by a series of parallel and anti-parallel helical domains in the central part of the protein and, in both cases, residues from the C-terminal region interact with the bound peptide. However, iCF10 interacts with different C-terminal PrgX residues than is the case for cCF10.
5. Multiple positive- and negative-regulatory loci can modulate expression of downstream conjugation functions following the initiation of the cCF10 pheromone response
While the molecular mechanism for initiation of the cCF10 pheromone response described previously might seem complex, it is noteworthy that several additional genetic loci located between the Qs-encoding region of pCF10 and the prgB gene (whose expression has frequently been used as an indicator of the induction state) have been shown to be involved in positive or negative post-transcriptional control of prgB expression (Chung et al. 1995; Bensing et al. 1997). These loci appear to encode products that act as novel RNAs to affect transcription extension or translation of prgB (Bensing & Dunny 1997; Bensing et al. 1997). The recent focus of the laboratory on PrgX and the initiation step has precluded analysis of these downstream determinants in the past few years, but our increased level of understanding of the initial stages of the pheromone response should make these downstream events more amenable to further study in the future. Although the results of recent microarray studies of pheromone induction are consistent with the possibility that the entire set of conjugation genes are expressed from transcripts initiating at the prgQ promoter (Hirt et al. 2005), it is very plausible that there could be separate functional coordinately regulated promoters for the more distal genes in this region, and it will be of interest to address this question experimentally.
6. Can the evolution of the pCF10 transfer system by a modular assembly process serve as a model for the evolution of biological complexity?
The hallmark of ‘intelligent design’ for a computer program, an electronic circuit or a mechanical device is that it is the simplest entity that efficiently and accurately performs its given function. In the lore engineering and computer science, there is a term known as the kluge (or kludge) referring to a process by which a simple and efficient device to accomplish one task gets converted in a stepwise fashion into a Heath Robinson- or Rube Goldberg-like contraption in an effort to adapt the device for alternative uses (http://catb.org/∼esr/jargon/html/K/kluge.html). The regulatory circuits controlling expression of pCF10 conjugation functions seem to be much more complex than one might expect to specifically activate the conjugation functions of the donor cells in the presence of recipients. Thus, it could be stated that the system resembles a biological kluge. One explanation for this Byzantine regulation system is that the components have been cobbled together from different ancestral sources and could still be in a fairly early stage of evolution to the optimal system. Alternatively, it is probable that the selective pressures shaping the evolution of the system have been for more functions than simply the optimal regulation of conjugation in response to recipient cell density. Recent experimental data provide support for both of these possibilities.
Complete sequence information is now available for several pheromone-responsive plasmids and predicted pheromone-responsive plasmids (the V583 strain whose genome was sequenced by TIGR (Paulsen et al. 2003) contains two large plasmids, each encoding a predicted pheromone-inducible conjugation system, but the functionality of these genes has not been demonstrated). The availability of these data allows for careful comparison of the relatedness of these mobile elements, and for some assessment of their possible modes of evolution. In the case of the three most extensively studied pheromone plasmids, pCF10, pAD1 and pPD1, genetic and molecular analyses strongly suggest that the region of each element corresponding to the prgN–prgC segment of pCF10 shown in figure 1, and encoding replication and pheromone-inducible aggregation functions, shows significant overall conservation, consistent with evolution from a common ancestor. However, once the sequencing of these elements was completed, it became clear that this sequence conservation did not extend through the rest of the putative mating pair formation and DNA processing genes, even though all of these genes are coordinately regulated with the conserved upstream genes. In the case of pCF10, the genes predicted to function in formation of the mating channel show much higher relatedness to a putative conjugative element from group B streptococci than to pAD1 or pPD1 (Hirt et al. 2005). Furthermore, the pCF10 region encoding the conjugative DNA processing machinery is highly similar to that of the lactococcal conjugative element pRS01 (Staddon et al. 2004; Hirt et al. 2005) and completely unrelated to the corresponding region of other pheromone plasmids like pAD1 (Hirt et al. 2005). From these data, it can be inferred that a primordial pheromone-inducible aggregation module, possibly an autonomously replicating plasmid, became linked in separate evolutionary events to several different mating channels and DNA processing modules to generate the current set of pheromone plasmids. The pheromone-inducible aggregation module has probably been in an enterococcal host for a longer time than the other modules. The fact that there has clearly been a coevolution between pheromone and at least four components of this segment of pCF10 (PrgX, PrgY, PrgZ and iCF10) is consistent with this idea. It would be of considerable interest to discover a pheromone-inducible (non-conjugative) enterococcal plasmid among current E. faecalis strains. Expression of the virulence factor aggregation substance (from prgB) is induced during growth of pCF10-carrying E. faecalis in blood by endogenously produced cCF10 (Hirt et al. 2002). The selective pressure for in vivo induction of virulence may have been as important as, or more important than, that for the ability of donor cells or detect recipients in the evolution of the cCF10 sensing system. The relatively recent discovery of the in vivo induction phenomenon and its possible role in evolution of the sensing system also illustrates how little we know about the ecology and evolution of cell–cell signalling in bacteria, even for a relatively well-studied organism like E. faecalis.
