Abstract
The tsetse thrombin inhibitor, a potent and specific low molecular mass (3,530 Da) anticoagulant peptide, was purified previously from salivary gland extracts of Glossina morsitans morsitans (Diptera: Glossinidae). A 303-bp coding sequence corresponding to the inhibitor has now been isolated from a tsetse salivary gland cDNA library by using degenerate oligonucleotide probes. The full-length cDNA contains a 26-bp untranslated segment at its 5′ end, followed by a 63-bp sequence corresponding to a putative secretory signal peptide. A 96-bp segment codes for the mature tsetse thrombin inhibitor, whose predicted molecular weight matches that of the purified native protein. Based on its lack of homology to any previously described family of molecules, the tsetse thrombin inhibitor appears to represent a unique class of naturally occurring protease inhibitors. Recombinant tsetse thrombin inhibitor expressed in Escherichia coli and the chemically synthesized peptide are both substantially less active than the purified native protein, suggesting that posttranslational modification(s) may be necessary for optimal inhibitory activity. The tsetse thrombin inhibitor gene, which is present as a single copy in the tsetse genome, is expressed at high levels in salivary glands and midguts of adult tsetse flies, suggesting a possible role for the anticoagulant in both feeding and processing of the bloodmeal.
Keywords: serine protease inhibitor/trypanosomiasis
The saliva of hematophagous invertebrates contains a diverse array of potent antithrombotic molecules, including anticoagulants, vasodilators, and inhibitors of platelet function (1–3). In the case of certain “pool-feeding” insects, particularly biting flies, these antihemostatic agents are thought to facilitate the rapid formation of a subcutaneous hematoma, from which the insect can suck blood efficiently. By inhibiting the critical steps through which the mammalian thrombotic response limits blood loss at the site of tissue damage, these insects are able to feed quickly, often requiring only seconds to complete their meal. It is during feeding that many of these arthropods also transmit important infectious pathogens, including protozoa, nematodes, and viruses, which are injected into or deposited on the skin as the insect probes for blood (4, 5).
Tsetse flies (Diptera: Glossinidae) are the invertebrate hosts of African trypanosomes, protozoan parasites that cause sleeping sickness in humans and related diseases in cattle. These bloodfeeding insects have been known for decades to produce a potent salivary anticoagulant (6). First characterized as an “antikinase” (7), this activity was ultimately identified as an inhibitor of thrombin (8, 9). Recently, the tsetse thrombin inhibitor (TTI), a potent (Ki* = 584 fM) 32-aa peptide, was purified to homogeneity from salivary gland extracts of Glossina morsitans morsitans (10). This inhibitor is highly specific for thrombin, showing no activity against a panel of 10 serine proteases, including components of the human coagulation/thrombolytic cascade, as well as trypsin and chymotrypsin. In addition to its remarkable anticoagulant effect in vitro, TTI is also a potent inhibitor of thrombin-induced platelet aggregation. Of interest, preliminary N-terminal amino acid sequence data from the purified protein did not show homology to any previously identified anticoagulants or class of protease inhibitors.
Here we report the cloning and molecular characterization of TTI-encoding sequences from G. morsitans morsitans. We describe the structure of its full-length cDNA and its genomic organization, as well as its developmental and tissue-specific regulation of expression in vivo. This anticoagulant, which is to our knowledge the first tsetse-specific gene ever cloned, is also expressed in the gut of tsetses following a bloodmeal, suggesting a biologic role for TTI in both feeding and digestion. As such, it may represent a unique target for future vector-based strategies aimed at limiting tsetse survival, thereby reducing transmission of African trypanosomes.
MATERIALS AND METHODS
Insects.
The G. morsitans morsitans puparia obtained from the Tsetse Research Laboratory (Bristol University, United Kingdom) were originally established from insects collected in Zimbabwe (11). The colony has been maintained for the past 6 years in the insectary at Yale University’s Laboratory of Epidemiology and Public Health at 24–26°C with 55% humidity. The flies are fed daily on defibrinated bovine blood (Crane Laboratories, Syracuse, NY) by using an artificial membrane system (12).
cDNA Synthesis and Construction of a Salivary Gland cDNA Library.
Salivary glands from 500 1- to 2-week-old male and female adult tsetses were dissected manually, and total RNA was purified by using a guanidinium-extraction protocol (13). The poly(A)+ RNA was used to construct a cDNA library by using the Uni-ZAP XR vector cDNA kit (Stratagene). First-strand synthesis was primed with an oligo(dT) primer/linker that contains an XhoI site and transcribed by using avian myeloblastosis virus reverse transcriptase and 5-methyl-CTP. DNA polymerase I was used for the second-strand synthesis in association with RNase, and EcoRI adaptors were ligated to the blunted ends. The cDNA was cleaved with EcoRI and XhoI, ligated to vector arms, packaged (Gigapack-III Gold packaging extract, Stratagene), and plated on the E. coli cell line SURE.
