Abstract
Two-hybrid technology provides a simple way to isolate small peptide aptamers that specifically recognize and strongly bind to a protein of interest. These aptamers have the potential to dominantly interfere with specific activities of their target proteins and, therefore, could be used as in vivo inhibitors. Here we explore the ability to use peptide aptamers as in vivo inhibitors by expressing aptamers directed against cell cycle regulators in Drosophila. We expressed two peptide aptamers, each of which specifically recognizes one of the two essential cyclin-dependent kinases (Cdks), DmCdk1 and DmCdk2, in Drosophila. Expression of each Cdk aptamer during organogenesis caused adult eye defects typical of those caused by cell cycle inhibition. Co-overexpression of DmCdk1 or DmCdk2 resulted in suppression of the eye phenotypes, indicating that each aptamer interacts with a Cdk target in vivo and suggesting that these peptides disrupt normal eye development by inhibiting Cdk function. Moreover, the specificity of each aptamer for one of the two Cdks as determined in two-hybrid assays was retained in Drosophila. Combined, our results demonstrate that peptide aptamers generated by yeast two-hybrid methods can serve as inhibitory reagents to target specific proteins in vivo.
Often the most effective way to determine the function of a protein is to analyze the phenotype that results from its inactivation in vivo. The traditional methods for inactivating the function of a protein involve knocking-out or otherwise obtaining loss-of-function mutations in the gene encoding the protein. Although these approaches are generally the most informative, they are often cumbersome and time consuming. An alternative and sometimes complementary approach is to express a dominant inhibitor of a normal protein in a wild-type cell or organism (1). Recently, yeast two-hybrid technology has been developed to allow identification of potential dominant inhibitors of protein function in the form of small peptides that specifically recognize a protein of interest (2–4). In principle, some of these highly specific peptides, called peptide aptamers, could inhibit the function of a protein in vivo. For example, a high-affinity aptamer could block the ability of its target protein to interact with other proteins, substrates, or cofactors.
Colas et al. (3) described a two-hybrid strategy to identify high-affinity peptide aptamers from a combinatorial library encoding 109 random dodecapeptides. They isolated aptamers that interacted with human cyclin-dependent kinase 2 (HsCdk2), a serine/threonine protein kinase required for entry into S phase (5, 6). The aptamers bound HsCdk2 tightly, with Kd values in the nanomolar range; the high affinity was due at least in part to the fact that they were expressed as conformationally constrained loops from a stable platform molecule, Escherichia coli thioredoxin (trxA) (7). Some of the aptamers were shown to inhibit Cdk kinase activity in vitro (3), suggesting that they may be able to act as in vivo inhibitors of Cdk function. Controlled expression of such aptamers in a living organism could provide a powerful approach to probe protein function.
Here, we tested the ability of peptide aptamers to interfere with Cdk function during Drosophila development. Two Drosophila Cdks have been identified that are required for normal cell proliferation, DmCdc2 (here called DmCdk1) and DmCdc2c (here called DmCdk2) (8, 9). DmCdk1 is the functional homolog of the Cdk known to be required for entry into M phase in most eukaryotes. In Drosophila, DmCdk1 is required for cell divisions in embryos and in imaginal tissues, which differentiate to form adult structures (10, 11). DmCdk2, the functional homolog of mammalian Cdk2, complexes with cyclin E to promote entry into S phase in embryonic and imaginal cells (12–14). Inhibition of Cdk function during development causes cell cycle defects, which can result in easily detectable phenotypes. For example, ectopic expression of natural Cdk inhibitors such as Drosophila Dacapo (or human p21) in cells of the developing eye imaginal disc results in adult flies with rough eyes (13, 15). This phenotype is caused by a deficit in precursor cells that normally give rise to the differentiated cells that constitute the adult eye.
We report that expression of two peptides directed against DmCdk1 or DmCdk2 during Drosophila development causes eye deformations that are typical of a cell cycle defect. The eye phenotypes were peptide dosage dependent and were suppressed by overexpression of the corresponding Drosophila Cdks. The two-hybrid specificity of each aptamer for a particular Cdk was partially retained in vivo, suggesting that peptide aptamers may be used as selective in vivo inhibitors of specific proteins or protein interactions.
MATERIALS AND METHODS
Yeast Two-Hybrid Methods.
Interaction mating assays were performed as described (16) by mating on yeast extract/peptone/dextrose plates haploid bait and prey yeast strains that had been grown on selective medium, and subsequent replica plating of mated diploids to 5-bromo-4-chloro-3-indolyl β-d-thiogalactoside indicator plates. The prey strains for interaction mating assays were RFY231 (MATα trp1Δ∷hisG his3 ura3–1 leu2∷3Lexop-LEU2), a derivative of EGY48 (17) in which the trp1–1 allele was replaced by a trp1 deletion, containing either pJG4–5 (17) or pJM-1 (3), expressing either activation-tagged Cdks or trxA-pep fusions, respectively. The bait strains were RFY206 (16) containing a lacZ reporter plasmid, pSH18–34 (18) and pEG202 (19) expressing LexA-tagged Cdks or trxA-pep fusions.