As mentioned previously, the complexity of regulation in this system does not end with the initiation of the pheromone response; several downstream steps are also subject to post-transcriptional control, and we do not have a good idea about why this should be true. However, since both the mating channel and the DNA processing machinery are multi-subunit entities, whose components probably need to be produced in a particular stoichiometry in order to be functional (and to avoid deleterious effects on the host cell), it is not surprising that there could be multiple checkpoints controlling their expression. It is also possible that some components could function in more than one important cellular process (already demonstrated for Aggregation Substance), providing a selective pressure for the evolution of control circuits that allow some conjugation proteins to be produced in higher levels from others encoded within the same transcript.
One of the current challenges for evolutionary biology is to provide more detailed and experimentally testable models for the generation of complexity. In this regard, bacterial structures such as flagella and type III secretion systems have received some attention as useful model systems because it has been suggested that they have certain inherent features, such as ‘irreducible complexity’ which might impact formulation of models for their genesis by evolutionary processes, while being more amenable to empirical analysis using the rapidly expanding data from microbial genome sequencing and molecular biological analyses of these systems in selected bacteria (Nguyen et al. 2000; Paulsen et al. 2000). Much of the attention has been focused on the sequence analysis of the protein components of these systems. As we begin to consider the pCF10 pheromone-inducible conjugation system from a molecular evolutionary perspective, it is apparent that both the structural and the regulatory aspects of the sensing and DNA transfer machinery of this system might also prove to be fruitful subjects for more study in relation to the evolution of complexity.
Acknowledgments
I thank all of the past and present members of the laboratory for their hard work and intellectual contributions to pCF10 research, especially Dawn Manias, Jungwon Chung, Barbara Bensing, Helmut Hirt, Tina Buttaro, Ed Bryan, Taeok Bae, Chris Waters, Jack Staddon, Briana Kozlowicz and Josie Chandler. I also thank long-time collaborators Marty Dworkin, Pat Schlievert, Doug Ohlendorf, Cathy Earhart and Ke Shi. The pCF10 research in my laboratory is supported by the PHS grants GM49530 and HL51987 from the NIH.
Footnotes
One contribution of 12 to a Theme Issue ‘Bacterial conversations: talking, listening and eavesdropping’.
