Abstract
Topoisomerase poisons are chemotherapeutic agents that are used extensively for treating human malignancies. These drugs can be highly effective, yet tumors are frequently refractory to treatment or become resistant upon tumor relapse. Using a pool-based RNAi screening approach and a well characterized mouse model of lymphoma, we explored the genetic basis for heterogeneous responses to topoisomerase poisons in vitro and in vivo. These experiments identified Top2A expression levels as major determinants of response to the topoisomerase 2 poison doxorubicin and showed that suppression of Top2A produces resistance to doxorubicin in vitro and in vivo. Analogously, using a targeted RNAi approach, we demonstrated that suppression of Top1 produces resistance to the topoisomerase 1 poison camptothecin yet hypersensitizes cancer cells to doxorubicin. Importantly, lymphomas relapsing after treatment display spontaneous changes in topoisomerase levels as predicted by in vitro gene knockdown studies. These results highlight the utility of pooled shRNA screens for identifying genetic determinants of chemotherapy response and suggest strategies for improving the effectiveness of topoisomerase poisons in the clinic.
Keywords: Chk2, doxorubicin, RNAi screen, Top1, Top2A
A myriad of genetic factors influence the efficacy of cancer chemotherapy, including both somatic changes in the tumor itself as well as genetic polymorphisms present in the patient. These factors include increased expression of detoxification pumps that prevent access of the drug to its target (1), point mutations that disrupt the drug–target interaction (2, 3), and mutations in stress response pathways [e.g., p53 loss (4)]. To tailor treatment successfully to the individual patient, a more complete understanding of the genetic determinants of therapy response is necessary.
RNA interference (RNAi) exploits a mechanism of gene regulation whereby double-stranded RNAs are processed by a conserved cellular machinery to suppress the expression of genes containing homologous sequences (5). Importantly, libraries of DNA-based vectors encoding short hairpin RNAs (shRNAs) capable of targeting most genes in the human and mouse genomes have been produced and enable forward genetic screens to be performed in mammalian cells. Indeed, by using human tumor-derived cell lines treated in vitro, RNAi has been used to evaluate potential drug targets (6) or to investigate mechanisms of drug action and drug resistance by screening for new molecules that modulate the response of tumor-derived cell lines to a given chemotherapeutic agent (7–10).
Here, we evaluate the suitability of combining mouse models and RNAi to identify genetic modifiers of drug action in tumors in their natural site. Initially, we chose to investigate resistance to doxorubicin in the Eμ-Myc mouse lymphoma system. Doxorubicin (Adriamycin) is an anthracycline DNA-damaging agent that exerts its effects primarily by targeting of the topoisomerase 2 activity and DNA intercalation (11). Along with etoposide and the camptothecin derivatives, doxorubicin is one of several topoisomerase-targeted drugs currently used as front-line therapies for a wide variety of cancers. The Eμ-Myc lymphoma system has been a highly tractable model for studying the genetic determinants of chemotherapeutic response in vivo in an immunocompetent setting (12), and recently we have adapted RNAi-based loss-of-function technology for use this model (13–15). Here, we demonstrate that the Eμ-Myc system can successfully identify crucial mediators of the response to topoisomerase poisons. These genes validate for relevance in vivo, suggesting strategies for improved clinical use of these drugs.
Results
RNAi Screens Identify shRNAs Mediating Doxorubicin Resistance.
Because in vivo studies of drug sensitivity and resistance require stable gene knockdown, we performed our initial in vitro screens using retrovirally encoded shRNAs based on the MiR-30 microRNA (16). Importantly, these shRNAs can stably and efficiently knockdown target genes when expressed at single copy in the genome (13). We chose to survey shRNAs targeting the “cancer 1000,” a set of known or putative cancer-relevant genes compiled by manual curation, microarray expression data, and literature mining (17). To improve gene knockdown and facilitate in vivo experiments (13), all of the existing murine shRNAs targeting the cancer 1000 set (≈2,300 shRNAs, two to three shRNAs per gene) were cloned into a murine stem cell virus (MSCV)-based vector that coexpressed green fluorescent protein.
Our initial screens for shRNAs capable of conferring doxorubicin resistance used p19ARF−/−;Eμ-Myc lymphoma cells, which retain the p53 tumor suppressor and an intact DNA damage response (18, 19). shRNA pools were introduced into lymphoma cells by retroviral transduction, and infected cultures were treated with doxorubicin at doses that typically would kill 70–95% of cells in 24 h.