The interactor hunt for DmCdk2 aptamers was performed essentially as described (18) but by using less sensitive versions of the LEU2 and lacZ reporters for preferential isolation of high-affinity aptamers. The selection strain EGY189 (MATa trp1 his3 ura3 LYS2 leu2∷1Lexop-LEU2) containing a lacZ reporter plasmid, pRB18–40 (19), and the DmCdk2 bait-expressing plasmid was transformed with pJM-1 expressing random peptides (3). Approximately 2 × 106 library transformants were screened for interactors, and 18 DmCdk2 aptamers were isolated. The two-hybrid hunts for Drosophila interactors of pep4 and pep8 were performed by mating 108 colony-forming units of RFY231 pretransformed with 7.8 × 106 plasmids of a Drosophila embryo interaction library, RFLY1 (20), with 5 × 108 colony-forming units of RFY206/pSH18–34 expressing either LexA-trxA-pep4 or LexA-trxA-pep8 on single YPD plates for 12 h, and subsequent screening of 108 diploid colony-forming units for activation of LEU2 and lacZ reporters as described (18).
Plasmids.
Bait plasmids expressing LexA-fused DmCdk4 (21) or DmCdk5 (22) were made by inserting respective PCR-amplified Cdk-encoding fragments into the pEG202 backbone cut with EcoRI and XhoI. Plasmids expressing other Cdk baits were as described (17, 20). Bait plasmids expressing LexA-fused trxA-pep4 or trxA-pep8 were made by inserting EcoRI–SalI fragments encoding trxA-peps from the respective pJM1 prey plasmids (3) into the pEG202 backbone cut with EcoRI and SalI. For heat-shock-inducible expression of peptides in Drosophila under the control of the hsp70 promoter, we used the germ-line transformation vector, pCaSpeR-hs (23), or a derivative, pMK1, for expression of peptides with a nuclear localization signal at their N termini. pMK1 was created by inserting the following annealed oligonucleotides: 5′-TTGCAAAATGCCGACGAAGAAGCGCGTCAAGGAATTC-3′ and 3′-CGTTTTACGGCTGCTTCTTCGCGCAGTTCCTTAAGTT-5′ [encoding the nuclear localization signal from the Drosophila sry delta protein (24) downstream of a consensus ribosome binding site and start codon] into pCaSpeRhs cut with EcoRI and partially filled-in with dATP and the Klenow fragment of DNA polymerase I. To subclone trxApeps, we PCR-amplified regions encoding hemagglutinin epitope-tagged trxA-pep4, trxA-pep8, trxA-pepC2, or trxApepCa fusions from respective pJM1-trxA-pep plasmids by using the 5′ primer 5′-AGAAGATCTCAAAATGTCCTACCCTTATGATGTGCCAG-3′ (which includes a consensus ribosome binding site) and the 3′ primer, BCO2 (18), cut the products with BglII and XbaI, and inserted them into pCaSpeR-hs or pMK1 cut with BglII and XbaI. All PCR-generated constructs were sequenced.
Fly Stocks and Generation of Transgenic Flies.
pCaSpeR-hs or pMK1 derivatives were used to construct transgenic Drosophila lines expressing trxA-peps by P element-mediated transformation of w1118 embryos (25). Germ-line transformants were isolated by backcrossing to w1118 and subsequent balancing of the P element insertions over FM7C, CyO, or TM3 (26). The balanced transgenic stocks were used for construction of Drosophila homozygous for one or two independent P element insertions. Drosophila that carried the hs-DmCdk2 transgene (10) were provided by C. Lehner (Univ. of Bayreath, Germany); Drosophila that carried the hs-DmCdk1 transgene were provided by P. O’Farrell (Univ. of California, San Francisco).
Induction of Transgene Expression and Phenotypic Characterization.
We induced trxA-peptide expression by cyclic heat-shock treatment with pulses of 37°C for 10 min (or 25 min for the Cdk rescue experiments) and recovery at 25°C for 70 min. For each experiment, 25 females and 15 males were placed in vials with regular medium and transferred to new vials daily. Eggs were collected overnight, aged at 25°C in the original collection vials until an appropriate developmental stage (Table 1), and then incubated in a humidified programmable thermocycler until the beginning of eclosion (Table 1, protocols 2–4) or until the third larval instar (Table 1, protocol 1). Adults were inspected for phenotypic abnormalities with a dissecting microscope at ×25 magnification. Samples for scanning electron microscopy were prepared as described (27).
Table 1.