References
- An F.Y, Sulavik M.C, Clewell D.B. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 1999;181:5915–5921. doi: 10.1128/jb.181.19.5915-5921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antiporta M.H, Dunny G.M. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J. Bacteriol. 2002;184:1155–1162. doi: 10.1128/jb.184.4.1155-1162.2002. doi:10.1128/jb.184.4.1155-1162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T, Dunny G.M. Dominant negative mutants of prgX: evidence for a role of PrgX dimerization in negative regulation of pheromone-inducible conjugation. Mol. Microbiol. 2001;39:1307–1320. doi: 10.1111/j.1365-2958.2001.02319.x. doi:10.1111/j.1365-2958.2001.02319.x [DOI] [PubMed] [Google Scholar]
- Bae T, Clerc-Bardin S, Dunny G.M. Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. J. Mol. Biol. 2000;297:861–875. doi: 10.1006/jmbi.2000.3628. doi:10.1006/jmbi.2000.3628 [DOI] [PubMed] [Google Scholar]
- Bae T, Kozlowicz B, Dunny G.M. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J. Mol. Biol. 2002;315:995–1007. doi: 10.1006/jmbi.2001.5294. doi:10.1006/jmbi.2001.5294 [DOI] [PubMed] [Google Scholar]
- Bae T, Kozlowicz B.K, Dunny G.M. Characterization of cis-acting prgQ mutants: evidence for two distinct repression mechanisms by Qa RNA and PrgX protein in pheromone-inducible enterococcal plasmid pCF10. Mol. Microbiol. 2004;51:271–281. doi: 10.1046/j.1365-2958.2003.03832.x. doi:10.1046/j.1365-2958.2003.03832.x [DOI] [PubMed] [Google Scholar]
- Bensing B.A, Dunny G.M. Pheromone-inducible expression of an aggregation protein in Enterococcus faecalis requires interaction of a plasmid-encoded RNA with components of the ribosome. Mol. Microbiol. 1997;24:295–308. doi: 10.1046/j.1365-2958.1997.3311709.x. doi:10.1046/j.1365-2958.1997.3311709.x [DOI] [PubMed] [Google Scholar]
- Bensing B.A, Meyer B.J, Dunny G.M. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc. Natl Acad. Sci. USA. 1996;93:7794–7799. doi: 10.1073/pnas.93.15.7794. doi:10.1073/pnas.93.15.7794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing B.A, Manias D.A, Dunny G.M. Pheromone cCF10 and plasmid pCF10-encoded regulatory molecules act post-transcriptionally to activate expression of downstream conjugation functions. Mol. Microbiol. 1997;24:295–308. doi: 10.1046/j.1365-2958.1997.3301710.x. doi:10.1046/j.1365-2958.1997.3311709.x [DOI] [PubMed] [Google Scholar]
- Buttaro(Leonard) B.A, Antiporta M.H, Dunny G.M. Cell-associated pheromone peptide (cCF10) production and pheromone inhibition in Enterococcus faecalis. J. Bacteriol. 2000;182:4926–4933. doi: 10.1128/jb.182.17.4926-4933.2000. doi:10.1128/JB.182.17.4926-4933.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J.R, Dunny G.M. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides. 2004;25:1377–1388. doi: 10.1016/j.peptides.2003.10.020. doi:10.1016/j.peptides.2003.10.020 [DOI] [PubMed] [Google Scholar]
- Chandler J.R, Flynn A.R, Bryan E.M, Dunny G.M. Specific control of endogenous cCF10 pheromone by a conserved domain of the pCF10-encoded regulatory protein PrgY in Enterococcus faecalis. J. Bacteriol. 2005a;187:4830–4843. doi: 10.1128/JB.187.14.4830-4843.2005. doi:10.1128/JB.187.14.4830-4843.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J.R, Hirt H, Dunny G.M. A paracrine peptide sex pheromone also acts as an autocrine signal to induce plasmid transfer and virulence factor expression in vivo. Proc. Natl Acad. Sci. USA. 2005b;102:15 617–15 622. doi: 10.1073/pnas.0505545102. doi:10.1073/pnas.0505545102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.W, Dunny G.M. Cis-acting, orientation-dependent, positive control system activates pheromone-inducible conjugation functions at distances greater than 10 kilobases upstream from its target in Enterococcus faecalis. Proc. Natl Acad. Sci. USA. 1992;89:9020–9024. doi: 10.1073/pnas.89.19.9020. doi:10.1073/pnas.89.19.9020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.W, Dunny G.M. Transcriptional analysis of a region of the Enterococcus faecalis plasmid pCF10 involved in positive regulation of conjugative transfer functions. J. Bacteriol. 