Three independent approaches were used to identify shRNAs enriched after doxorubicin treatment [supporting information (SI) Fig. S1]. Specifically, the library was screened by using either (i) single treatments of lymphoma cells transduced with low-complexity shRNA pools or, alternatively, (ii) single or (iii) serial treatments of lymphoma cells transduced with the whole shRNA set. Standard DNA sequencing of amplified provirus shRNAs was used to identify constituent shRNAs and to determine their relative representation in the treated and untreated cell populations (Fig. S2). Similar results were also produced by using high-throughput shRNA deconvolution via DNA microarrays and Solexa deep sequencing, illustrating the potential for pooled screens of expanded scope (D.L.B., S.W.L., J. Zuber, and E. Hodges, unpublished work). Irrespective of the screening approach, shRNAs targeting p53, Chk2, and Top2A (two independent shRNAs) were repeatedly identified as being enriched upon doxorubicin treatment. Additional shRNAs were also identified as becoming enriched after drug treatment via one strategy or another (Fig. S2), and these should be the subject of future studies.
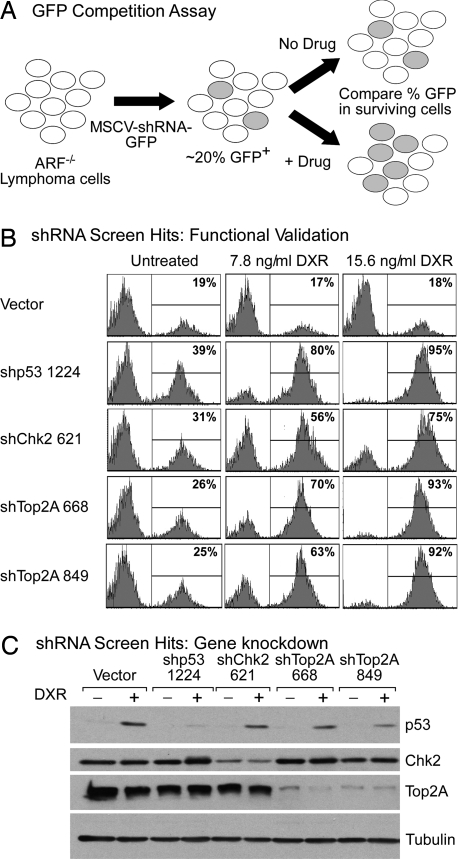
To validate the screening results, we retested the major shRNA hits in an “in vitro competition assay.” This assay examines the impact of specific shRNAs on therapy response in partially transduced cell populations, using GFP-based flow cytometry to track the survival advantage or disadvantage conferred by specific shRNAs (Fig. 1A). shRNAs targeting p53, Chk2, and Top2A successfully validated in the competition assay: the shRNAs were dramatically enriched in cell populations within 24 h after doxorubicin treatment (Fig. 1B). Additionally, these shRNAs effectively suppressed expression of their intended target (Fig. 1C).
Fig. 1.
A rapid RNAi enrichment screen identifies mediators of doxorubicin resistance. (A) The GFP competition assay. Differential survival of shRNA-transduced cells (green) relative to control cells (colorless) is assayed by changes in the percentage GFP in the surviving cell population. (B) GFP competition assay data from lymphoma cells infected with the indicated shRNA either untreated or 24 h after doxorubicin (DXR) treatment at the indicated doses. (C) Immunoblotting of lysates from lymphoma cells transduced with shRNAs targeting p53, Chk2, and Top2A either untreated or treated for 8 h with 31 ng/ml doxorubicin to stabilize p53. Tubulin serves as a loading control.
p53 and Chk2 are key components of DNA damage response pathways and, indeed, p53 loss confers resistance to doxorubicin in the Eμ-Myc transgenic model (13, 20). Importantly, multiple shRNAs targeting Chk2 promoted doxorubicin resistance, suggesting that the effects of these shRNAs were “on target,” i.e., specifically due to Chk2 gene knockdown (Fig. 1B and Fig. S3). Although Chk2 can sensitize cells to DNA-damaging agents in some contexts (21, 22), our results are consistent with a role for Chk2 in signaling p53-dependent apoptosis in lymphoid cells (20, 23). These results suggest we can identify relevant mediators of drug resistance using pool-based RNAi screening approaches.