Incidence of rough eye phenotypes determined for Drosophila expressing peptide transgenes
| Line | Chromo-some | Heat-shock administration
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| None
|
Protocol 1 (L1-L3)
|
Protocol 2 (L3-P)
|
Protocol 3 (L2-P)
|
Protocol 4 (L1-P)
|
|||||||
| No. scored | % rough | No. scored | % rough | No. scored | % rough | No. scored | % rough | No. scored | % rough | ||
| w1118 | – | >200 | 0 | 41 | 0 | 703 | 1 | 122 | 0 | 77 | 0 |
| hs-pep4 | |||||||||||
| 1 | 2d | >200 | 0 | 140 | 2 | 567 | 51 | 68 | 73 | 66 | 56 |
| 2 | 2d | >200 | 0 | 18 | 28 | 16 | 31 | 77 | 39 | 28 | 89 |
| 3 | X | >200 | 0 | 145 | 3 | 94 | 52 | 136 | 30 | 118 | 15 |
| 4 | 3d | >200 | 0 | 13 | 0 | 34 | 20 | 54 | 4 | 21 | 14 |
| 5 | 2d | >200 | 0 | 104 | 2 | 20 | 15 | 14 | 36 | 83 | 56 |
| 6 | 2d | >200 | 0 | 60 | 40 | 11 | 27 | 12 | 67 | 159 | 8 |
| 7 | X | 296 | 28 | 36 | 56 | 35 | 60 | 48 | 67 | 148 | 62 |
| hs-NLS-pep4 | |||||||||||
| 1 | 2d | 77 | 1 | ND | ND | 548 | 49 | ND | ND | 158 | 28 |
| 2 | X | 97 | 1 | ND | ND | 158 | 15 | 49 | 39 | 78 | 58 |
| hs-pep8 | |||||||||||
| 1 | X | >200 | 0 | 169 | 4 | 544 | 31 | 136 | 30 | 48 | 19 |
| 2 | X | >200 | 0 | 87 | 0 | 185 | 13 | 35 | 57 | 129 | 74 |
| 3 | 2d | >200 | 0 | 16 | 13 | 41 | 10 | 62 | 72 | 4 | 50 |
| 4 | X | >200 | 0 | 119 | 2 | 127 | 15 | 11 | 36 | 127 | 15 |
| 5 | 3d | >200 | 0 | ND | ND | 112 | 0 | 66 | 45 | ND | ND |
| hs-NLS-pep8 | |||||||||||
| 1 | 3d | >200 | 0 | ND | ND | 501 | 21 | 89 | 31 | 81 | 25 |
| 2 | 2d | >200 | 0 | ND | ND | 170 | 14 | 47 | 30 | 109 | 30 |
| hs-pepC2 | |||||||||||
| 1 | 3d | >200 | 0 | 35 | 0 | 847 | 2 | 16 | 0 | 5 | 0 |
| 2 | X | >200 | 0 | 96 | 3 | 100 | 2 | 115 | 0 | 23 | 0 |
| 3 | 2d | >200 | 0 | 38 | 3 | 56 | 2 | 144 | 3 | 32 | 9 |
| 4 | 3d | >200 | 0 | 37 | 0 | 118 | 0 | 12 | 0 | 23 | 0 |
| 5 | 2d | >200 | 0 | 87 | 0 | 106 | 3 | 27 | 7 | 101 | 2 |
| 6 | 2d | >200 | 0 | 63 | 0 | ND | ND | ND | ND | 11 | 0 |
| hs-NLS-pepCa | |||||||||||
| 1 | 2d | 83 | 2 | ND | ND | 852 | 7 | 84 | 5 | 52 | 0 |
| 2 | X | ND | ND | ND | ND | 111 | 7 | 76 | 5 | 83 | 9 |
| 3 | 2d | 80 | 34 | ND | ND | ND | ND | ND | ND | 144 | 27 |
Progeny of lines carrying single independent homozygous hs-pep insertions were scored for frequency of adults with rough eyes either with or without induction of transgene expression by heat-shock administration. Combined data are shown from a number of experiments that involved heat-shock treatment of semisynchronized developing Drosophila beginning at the third larval instar (L3) for protocol 2, second larval instar (L2) for protocol 3, and first larval instar (L1) for protocol 4, throughout pupariation (P). In protocol 1 heat treatment began at L1 and was suspended during L3. The indicated numbers of adults were scored for rough eyes. Line 7 of hs-pep4 and line 3 of hs-pepCa produced flies with rough eyes at noninducible temperature and, therefore, were not included in calculation of average rough eye frequencies. ND, not determined. NLS, nuclear localization signal.
RESULTS
Drosophila Cdk Aptamers.