Identification and Characterization of the TTI Encoding Gene.
Two degenerate overlapping DNA oligonucleotides were designed and synthesized according to the previously identified N-terminal amino acid sequence from purified TTI (1). TTI-1 (5′- GGIIGARCCIGGIGCTCCIATIGAYTA-3′) corresponds to residues 1–8 and TTI-2 (5′-ATIGAYTAYGAYGARTAYGGIGGIGA-3′) corresponds to residues 6–16 of the mature TTI protein sequence. The oligonucleotides were radioactively end-labeled with [γ-32P]ATP and hybridized to approximately 10,000 plaques from the tsetse salivary cDNA library. The hybridization conditions were 50% formamide/5× Denhardt’s solution (1× Denhardt’s solution = 0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA)/5× standard saline citrate (SSC) (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/0.5% SDS at 42°C overnight, and the washing steps were carried out in 5× SSC at 42°C. The plaques that hybridized to both oligonucleotide probes were purified, and the phagemids containing the cDNA inserts were excised by using the helper phage f1 according to the Stratagene kit instructions. The DNA sequences of five inserts from independently isolated phagemid clones were obtained from both strands by manual sequencing by using the Sequenase version 2.0 DNA sequencing kit (United States Biochemical).
Characterization of the Full-Length TTI mRNA by 5′ cDNA-Rapid Amplification of cDNA Ends (RACE) Analysis.
The 5′ end of the TTI mRNA was confirmed by using the 5′ RACE System (version 2.0) from GIBCO/BRL with TTI specific primers. Briefly, a specific minus strand primer corresponding to the 3′ end of the mature TTI-encoding sequence (TTIrev1: 5′-GCATGAGATTCCTGGCATAAG-3′) was annealed to poly(A)+ salivary gland mRNA (50–100 ng), followed by reverse transcription of cDNA by using SuperScript II reverse transcriptase (GIBCO/BRL) at 42°C. The RNA was degraded with RNase, and the cDNA was purified by using a MAX Spin (GIBCO/BRL) cartridge. The cDNA was modified by the addition of a dCTP tail by using terminal transferase, and this product was PCR amplified by using a second TTI-specific internal primer (TTIrev2: 5′-CTATGGGTGCACCTGGTTCAC) and an Abridged Anchor Primer. The generated PCR product(s) were reamplified by using a third specific internal oligonucleotide as the 3′ end primer (TTIrev3: 5′-GGTGCGGCAACTATCAGATA-3′) and the Universal Amplification Primer (GIBCO/BRL). The resulting 150-bp amplification product(s) from two independent PCR-reactions were cloned into the pGEM-T vector (Promega) and two clones from each ligation product were sequenced. The complete TTI cDNA sequence has been submitted to GenBank (accession no. AF054616).
Genomic Organization.
The organization of TTI-encoding sequences in the tsetse genome was compared with that found in the cDNA by using a PCR-amplification approach. As cDNA template, we used plasmid TTI1 (pTTI1), which was constructed by subcloning bases 27 to 216 of the full TTI-encoding sequence by PCR-amplification into the pBSK vector (Stratagene). For PCR amplifications, we used the following primers: TTI5′: 5′- CGGAATTCAACATGAAGTTTTTCACTGTACTATTTTTCTTGCTCAG-3′ and TTI3′: 5′-AGGCTGCAGTTAATCAATTCTTCTTCTGG-3′. The PCR reactions were treated for 1 min at 94°, 55°, and 72°C, respectively, for 35 cycles in a buffer containing 2.5 mM MgCl2/0.25 mM dNTPs/500 nM concentrations of each primer. The reactions were carried out in a Perkin-Elmer DNA Thermal Cycler, and the PCR products were analyzed by SDS/PAGE on a 12% acrylamide gel. The products obtained from genomic DNA amplification were subsequently cloned into the T-tailed pGEM vector (Promega) and subjected to DNA sequence analysis.
For Southern analysis, total G. morsitans DNA (approximately 1–2 μg) was restricted with the enzymes EcoRI, HindIII, XhoI, and PstI, respectively. The digestion products were analyzed by electrophoresis on a 0.8% agarose gel and transferred to a Zeta-Probe membrane (Bio-Rad). For hybridization, an antisense [γ-32P]UTP-labeled RNA probe was generated by using the linearized pTTI1 described above, according to the instructions from the RNA labeling kit (Amersham).
Expression of Recombinant TTI in Escherichia coli.