Top2A shRNAs Cause Resistance Specifically to Topoisomerase 2 Poisons.
shRNAs targeting Topoisomerase 2α (Top2A) were the most frequently recovered shRNAs from doxorubicin-treated cells, with at least two independent shRNAs isolated per screen. Top2A is a target of the drug doxorubicin (11) and is an essential gene in mammals (24). Unlike typical enzyme inhibitors where knockdown of the drug target would be expected to mimic drug action and promote cell death, doxorubicin is a topoisomerase poison that stabilizes the cleavable complex consisting of double-stranded DNA breaks to which the enzyme is covalently attached. Doxorubicin therefore causes excessive double-stranded DNA breaks via unresolved cleavable complexes in a topoisomerase-dependent manner, thereby explaining how Top2A down-regulation might confer doxorubicin resistance (25). Remarkably, even very potent knockdown of Top2A (Fig. 1C) had little, if any, impact on cell proliferation in the absence of drug treatment, suggesting that normal cell proliferation can proceed with relatively low Top2A expression (data not shown).
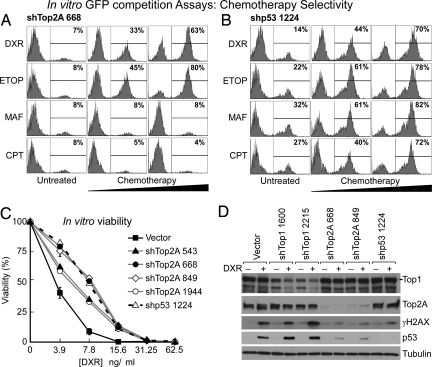
Although previous work has suggested a relationship between Top2A levels and doxorubicin sensitivity (26), the effect has not been studied extensively or validated in vivo. The effects of Top2A knockdown were specific to topoisomerase 2 poisons: shTop2A caused resistance to another, structurally unrelated TOP2A poison, etoposide, but not to the alkylating agent maphosphamide (an active metabolite of cyclophosphamide) nor the topoisomerase 1 poison camptothecin (Fig. 2A). In contrast, an shRNA targeting p53 caused cross-resistance to these different agents (Fig. 2B). The drug response-modifying effects of Top2A knockdown were likely “on target”: four of four Top2A shRNAs mediated resistance specifically to topoisomerase 2 poisons (Fig. 2C and Fig. S4 A and C). As expected, cells with reduced TOP2A levels displayed a diminished DNA damage signal and response, as shown by lower γ-H2AX signal, less p53 stabilization, and less apoptosis upon doxorubicin treatment (Fig. 2D and Fig. S5). Accordingly, the ability of Top2A shRNAs to promote doxorubicin resistance was attenuated in p53-null Eμ-Myc lymphoma cells (Fig. S4B), although clearly some signals downstream of chemotherapy-induced DNA damage are p53-independent (27).
Fig. 2.
Suppression of Top2A expression causes resistance to topoisomerase 2 poisons in vitro. (A and B) Flow cytometric analyses of lymphoma cells expressing shTop2A 668 (A) or shp53 1224 (B) after 24 h of the indicated drug treatments. DXR, doxorubicin; ETOP, etoposide; MAF, maphosphamide; CPT, camptothecin. (C) Lymphoma cells, transduced singly with four independent Top2A shRNAs, were puromycin-selected and treated with doxorubicin for 24 h at the indicated doses. Viability was assayed by flow cytometry (FSC versus SSC) and plotted relative to untreated controls. Error bars are ±SEM from three replicates. (D) Immunoblotting of lymphoma cell lysates expressing no short hairpin (Vector) or Top1, Top2A, or p53 shRNAs in the presence or absence of doxorubicin (DXR; 15.6 ng/ml for 8 h).
Top2A shRNAs Confer Resistance to Doxorubicin in Vivo.
To test the role of Top2A in doxorubicin resistance in vivo, Eμ-Myc;Arf−/− lymphoma cells were infected in vitro with shTop2A or a control vector and transplanted via tail vein injection into multiple syngeneic recipient mice. Tumor-bearing recipient mice were then treated with the maximum tolerated dose of doxorubicin (Fig. S6). Top2A knockdown caused doxorubicin resistance in vivo as measured by an in vivo competition assay (an increase in the percentage of GFP-positive cells after drug treatment; Fig. 3A) and reduced tumor-free (Fig. 3B) and overall survival (data not shown). These results demonstrate that reduced Top2A expression is a bona fide mechanism of drug resistance in vivo.