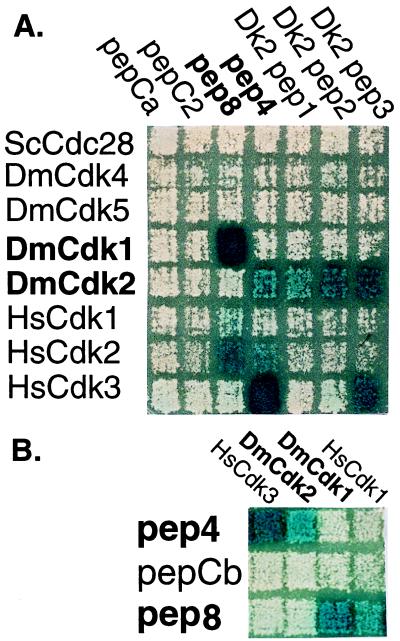
A number of peptides that tightly bind HsCdk2 were previously isolated in a two-hybrid screen of a random peptide library (3). We chose to express in Drosophila two HsCdk2 aptamers, pep4 and pep8, that cross-reacted with DmCdk2 or DmCdk1, respectively. Both aptamers had been preliminarily characterized, and one of them (pep8) had been tested and shown to inhibit Cdk kinase activity in vitro (3). We further tested the specificity of pep4 and pep8 by performing two-hybrid interaction mating assays with a number of Drosophila Cdks (Fig. 1A). Activation domain-tagged pep4 and pep8 each interacted more strongly with a Drosophila Cdk than with human Cdk2. Moreover, each interacted with only one Drosophila Cdk: pep8 interacted with DmCdk1, and pep4 interacted with DmCdk2 (Fig. 1A). Neither peptide interacted with Drosophila Cdk4 or Cdk5 (Fig. 1A), or with more than 100 other bait proteins tested (data not shown). We also showed that aptamers highly selective for DmCdk2 could be directly obtained by screening the random peptide library using DmCdk2 as the bait (Fig. 1A). For example, two peptides (Dk2pep1 and Dk2pep2) interacted exclusively with DmCdk2 and not with any of the other Cdk baits. Two randomly selected control peptides (pepC2 and pepCa) did not interact with any of the Cdks.
Figure 1.
Specificity of Cdk aptamers in yeast two-hybrid assays. Interaction corresponds to 5-bromo-4-chloro-3-indolyl β-d-thiogalactoside staining (blue) in response to expression of the lacZ reporter. Strains expressing the indicated LexA-fused baits in rows were mated with strains expressing the indicated activation domain-fused preys in columns as described (16). (A) Activation domain-tagged trxA-peps isolated in a hunt for DmCdk2 interactors (Dk2pep1, Dk2pep2, and Dk2pep3), trxA-peps [pep4, pep8, and pepC2 (3)], or a control trxA-pep (pepCa) were tested for interaction with LexA-fused yeast Cdc28 (ScCdc28), human Cdks (HsCdk1, HsCdk2, and HsCdk3), and Drosophila Cdks (DmCdk1, DmCdk2, DmCdk4, and DmCdk5). (B) The indicated activation domain-tagged Cdks were tested for interaction with LexA-fused trxA-pep4 (pep4), trxA-pep8 (pep8), or a control trxA-pep (pepCb). The DmCdk2 and DmCdk1 preys were isolated with the pep4 and pep8 baits in the respective yeast two-hybrid hunts from a Drosophila embryo cDNA library.
To further verify the specificities of pep4 and pep8 for their Cdk targets, we performed interaction mating assays with the fusion moieties on the opposite molecules: trxA-pep4 and trxA-pep8 were expressed as LexA-fused baits, and the Drosophila Cdks and other proteins were expressed as activation domain fusions. In this orientation the specificity of pep8 for DmCdk1 and pep4 for DmCdk2 was verified (Fig. 1B). A randomly selected control peptide (pepCb) did not interact with the Cdks. We also performed interactor hunts using trxA-pep4 and trxA-pep8 as baits to screen a Drosophila embryo cDNA library. In these hunts we isolated DmCdk1 but not DmCdk2 with the pep8 bait, and DmCdk2 but not DmCdk1 with the pep4 bait (Fig. 1B and data not shown). These results demonstrate that the specificity of pep4 for DmCdk2 and of pep8 for DmCdk1 is independent of the domain fused at the amino terminus of the trxA-pep.
Expression of Cdk Aptamers Causes Eye Defects.
To test whether pep4 or pep8 expression could interfere with Cdk function in vivo, we expressed each aptamer in Drosophila larvae at times when patterned cell divisions are required for normal development of the adult eye (28, 29). Because the affinity of aptamers for their targets is optimal when they are expressed as conformationally constrained loops displayed from the trxA molecule (3), we chose to express the peptides in Drosophila by using the trxA platform. We expressed the trxA-fused peptides, either with or without an amino-terminal nuclear localization signal, using the inducible hsp70 promoter (23). After inducing expression of the peptides at different intervals of larval development, we examined the adults for “rough” eye phenotypes; such phenotypes can arise when cell divisions are improperly regulated in the eye imaginal disc (15, 29–33). Table 1 shows the results from a number of different experiments with multiple independent lines, each homozygous for a single heat-shock-inducible transgene (hs-pep) that expresses a peptide. The data from six separate experiments that involved scoring more than 500 heat-shocked progeny for a representative line of each transgenic genotype (Table 1) are graphically represented in Fig. 2.