1995;177:2118–2124. doi: 10.1128/jb.177.8.2118-2124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.W, Bensing B.A, Dunny G.M. Genetic analysis of a region of the Enterococcus faecalis plasmid pCF10 involved in positive regulation of conjugative transfer functions. J. Bacteriol. 1995;177:2107–2117. doi: 10.1128/jb.177.8.2107-2117.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D.B, Dunny G.M. Conjugation and genetic exchange in enterococci. In: Gilmore M.S, Clewell D.B, Courvalin P, Dunny G.M, Murray B.E, Rice L.B, editors. The enterococci: pathogenesis, molecular biology and antibiotic resistance. American Society for Microbiology Press; Washington, DC: 2002. pp. 265–300. [Google Scholar]
- Clewell D.B, An F.Y, Flannagan S.E, Antiporta M, Dunny G.M. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol. Microbiol. 2000;35:246–248. doi: 10.1046/j.1365-2958.2000.01687.x. doi:10.1046/j.1365-2958.2000.01687.x [DOI] [PubMed] [Google Scholar]
- Dunny G.M, Brown B.L, Clewell D.B. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl Acad. Sci. USA. 1978;75:3479–3483. doi: 10.1073/pnas.75.7.3479. doi:10.1073/pnas.75.7.3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G.M, Craig R.A, Carron R.L, Clewell D.B. Plasmid transfer in Streptococcus faecalis: production of multiple pheromones by recipients. Plasmid. 1979;2:454–465. doi: 10.1016/0147-619x(79)90029-5. doi:10.1016/0147-619X(79)90029-5 [DOI] [PubMed] [Google Scholar]
- Dunny G.M, Zimmerman D.L, Tortorello M.L. Induction of surface exclusion (entry exclusion) by Streptococcus faecalis sex pheromones: use of monoclonal antibodies to identify an inducible surface antigen involved in the exclusion process. Proc. Natl Acad. Sci. USA. 1985;82:8582–8586. doi: 10.1073/pnas.82.24.8582. doi:10.1073/pnas.82.24.8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedberg P.J, Leonard B.A.B, Ruhfel R.E, Dunny G.M. Identification and characterization of the genes of Enterococcus faecalis plasmid pCF10 involved in replication and in negative control of pheromone-inducible conjugation. Plasmid. 1996;35:46–57. doi: 10.1006/plas.1996.0005. doi:10.1006/plas.1996.0005 [DOI] [PubMed] [Google Scholar]
- Hirt H, Schlievert P.M, Dunny G.M. In vivo induction of virulence and antibiotic resistance transfer in Enterococcus faecalis mediated by the sex pheromone-sensing system of pCF10. Infect. Immun. 2002;70:716–723. doi: 10.1128/iai.70.2.716-723.2002. doi:10.1128/IAI.70.2.716-723.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt H, Manias D.A, Bryan E.M, Klein J.R, Marklund J.K, Staddon J.H, Paustian M.L, Kapur V, Dunny G.M. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J. Bacteriol. 2005;187:1044–1054. doi: 10.1128/JB.187.3.1044-1054.2005. doi:10.1128/JB.187.3.1044-1054.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowicz B.K, Shi K, Gu Z.-Y, Ohlendorf D.H, Earhart C.A, Dunny G.M. Molecular basis for the control of conjugation by bacterial pheromone and inhibitor peptides. Mol. Microbiol. 2006;62:958–969. doi: 10.1111/j.1365-2958.2006.05434.x. doi:10.1111/j.1365-2958.2006.05434.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowicz B.K, Bae T, Dunny G.M. Enterococcus faecalis pheromone-responsive protein PrgX: genetic separation of positive autoregulatory functions from those involved in negative regulation of conjugative plasmid transfer. Mol. Microbiol. 2004;54:520–532. doi: 10.1111/j.1365-2958.2004.04286.x. doi:10.1111/j.1365-2958.2004.04286.x [DOI] [PubMed] [Google Scholar]