Fig. 3.
Top2A knockdown causes doxorubicin resistance in vivo. In vivo competition assay is shown. (A) GFP flow cytometry plots. Lymphoma cells were infected in vitro with GFP-tagged shTop2A 668 or 849, shp53, or vector control constructs (A Left). These cells were injected into the tail vein of syngeneic recipient mice (five mice per cohort) and were monitored daily for tumors by palpation. Upon tumor onset (day 0), one mouse from each cohort was killed, and lymphoma cells were assayed for percentage GFP+ (A Middle). The remaining mice were treated with doxorubicin (10 mg/kg i.p. injection), and tumors were harvested upon relapse and assayed for percentage GFP (A Right). (B) Kaplan–Meier tumor-free survival curves. Vector, shTop2A, and shp53 tumors were FACS-sorted to 100% GFP+ before injection into recipient mice and DXR-treated as for A at day 0.
Top1 shRNAs Confer Resistance to Topoisomerase 1 Poisons in Vitro and in Vivo.
TOP2A is not the only topoisomerase targeted by front-line anticancer therapeutics. Topoisomerase 1 (TOP1) is the target of camptothecin (28, 29) and its derivatives irinotecan (Camptosar/CPT-11) and topotecan (Hycamtin). TOP1-deficient yeast are viable and resistant to camptothecin (30), but complete knockout of Top1, like Top2A, is lethal in mammals (31). Prompted by our studies on doxorubicin and Top2A, we tested whether Top1 knockdown could induce camptothecin resistance in cancer cells. Indeed, Top1 knockdown in Eμ-Myc;Arf−/− lymphomas caused resistance specifically to camptothecin (Fig. 4A), and the effects were reproducible by using multiple independent Top1 shRNAs (Fig. 4B, Fig. S7, and Fig. S8). Even modest Top1 knockdown achieved this cytoprotective effect (Fig. 4C). Importantly, this effect was also seen in human cells expressing a TOP1 shRNA (Fig. S8C).
Fig. 4.
Top1 knockdown causes camptothecin resistance in vitro and in vivo. (A) Top1 knockdown causes resistance to camptothecin but hypersensitizes to the topoisomerase 2 poisons, doxorubicin and etoposide, as shown by a GFP competition assay 24 h after drug treatment. (B) In vitro viability assays of puromycin-selected (shRNA-containing) cells for four independent shRNAs targeting Top1, after 24-h camptothecin treatment. Error bars are ±SEM from three replicates. (C) Immunoblotting of Eμ-Myc;Arf−/− lymphoma cell lysates with or without camptothecin (31 nM CPT, 8 h). (D) Kaplan–Meier survival curve. Eμ-Myc;Arf−/− lymphomas were infected in vitro with vector control or shTop1 2215 and were FACS-sorted to 100% GFP+ before injection into recipient mice. Upon lymphoma onset (day 0) mice were treated with irinotecan (CPT-11), a clinically relevant camptothecin derivative (50 mg/kg intraperitoneal injection daily for 2 days) and monitored for survival.
p53 induction was compromised in shTop1-expressing lymphoma cells treated with camptothecin, suggesting that these cells mounted a weaker DNA damage response (Fig. 4C). Accordingly, resistance was also attenuated in an Eμ-Myc;p53−/− background (Fig. S7B). Mice harboring shTop1-expressing lymphomas displayed a reduced tumor-free survival compared with controls after treatment with irinotecan, indicating that reduced Top1 expression promotes resistance to topoisomerase 1 poisons in vivo (Fig. 4D). Therefore, sufficient expression of Top2A or Top1 is required to achieve a potent response to chemotherapeutic agents targeting each particular topoisomerase.
Top1 shRNAs Enhance Sensitivity to Topoisomerase 2 Poisons.