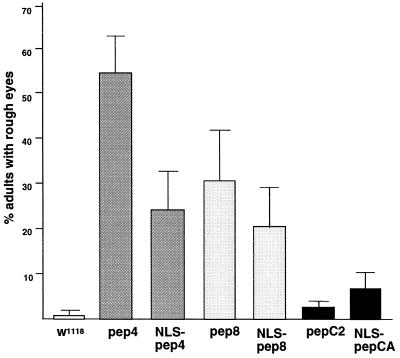
Figure 2.
Frequency of peptide-induced eye deformations. Representative Drosophila lines that gave rise to affected adults with frequencies within two SDs of the mean calculated for all lines of that genotype (Line 1 of each transgenic genotype in Table 1) were tested. Animals were exposed to cyclic heat shock from the third larval instar until eclosion. For each line more than 500 adults were scored for eye abnormalities in six experiments. Error bars show SD for the six experiments.
Heat-shock-induced expression of pep4 or pep8 throughout the third larval instar (L3) resulted in significant numbers of adults with rough eyes in most lines (Table 1). A significantly lower frequency of rough eyes resulted from expression of the two aptamers during only part of L3 (Table 1, protocol 1). The same lines did not display detectable eye phenotypes in the absence of induction, with the exception of one hs-pep4 line (Table 1). The heat-shock-induced eye abnormalities were similar in pep4- and pep8-expressing Drosophila and varied from rough eyes with disorganized ommatidia and bristles (Fig. 3 C and D) to rough eyes reduced in size due to ommatidium fusion and bristle underrepresention (Fig. 3 E and F). Cdk aptamers containing a nuclear localization signal caused eye defects similar in penetrance and severity to those induced by pep4 and pep8 lacking localization signals (Table 1 and data not shown).
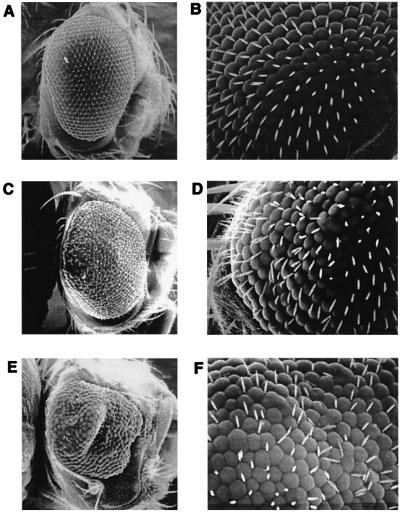
Figure 3.
Disruption of Drosophila eye development by expression of pep4 and pep8 Cdk aptamers. Scanning electron micrographs of adult Drosophila eyes developed from larvae exposed to heat shock as in Table 1, protocol 4. (A and B) Heat-shocked control hs-pepC2 (Line 1) fly showing the typical wild-type organized array of ommatidia. (C and D) A typical rough eye from a heat-shocked hs-pep4 (Line 2) fly. (E and F) A case of severe deformation of an eye from a heat-shocked hs-pep8 (Line 2) fly. (A, C, and E, ×100; B, D, and F, ×500.)
Drosophila expressing each of the different control trxA-fused peptides, pepC2 and pepCa, predominantly developed into adults with wild-type eyes (Fig. 3 A and B). Although the frequencies of rough eyes in some lines expressing the control peptides were elevated compared with the parental line, both the frequency and the severity of eye defects in these lines were significantly lower than those produced by pep4 or pep8 (Table 1). For example, the average frequency of the rough eye phenotype from all of the experiments in which heat treatment was administered throughout L3 was 36.7% for hs-pep4 lines and 31.9% for hs-pep8 lines, but was only 2.9% for lines expressing the control peptides. Moreover, in 39 of 42 of these experiments, the frequency of rough eyes induced by pep4 or pep8 was higher than the highest frequency induced by the control peptides (Table 1). Combined, these results demonstrate that expression of each of the two Cdk aptamers during eye morphogenesis considerably disrupts normal eye development in a manner consistent with inhibition of cell proliferation. They also demonstrate that expression of control peptides from the trxA platform only weakly interferes with normal eye development and that this level of interference can be easily distinguished from that induced by the Cdk aptamers. Finally, these results suggest that nuclear localization of the Cdk aptamers may not be critical for their ability to disrupt Cdk functions.
Phenotypes Induced by Expression of Cdk1 and Cdk2 Aptamers Are Dosage Dependent.
To test whether the severity of the phenotypes produced by expression of pep4 and pep8 depends on the level of peptide expression, we compared the incidence of rough eyes in adults with different numbers of hs-pep transgene insertions. Males that were hemizygous for a hs-pep insertion on the X chromosome and homozygous for a hs-pep insertion on chromosome 2 were crossed to females homozygous for the same two transgene insertions or females that carried no transgene, and the frequencies of rough eyes among the heat-shocked progeny of these crosses were compared (Fig. 4).