- Leonard B.A, Podbielski A, Hedberg P.J, Dunny G.M. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl Acad. Sci. USA. 1996;93:260–264. doi: 10.1073/pnas.93.1.260. doi:10.1073/pnas.93.1.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick J.K, Hirt H, Dunny G.M, Schlievert P.M. Pathogenic mechanisms of enterococcal endocarditis. Curr. Infect. Dis. Rep. 2000;2:315–321. doi: 10.1007/s11908-000-0009-9. [DOI] [PubMed] [Google Scholar]
- Mori M, Sakagami Y, Narita M, Isogai A, Fujino M, Kitada C, Craig R.A, Clewell D.B, Suzuki A. Isolation and structure of the bacterial sex pheromone, cAD1, that induces plasmid transfer in Streptococcus faecalis. FEBS. 1984;178:97–100. doi: 10.1016/0014-5793(84)81248-x. doi:10.1016/0014-5793(84)81248-X [DOI] [PubMed] [Google Scholar]
- Mori M, Isogai A, Sakagami Y, Fujino M, Kitada C, Clewell D.B, Suzuki A. Isolation and structure of the Streptococcus faecalis sex pheromone inhibitor, iAD1, that is excreted by the donor strain harboring plasmid pAD1. Agr. Biol. Chem. 1986;50:539–541. [Google Scholar]
- Mori M, Sakagami Y, Ishii Y, Isogai A, Kitada C, Fujino M, Adsit J.C, Dunny G.M, Suzuki A. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J. Biol. Chem. 1988;263:14 574–14 578. [PubMed] [Google Scholar]
- Nakayama J, Isogai A, Clewell D.B, Suzuki A. Molecular and genetic analysis of a region of Enterococcus faecalis plasmid, pPD1 containing sex pheromone sensitivity (traC), pheromone shutdown (traB) and pheromone inhibitor (ipd) genes. GB. 1994a;1:1–2. doi: 10.1128/jb.177.19.5567-5573.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Ruhfel R.E, Dunny G.M, Isogai A, Suzuki A. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid, pCF10, encodes a peptide inhibitor, iCF10. J. Bacteriol. 1994b;176:2003–2004. doi: 10.1128/jb.176.23.7405-7408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L, Paulsen I.T, Tchieu J, Hueck C.J, Saier M.H., Jr Phylogenetic analyses of the constituents of type III protein secretion systems. J. Mol. Microbiol. Biotechnol. 2000;2:125–144. [PubMed] [Google Scholar]
- Olmsted S.B, Kao S.-M, van Putte L.J, Gallo J.C, Dunny G.M. Role of the pheromone-inducible surface protein Asc10 in mating aggregate formation and conjugal transfer of the Enterococcus faecalis plasmid pCF10. J. Bacteriol. 1991;173:7665–7672. doi: 10.1128/jb.173.23.7665-7672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I.T, Nguyen L, Sliwinski M.K, Rabus R, Saier M.H., Jr Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 2000;301:75–100. doi: 10.1006/jmbi.2000.3961. doi:10.1006/jmbi.2000.3961 [DOI] [PubMed] [Google Scholar]
- Paulsen I, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. doi:10.1126/science.1080613 [DOI] [PubMed] [Google Scholar]
- Shi K, Brown C.K, Gu Z.Y, Kozlowicz B.K, Dunny G.M, Ohlendorf D.H, Earhart C.A. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc. Natl Acad. Sci. USA. 2005;102:18 596–18 601. doi: 10.1073/pnas.0506163102. doi:10.1073/pnas.0506163102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staddon J.H, Bryan E.M, Manias D.A, Chen Y, Dunny G.M. Genetic characterization of the conjugative DNA processing system of enterococcal plasmid pCF10. Plasmid. 2006;56:102–111. doi: 10.1016/j.plasmid.2006.05.001. doi:10.1016/j.plasmid.2006.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staddon J.H, Bryan E.M, Manias D.A, Dunny G.M. Conserved target for group II intron insertion in relaxase genes of conjugative elements of gram-positive bacteria. J. Bacteriol. 2004;186:2393–2401. doi: 10.1128/JB.186.8.2393-2401.2004. doi:10.1128/JB.186.8.2393-2401.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver K.E, Clewell D.B. Regulation of the pAD1 sex pheromone response in Enterococcus faecalis: effects of host strain and traA, traB, and C region mutants on expression of an E region pheromone-inducible lacZ fusion. J. Bacteriol. 1990;172:2633–2641. doi: 10.1128/jb.172.5.2633-2641.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]