The drug resistance phenotypes conferred by Top1 shRNAs were specific for topoisomerase 1 poisons. For example, Top1 knockdown had little effect on tumor cell sensitivity to the alkylating agent maphosphamide (Fig. 4A). Unexpectedly, Top1 knockdown hypersensitized cells to the topoisomerase 2 poisons doxorubicin and etoposide (Fig. 4A), an effect reproduced with nine independent Top1 shRNAs (Fig. S7 and Fig. S8). Furthermore, mice harboring transplanted lymphomas expressing Top1 shRNAs showed an improved tumor-free survival compared with controls after irinotecan treatment (Fig. 5A). Therefore, in this tumor model, suppression of Top1 synergizes with topoisomerase 2 poisoning by chemotherapeutic agents.
Fig. 5.
Top1 knockdown can sensitize to doxorubicin treatment in vivo. (A) Top1 knockdown sensitizes Eμ-Myc;Arf−/− lymphomas to doxorubicin in vivo, as shown by an increased in vivo tumor-free survival after doxorubicin treatment (10 mg/kg, day 0). shTop1 data are pooled from four shTop1 1600 and four shTop1 2215 mice. (B) Predicted changes in topoisomerase expression levels occur spontaneously during treatment failure in vivo. Immunoblotting analysis of untreated lymphomas and postdoxorubicin-treated relapses from A.
Spontaneous Changes in Topoisomerase Levels Accompany Relapse After Doxorubicin Therapy.
To examine the relevance of topoisomerase status to resistance mechanisms spontaneously occurring in treated lymphomas, primary tumors and postdoxorubicin treatment relapses from Fig. 5A were analyzed for Top1 and Top2A expression levels (Fig. 5B). The relevance of Top2A levels to the emergence of tumor relapses was supported by the fact that half of the relapsed tumors displayed dramatically reduced Top2A levels (one of two control tumors and two of four shTop1-expressing tumors) without experimental manipulation via Top2A shRNAs. As further evidence that Top1 knockdown can sensitize to the topoisomerase 2 poison doxorubicin, one shTop1 relapse (relapse 3) recovered expression of Top1 to approximately wild-type levels. Relapsed tumors treated ex vivo showed resistance to doxorubicin, but not cisplatin, suggesting that the resistance mechanisms were topoisomerase-specific (Fig. S9). Together, these results indicate that although alterations in topoisomerase expression levels represent one of undoubtedly many therapy resistance mechanisms, these changes can play a substantial role in chemotherapy response in vivo.
Discussion
In this study we document the utility of combining RNAi screens with mouse cancer models to identify and characterize molecular determinants of therapeutic response that are relevant to treatment outcome in vivo. This approach is ideal for rapid in vivo validation of candidate genes and may serve as a relevant setting for conducting in vivo RNAi-based screens for genetic determinants of drug resistance. Such methodology is easily extendable to other chemotherapeutics and tumor systems to allow a more global view of therapy response mediators, including their context-dependence across different tumor and host genotypes.
The mechanism whereby Top1 and Top2A down-regulation produces resistance to their cognate poisons is probably due to a reduction in topoisomerase–DNA cleavage complexes, resulting in less DNA damage (see Figs. 2D and 4C; ref. 32). By contrast, the mechanism whereby Top1 down-regulation hypersensitizes to topoisomerase 2 poisons remains to be precisely determined. However, this effect is not simply due to a compensatory up-regulation of Top2A because Top2A levels did not increase in response to Top1 knockdown in our system (Figs. 2D and 4C). Studies in yeast suggest that an overall amount of topoisomerase activity may be required for cell viability because topoisomerase I/II double mutants exhibit more serious defects in DNA unwinding, chromatin structure, and cell cycle progression compared with either single mutant (33, 34). If so, therapeutic poisoning of Top2A with simultaneous down-regulation of Top1 could cause cellular topoisomerase activity to fall below this crucial threshold, triggering cell death. Consistent with this model, shTop1 lymphoma cells treated with doxorubicin show an impaired progression through S phase compared with control cells (Fig. S10).
The relative importance of various mechanisms to clinical drug resistance is an area of active debate. In some settings efflux pump overexpression may predominate (35), whereas in other settings, blocked apoptosis or senescence may be largely responsible for resistance (18, 36). Our studies using RNAi in vivo, together with our observation that relapsed tumors frequently display altered topoisomerase levels compared with the parental tumor, suggest that topoisomerase expression levels are relevant determinants of therapeutic response. In fact, TOP2A amplification [linked to the ERBB2 locus and thus common in human breast cancer (37)] predicts a favorable response to anthracycline therapy, if ERBB2 status is appropriately controlled for (38). Surprisingly, hemizygous deletion of TOP2A is also common in breast cancer (39), and our results suggest that patients with such deletions in TOP2A may be less responsive to doxorubicin therapy, a possibility that is readily testable.