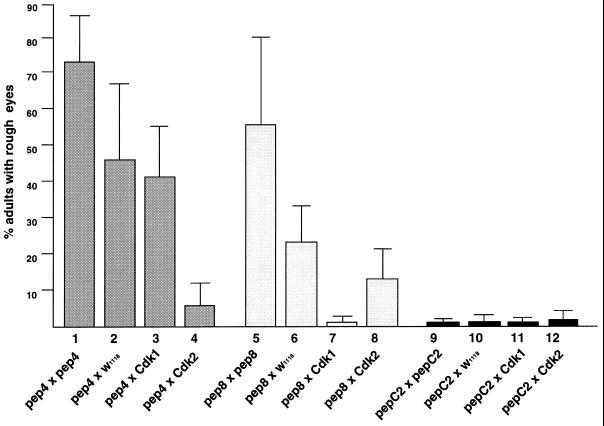
Figure 4.
Rough eye phenotypes are peptide-dosage-dependent and suppressed by overexpression of specific Cdks. Frequencies of eye defects were determined by scoring more than 300 adult progeny of each indicated cross in three experiments. Females carrying either no transgene (crosses 2, 3, 4, 6, 7, 8, 10, 11, and 12) or a homozygous hs-pep transgene (crosses 1, 5, and 9) on the X chromosome and either no transgene (crosses 2, 6, and 10), a homozygous hs-pep transgene (crosses 1, 5, and 9), a homozygous hs-DmCdk1 transgene (crosses 3, 7, and 11) or a homozygous hs-DmCdk2 transgene (crosses 4, 8, and 12) on chromosome 2 were crossed to males carrying a hemizygous hs-pep insertion on the X chromosome and a homozygous hs-pep insertion on chromosome 2. The following transgene insertions corresponding to Table 1 were used in these crosses: hs-pep4 from lines 2 and 3, hs-pep8 from lines 2 and 3, and hs-pepC2 from lines 2 and 3. Error bars show SD for the three experiments.
As shown in Fig. 4, an increase in the copy number of the control hs-pepC2 transgene (by 2-fold in females and 3-fold in males) did not influence the frequency of the rough eye phenotype. In contrast, increasing the aptamer transgene copy number by 2- to 3-fold elevated the frequency of pep4-induced rough eyes from an average of 46% to an average of 73% (1.6-fold) and the frequency of pep8-induced rough eyes from an average of 24% to an average of 55% (2.3-fold). These data suggest that the phenotypes produced by expression of the Cdk aptamers are dosage-dependent. Furthermore, the high frequencies of eye defects detected in Drosophila heterozygous for hs-pep4 and hs-pep8 transgenes confirm that the observed phenotypes are due to expression of the aptamers rather than to disruption of a gene involved in eye development by transgene insertion.
Cdk Aptamer Phenotypes Are Suppressed by Co-Overexpression of Cdk1 and Cdk2.
To investigate whether the eye phenotypes induced by expression of pep4 and pep8 result from inhibition of Cdk function, we determined the aptamer-induced rough eye frequency in lines also expressing hs-DmCdk1 or hs-DmCdk2 transgenes inducible from the hsp70 promoter. As shown in Fig. 4, overexpression of DmCdk2 resulted in a more than 8-fold reduction in the frequency of pep4-induced rough eyes. In contrast, overexpression of DmCdk1 had virtually no effect (1.1-fold reduction) on the incidence of rough eyes induced by pep4. Similarly, the frequency of pep8-induced rough eyes was reduced more than 13-fold by overexpression of DmCdk1 and only 1.8-fold by overexpression of DmCdk2 (Fig. 4). These results suggest that the eye defects caused by expression of each Cdk aptamer during late Drosophila development result, at least in part, from disruption of Cdk function. They also indicate that both Cdk aptamers interact with their in vitro targets in living Drosophila. Moreover, the rescue experiments suggest that in Drosophila pep4 and pep8 target the same Cdks that each of them binds in the two-hybrid assays.
DISCUSSION
Peptides that interfere with a specific protein offer several advantages for studying protein function in vivo. (i) Peptides have the potential to be highly selective; they could be used to target not only specific proteins but also specific functions of a given protein, for example, by disrupting individual interactions with other proteins. (ii) Inhibitory peptides would act dominantly, allowing analysis of protein function in cases where loss-of-function mutants are not available or informative. (iii) Peptides could be expressed in a controlled fashion in living cells. For example, peptide expression could be directed by conditional promoters in transfected cells, or in specific spatial and temporal patterns during development. (iv) Peptides that interfere with protein function can be easily identified and characterized by yeast two-hybrid methods.
Previous studies have shown that peptide aptamers that bind with high affinity and specificity to a target protein can be isolated from random peptide libraries by two-hybrid screens (Fig. 1) (3, 4). Some of the aptamers directed against a target protein might be expected to interfere with its function. For example, Colas et al. (3) found that some of the peptides directed against human Cdk2 inhibited its kinase activity in vitro, and B. Cohen and R. Brent (38) showed that this inhibition could be substrate-specific. The two-hybrid system also can be used to test aptamers for their ability to disrupt specific protein interactions, which allows identification of the best candidates for expression in vivo (unpublished data).