Similarly, TOP1 levels may also influence the response to topoisomerase poisons and thus serve as a useful biomarker to guide the use of these agents in the clinic. The TOP1 gene is located on chromosome 20q12, a locus that is often amplified in colon carcinoma (40). The enhanced sensitivity to these drugs predicted to arise from higher TOP1 levels may explain, in part, why topoisomerase 1 poisons are a mainstay therapy for this disease. In contrast, hemizygous deletion of chromosome 20q12 is observed in a subset of acute myeloid leukemia samples [M. Spector, Cold Spring Harbor Laboratory (CSHL), personal communication], a leukemia where patients are typically treated with doxorubicin plus cytarabine. Consistent with our work, 20q deletions, when found as the sole chromosomal aberration, are associated with a more favorable clinical outcome (41). Although more detailed functional and clinical studies remain to be performed, our results highlight the potential of combining RNAi and in vivo mouse models to identify potential therapeutic targets as well as biomarkers for predicting treatment response.
Materials and Methods
Short Hairpin RNA Vectors.
A MiR-30-based shRNA library (16) targeting the cancer 1000 gene set (≈2,300 shRNAs) was subcloned into LTR-driven MiR30 Puro-IRES-GFP (LMP) and LTR-driven MiR30 SV40-GFP (LMS) (MSCV-based vectors) (13) in pools of 96 or 48 shRNAs, respectively. Individual shRNA constructs were generated as described previously. Targeting sequences were selected based on RNAi Codex algorithms (16) or BIOPREDsi design (42) and are available upon request.
RNAi Screens.
Lymphoma cells were cultured and infected as described previously. Eμ-Myc;Arf−/− lymphoma cells, 2 days after infection with shRNA libraries (infected to ≈30%), were treated for 24 h with 7.8 ng/ml and 15.6 ng/ml doxorubicin for lenient and stringent selection conditions, respectively. Ninety percent of the culture was removed and replaced with fresh B cell medium on day 2 and day 5 after infection to allow recovery and proliferation of surviving cells. Final samples were taken on day 8 for GFP competition assay/shRNA representation determination. Pool-by-pool screens (Fig. S1A) were performed in a 12-well format by using ≈500,000 cells per experimental condition (pool sizes 96 or 48 shRNAs). The single treatment, whole cancer 1000 library screen (Fig. S1B), was performed in six biological replicates, using 1 million live, infected cells per treatment. Serial enrichment screening (Fig. S1C) was performed by infecting 1 × 107 cells with the entire cancer 1000 shRNA library to a final infection rate of ≈20%. Unsorted populations of infected cells were treated for 24 h with 7.8 ng/ml doxorubicin and then surviving cells were allowed to regrow for 4 days in fresh medium. shRNAs from GFP-sorted surviving cells were recloned into the LMS parent vector and used to infect naïve lymphoma cells. This process was repeated until GFP enrichment was detectable acutely (at 24 h) after doxorubicin treatment. This occurred consistently after three rounds of treatment.
To identify constituent shRNAs, genomic shRNA integrants were PCR-amplified and subcloned into the LMP vector. Constituent shRNAs were identified by using the MSCV-specific 5′ primer, CCCTTGAACCTCCTCGTTCGACC.
Immunoblotting.
Western blotting was performed as described in ref. 13. Proteins were detected by using the following antibodies: anti-p53 (clone 505, 1:500; Novacastra); anti-CHK2 (clone 151-176, in-house monoclonal, 1:100); anti-TOP1 (human scleroderma serum, 1:1,000; Topogen); anti-TOP2A (rabbit polyclonal, 1:1,000; Topogen); anti-γH2AX (monoclonal clone JBW301, 1:1000; Upstate/Millipore); and anti-tubulin (B5-1-2, 1:5,000; Sigma). Secondary antibodies were horseradish peroxidase-conjugated anti-mouse/rabbit/human IgG (GE Healthcare; 1:5,000). p53 was stabilized by using 31 ng/ml doxorubicin for 8 h (Fig. 1C), 16 ng/ml doxorubicin for 8 h (Fig. 2D), or 31 nM camptothecin for 8 h (Fig. 4C).
Competition and Viability Assays.
Two days after infection, lymphoma cells were split into replicate wells of ≈500,000 cells in 12-well plates. After 24-h treatments with a range of drug doses, the GFP-positive percentage was quantified in the surviving cell population by using a BD Biosciences LSRII flow cytometer. The live cell population was gated via a forward scatter (FSC) versus side scatter (SSC) plotting. For in vivo competition assays, lymphoma cells were infected in vitro, as described above. Lymphoma cells, GFP+ FACS sorted or unsorted, as indicated, were tail vein-injected into syngeneic recipient mice. Upon tumor onset (day 0), mice were treated with doxorubicin (10 mg/kg intraperitoneal injection) or irinotecan (CPT-11, 50 mg/kg intraperitoneal injection, daily for 2 days) and monitored for overall survival and tumor-free survival. Isolation of lymphomas for the GFP competition assay was carried out as described (13, 36). For in vitro cell viability assays, lymphoma cells were treated in triplicate at the indicated doses of doxorubicin/camptothecin. Viability was determined after 24 h by an FSC versus SSC gate and plotted relative to untreated viability.
Supplementary Material
Acknowledgments.
We thank Lidia Nascimento, Beth Miller, and Stephanie Muller (CSHL) for analysis of shRNA representation by DNA sequencing, Carmelita Bautista (CSHL) for Chk2 antibodies, Holly Thompson for H2AX analysis, and Luke Gilbert for help with sensitization studies. We thank Mona Spector for critical reading and editing of the manuscript and members of the Lowe and Hannon laboratories for helpful advice and discussions. D.J.B. is an Engelhorn Scholar of the Watson School of Biological Sciences (CSHL). W.X. is in the Molecular and Cellular Biology graduate program at Stony Brook University (Stony Brook, NY). This work was supported by Alan and Edith Seligson (to L.Z.), the V Foundation for Cancer Research (to M.T.H.), a Mouse Models of Human Cancer Consortium grant, a program project grant from the National Cancer Institute (to G.J.H. and S.W.L.), and the Don Monti Memorial Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803513105/DCSupplemental.
References
- 1.Gros P, Ben Neriah YB, Croop JM, Housman DE. Isolation and expression of a complementary DNA that confers multidrug resistance. Nature. 1986;323:728–731. doi: 10.1038/323728a0. [DOI] [PubMed] [Google Scholar]
- 2.Mao Y, et al. Mutations of human topoisomerase IIα affecting multidrug resistance and sensitivity. Biochemistry. 1999;38:10793–10800. doi: 10.1021/bi9909804. [DOI] [PubMed] [Google Scholar]
- 3.Shah NP, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 4.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 5.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 6.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 7.Brummelkamp TR, et al. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol. 2006;2:202–206. doi: 10.1038/nchembio774. [DOI] [PubMed] [Google Scholar]
- 8.Berns K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 9.Whitehurst AW, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 10.Iorns E, Lord CJ, Turner N, Ashworth A. Utilizing RNA interference to enhance cancer drug discovery. Nat Rev Drug Discov. 2007;6:556–568. doi: 10.1038/nrd2355. [DOI] [PubMed] [Google Scholar]
- 11.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: When enzymes stop being nice. Prog Nucleic Acid Res Mol Biol. 2000;64:221–253. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 12.Schmitt CA, et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 13.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 14.Hemann MT, et al. Suppression of tumorigenesis by the p53 target PUMA. Proc Natl Acad Sci USA. 2004;101:9333–9338. doi: 10.1073/pnas.0403286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemann MT, et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 16.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 17.Witt AE, et al. Functional proteomics approach to investigate the biological activities of cDNAs implicated in breast cancer. J Proteome Res. 2006;5:599–610. doi: 10.1021/pr050395r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitt CA, et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002;109:335–346. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 19.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 20.Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Q, Rose JH, Zhang H, Pommier Y. Antisense inhibition of Chk2/hCds1 expression attenuates DNA damage-induced S and G2 checkpoints and enhances apoptotic activity in HEK-293 cells. FEBS Lett. 2001;505:7–12. doi: 10.1016/s0014-5793(01)02756-9. [DOI] [PubMed] [Google Scholar]