Here we tested the application of peptide aptamers to probe protein function in a living organism. We targeted two Drosophila Cdks known to be important cell cycle regulators, DmCdk1 and DmCdk2, with two specific aptamers directed against them. Expression of each Cdk aptamer during larval development caused a rough eye phenotype in adults marked by missing eye bristles and fused ommatidia. This phenotype can arise as the result of a deficit in the number of precursor cells that normally give rise to the differentiated cells that constitute the adult eye (15, 29). Moreover, it is characteristic of Drosophila defective in the function of either DmCdk1 or DmCdk2. For example, ectopic expression of natural Cdk inhibitors such as human p21 or the Drosophila p21 homolog Dacapo in the developing eye results in a rough eye phenotype (13, 15). Similar phenotypes are caused by compromised Cdk1 activity resulting from either a Cdk1 deficiency (S. Hayashi, personal communication) or from a mitotic arrest induced by constitutive activation of Cdk1 (34). Suppression of each peptide-induced rough eye phenotype by overexpression of a specific Cdk (Fig. 4) further suggests that the phenotypes we observed were caused by targeting of the Cdks and not by interference with other cell cycle regulators or cell signaling molecules required for normal eye development (27, 29, 32, 35, 36). Interestingly, the two Cdk aptamers that we tested caused no obvious developmental defects other than the eye deformations, even though DmCdk1 and DmCdk2 are essential for cell proliferation throughout Drosophila development (10, 13, 14). This could be explained in part by the fact that eye development appears to be particularly sensitive to perturbations in the pattern of cell divisions (29, 32, 33, 36). For example, overexpression of such critical cell cycle regulators as cyclin E or dE2F from the same promoter used in this study can also exclusively affect eye development (30, 31).
Our results with the peptide aptamers, combined with the minimal effects we observed from expressing two control trxA-pep fusions, suggest that trxA may be an appropriate platform for expression of aptamers in Drosophila. However, the dosage dependence of the Cdk aptamer-induced rough eye phenotypes indicates that peptide levels may be limiting and suggests that increasing aptamer expression may be useful. One approach would be to optimize the codon bias of trxA, which is particularly poor for expression in Drosophila (data not shown). Alternatively, aptamers could be expressed by using other platforms that would allow exposure of conformationally-constrained peptides. For example, green fluorescent protein, which is suitable for display of small peptide loops (ref. 37 and M. Bolin and R.L.F., unpublished results) and neutral in a variety of organisms, is an attractive candidate for the aptamer platform. Finally, peptide aptamers could readily be expressed from promoters stronger than the hsp70 promoter used herein.
Our results demonstrate that peptide aptamers isolated and characterized by two-hybrid methods may be used as reagents to disrupt protein functions during Drosophila development. The ability of the aptamers to distinguish between the two closely related Cdks, both in the yeast assays and in Drosophila, suggests that this could be a generally useful approach to target specific proteins and protein interactions in vivo. Expression of aptamers using tissue-specific and developmental-stage-specific expression systems should make them highly selective probes for protein function.
Acknowledgments
We thank Roger Brent, Barak Cohen, Erica Golemis, John Tomkiel, and Mark VanBerkum for critically reading the manuscript and John Tomkiel, Mark VanBerkum, Gerard Tromp, and members of the Finley Lab for many helpful discussions. We particularly thank Roger Brent for providing pep4, pep8, and the random peptide library and for helpful advice. We also thank Mark VanBerkum for help with injections and James Lee for technical assistance with electron microscopy.