- 22.Mukhopadhyay UK, Senderowicz AM, Ferbeyre G. RNA silencing of checkpoint regulators sensitizes p53-defective prostate cancer cells to chemotherapy while sparing normal cells. Cancer Res. 2005;65:2872–2881. doi: 10.1158/0008-5472.CAN-04-2502. [DOI] [PubMed] [Google Scholar]
- 23.Hirao A, et al. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol Cell Biol. 2002;22:6521–6532. doi: 10.1128/MCB.22.18.6521-6532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akimitsu N, et al. Enforced cytokinesis without complete nuclear division in embryonic cells depleting the activity of DNA topoisomerase IIalpha. Genes Cells. 2003;8:393–402. doi: 10.1046/j.1365-2443.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang JC. Cellular roles of DNA topoisomerases: A molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 26.Gudkov AV, et al. Isolation of genetic suppressor elements, inducing resistance to topoisomerase II-interactive cytotoxic drugs, from human topoisomerase II cDNA. Proc Natl Acad Sci USA. 1993;90:3231–3235. doi: 10.1073/pnas.90.8.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strasser A, Harris AW, Jacks T, Cory S. DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell. 1994;79:329–339. doi: 10.1016/0092-8674(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 28.Hsiang YH, Liu LF. Identification of mammalian DNA topoisomerase I as an intracellular target of the anticancer drug camptothecin. Cancer Res. 1988;48:1722–1726. [PubMed] [Google Scholar]
- 29.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 30.Nitiss J, Wang JC. DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc Natl Acad Sci USA. 1988;85:7501–7505. doi: 10.1073/pnas.85.20.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morham SG, Kluckman KD, Voulomanos N, Smithies O. Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol Cell Biol. 1996;16:6804–6809. doi: 10.1128/mcb.16.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miao ZH, et al. 4-nitroquinoline-1-oxide induces the formation of cellular topoisomerase I-DNA cleavage complexes. Cancer Res. 2006;66:6540–6545. doi: 10.1158/0008-5472.CAN-05-4471. [DOI] [PubMed] [Google Scholar]
- 33.Trigueros S, Roca J. Failure to relax negative supercoiling of DNA is a primary cause of mitotic hyper-recombination in topoisomerase-deficient yeast cells. J Biol Chem. 2002;277:37207–37211. doi: 10.1074/jbc.M206663200. [DOI] [PubMed] [Google Scholar]
- 34.Uemura T, Yanagida M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: Single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 1984;3:1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rottenberg S, et al. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc Natl Acad Sci USA. 2007;104:12117–12122. doi: 10.1073/pnas.0702955104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt CA, Rosenthal CT, Lowe SW. Genetic analysis of chemoresistance in primary murine lymphomas. Nat Med. 2000;6:1029–1035. doi: 10.1038/79542. [DOI] [PubMed] [Google Scholar]
- 37.Mano MS, Rosa DD, de Azambuja E, Ismael GF, Durbecq V. The 17q12–q21 amplicon: Her2 and topoisomerase-IIα and their importance to the biology of solid tumours. Cancer Treat Rev. 2007;33:64–77. doi: 10.1016/j.ctrv.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 38.Tanner M, et al. Topoisomerase IIα gene amplification predicts favorable treatment response to tailored and dose-escalated anthracycline-based adjuvant chemotherapy in HER-2/neu-amplified breast cancer: Scandinavian Breast Group Trial 9401. J Clin Oncol. 2006;24:2428–2436. doi: 10.1200/JCO.2005.02.9264. [DOI] [PubMed] [Google Scholar]
- 39.Jarvinen TA, Liu ET. Topoisomerase IIα gene (TOP2A) amplification and deletion in cancer: More common than anticipated. Cytopathology. 2003;14:309–313. doi: 10.1046/j.0956-5507.2003.00105.x. [DOI] [PubMed] [Google Scholar]
- 40.Boonsong A, et al. Characterization of the topoisomerase I locus in human colorectal cancer. Cancer Genet Cytogenet. 2000;121:56–60. doi: 10.1016/s0165-4608(00)00242-9. [DOI] [PubMed] [Google Scholar]
- 41.Brezinova J, et al. Prognostic significance of del(20q) in patients with hematological malignancies. Cancer Genet Cytogenet. 2005;160:188–192. doi: 10.1016/j.cancergencyto.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 42.Huesken D, et al. Design of a genome-wide siRNA library using an artificial neural network. Nat Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.