ABBREVIATIONS
- Cdk
cyclin-dependent kinase
- trxA
thioredoxin
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Herskowitz I. Nature (London) 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 2.Yang M, Wu Z, Fields S. Nucleic Acids Res. 1995;23:1152–1156. doi: 10.1093/nar/23.7.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Nature (London) 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 4.Xu C W, Mendelsohn A R, Brent R. Proc Natl Acad Sci USA. 1997;94:12473–12478. doi: 10.1073/pnas.94.23.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pagano M, Pepperkok R, Lukas J, Baldin V, Ansorge W, Bartek J, Draetta G. J Cell Biol. 1993;121:101–111. doi: 10.1083/jcb.121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai L H, Lees E, Faha B, Harlow E, Riabowol K. Oncogene. 1993;8:1593–1602. [PubMed] [Google Scholar]
- 7.Lu Z, Murray K S, Van Cleave V, LaVallie E R, Stahl M L, McCoy J M. Biotechnology. 1995;13:366–372. doi: 10.1038/nbt0495-366. [DOI] [PubMed] [Google Scholar]
- 8.Jimenez J, Alphy L, Nurse P, Glover D M. EMBO J. 1990;9:3565–3571. doi: 10.1002/j.1460-2075.1990.tb07567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehner C F, O’Farrell P H. EMBO J. 1990;9:3573–3581. doi: 10.1002/j.1460-2075.1990.tb07568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stern B, Ried G, Clegg N J, Grigliatti T A, Lehner C F. Development (Cambridge, UK) 1993;117:219–232. doi: 10.1242/dev.117.1.219. [DOI] [PubMed] [Google Scholar]
- 11.Weigmann K, Cohen S M, Lehner C F. Development (Cambridge, UK) 1997;124:3555–3563. doi: 10.1242/dev.124.18.3555. [DOI] [PubMed] [Google Scholar]
- 12.Knoblich J A, Sauer K, Jones L, Richardson H, Saint R, Lehner C F. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 13.de Nooij J C, Letendre M A, Hariharan I K. Cell. 1996;87:1237–1247. doi: 10.1016/s0092-8674(00)81819-x. [DOI] [PubMed] [Google Scholar]
- 14.Lane M E, Sauer K, Wallace K, Jan Y N, Lehner C F, Vaessin H. Cell. 1996;87:1225–1235. doi: 10.1016/s0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- 15.de Nooij J C, Hariharan I K. Science. 1995;270:983–985. doi: 10.1126/science.270.5238.983. [DOI] [PubMed] [Google Scholar]
- 16.Finley R L, Jr, Brent R. Proc Natl Acad Sci USA. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 18.Finley R L, Jr, Brent R. In: DNA Cloning, Expression Systems: A Practical Approach. Hames B D, Glover D M, editors. Oxford, U.K.: Oxford Univ. Press; 1995. pp. 169–203. [Google Scholar]
- 19.Estojak J, Brent R, Golemis E A. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finley R L, Jr, Thomas B J, Zipursky S L, Brent R. Proc Natl Acad Sci USA. 1996;93:3011–3015. doi: 10.1073/pnas.93.7.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sauer K, Weigmann K, Sigrist S, Lehner C F. Mol Biol Cell. 1996;7:1759–1769. doi: 10.1091/mbc.7.11.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellmich M R, Kennison J A, Hampton L L, Battey J F. FEBS Lett. 1994;356:317–321. doi: 10.1016/0014-5793(94)01298-9. [DOI] [PubMed] [Google Scholar]
- 23.Pirrotta V. Biotechnology. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- 24.Noselli S, Vincent A. FEBS Lett. 1991;280:167–170. doi: 10.1016/0014-5793(91)80229-v. [DOI] [PubMed] [Google Scholar]
- 25.Spradling A C, Rubin G M. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 26.Lindsley D L, Zimm G G. The Genome of Drosophila melanogaster. San Diego: Academic; 1992. [Google Scholar]
- 27.Kimmel B E, Heberlein U, Rubin G M. Genes Dev. 1990;4:712–727. doi: 10.1101/gad.4.5.712. [DOI] [PubMed] [Google Scholar]
- 28.Tomlinson A, Ready D F. Dev Biol. 1987;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- 29.Thomas B J, Gunning D A, Cho J, Zipursky L. Cell. 1994;77:1003–1014. doi: 10.1016/0092-8674(94)90440-5. [DOI] [PubMed] [Google Scholar]
- 30.Richardson H, O’Keefe L V, Marty T, Saint R. Development (Cambridge, UK) 1995;121:3371–3379. doi: 10.1242/dev.121.10.3371. [DOI] [PubMed] [Google Scholar]
- 31.Asano M, Nevins J R, Wharton R P. Genes Dev. 1996;10:1422–1432. doi: 10.1101/gad.10.11.1422. [DOI] [PubMed] [Google Scholar]
- 32.Dong X, Zavitz K H, Thomas B J, Lin M, Campbell S, Zipursky S L. Genes Dev. 1997;11:94–105. doi: 10.1101/gad.11.1.94. [DOI] [PubMed] [Google Scholar]
- 33.Thomas B J, Zavitz K H, Dong X, Lane M E, Weigmann K, Finley R L, Jr, Brent R, Lehner C F, Zipursky S L. Genes Dev. 1997;11:1289–1298. doi: 10.1101/gad.11.10.1289. [DOI] [PubMed] [Google Scholar]
- 34.Rimmington G, Dalby B, Glover D M. J Cell Sci. 1994;107:2729–2738. doi: 10.1242/jcs.107.10.2729. [DOI] [PubMed] [Google Scholar]
- 35.Brook A, Xie J E, Du W, Dyson N. EMBO J. 1996;15:3676–3683. [PMC free article] [PubMed] [Google Scholar]
- 36.Royzman I, Whittaker A J, Orr-Weaver T L. Genes Dev. 1997;11:1999–2011. doi: 10.1101/gad.11.15.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abedi M R, Caponigro G, Kamb A. Nucleic Acids Res. 1998;26:623–630. doi: 10.1093/nar/26.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen B A, Colas P, Brent R. Proc Natl Acad Sci USA. 1998;95:14272–14277. doi: 10.1073/pnas.95.24.14272. [DOI] [PMC free article] [PubMed] [Google Scholar]