The DNA sequence encoding the mature TTI peptide was PCR amplified from pTTI1 by using the primer set TTI5′Nco (5′-GTACCATGGGGGAACCAGGTGCACCCATA-3′) and M13–20 (5′-GTAAAACGACGGCCAGT-3′). The 250-bp PCR product was digested with NcoI and XhoI, gel purified, and ligated into the pET-32 expression vector (Novagen). The recombinant plasmid (pET32-TTI) was used to transform the E. coli strain BL21(DE3) and expression of recombinant TTI (rTTI) was induced by the addition of isopropyl-d-thiogalactoside at 1 mM final concentration. After 4 hr of continuous shaking at 37°C, the cells were pelleted and resuspended in 1/10 the original culture volume of binding buffer (5 mM imidazole/0.5 M NaCl/20 mM Tris⋅HCl, pH 7.9). Triton X-100 was added to a final concentration of 0.1%, followed by incubation at 30°C for 30 min. After sonication, the cell lysate was centrifuged at 12,000 × g for 30 min at 4°C, and the supernatant was removed and tested for thrombin inhibition by using a single-stage chromogenic assay (see below).
The soluble cell lysate containing the rTTI was applied at 25°C to a His-Bind nickel resin (Novagen) affinity column (5 ml bed volume) previously equilibrated with binding buffer. After washing, the bound protein was eluted with 1 M imidazole. Approximately 5 mg of the affinity-purified rTTI was subsequently purified by RP-HPLC by using a C18 column (Vydac, Hesperia, CA) preequilibrated in 10% acetonitrile/0.1% trifluoroacetic acid. The bound protein was eluted from the C18 column under a linear gradient of 10–80% acetonitrile/0.1% trifluoroacetic acid. Pooled active fractions were lyophilized and stored at −80°C before use.
Chemical Synthesis of TTI.
A 32-aa peptide corresponding to the mature TTI protein was synthesized at the Protein Core Facility of the Centers for Disease Control (Atlanta, GA) by using fluorenylmethoxycarbonyl chemistry (14) on an Advanced MPS-396 peptide synthesizer (ChemTech). The synthetic TTI (sTTI) was purified by RP-HPLC and lyophilized before use.
Assay of Thrombin Inhibition.
A single-stage chromogenic assay (10) was used to measure the inhibition of thrombin by TTI. Briefly, 100 μl of human α-thrombin (Haematologic Technologies, Burlington, VT) in HBSA (25 mM Hepes, pH 7.5/0.1% BSA) was added to 50 μl of TTI in individual wells of a microtiter plate. After incubating at 25°C for 15 min, 50 μl of chromogenic peptide substrate S2238 (Chromogenix, Molndal, Sweden) was added, and the rate of hydrolysis (OD405/min) was measured over 5 min by using a kinetic microplate reader (Dynex, Chantilly, VA). The final concentrations of thrombin and S2238 were 500 pM and 250 μM, respectively. All experiments were carried out in duplicate.
Before determining the kinetics of thrombin inhibition, the lyophilized preparations of rTTI and sTTI were resuspended in 50 mM Tris⋅HCl, pH 7.5 and subjected to matrix-assisted laser desorption ionization (MALDI)-MS (14) to determine molecular mass and degree of purity. The molar concentration of each was then estimated by using quantitative amino acid analysis, and further dilutions of the inhibitors were made in HBSA. By using the single-stage chromogenic assay described above, a curve of the inhibited rate (Vi) of thrombin-mediated S2238 hydrolysis (OD405/min) vs. TTI concentration was constructed over a broad range of inhibitor concentrations (0–12.5 μM). These data were fit as previously described to an equation for tight-binding protease inhibitors (10, 15), to derive the apparent Ki (Ki*) for both rTTI and sTTI.
Tissue-Specific Expression of TTI.
The tissue specificity and regulation of TTI expression was determined by Northern analysis. Total RNA was prepared from salivary glands, guts, and carcasses of 50 teneral (newly emerged) and nonteneral (fed) adults, as well as from puparia (2 weeks old) and dissected second instar larvae. Expression of TTI in the anterior and midgut tissues was evaluated 12 hr after a bloodmeal. For hybridization, the antisense TTI RNA probe was used as described earlier. Hybridization was carried out in a buffer containing 7% SDS/0.5 M sodium phosphate, pH 7.2/1% BSA/1 mM EDTA at 60°C. The filters were washed at 60°C for 20 minutes in low-stringency buffer (5% SDS/40 mM phosphate buffer, pH 7.2) and subsequently in high-stringency buffer (40 mM phosphate buffer, pH 7.2/1% SDS) (15). The filters were stripped, and a radiolabeled tsetse mitochondrial RNA (mtRNA) probe (GenBank accession no. AF072373) was hybridized to quantitate the input RNA in each sample. The autoradiograms were scanned by using the computer software program nih-image 1.61, and the amount of TTI RNA in each sample was standardized according to the hybridization signal obtained with the mtRNA probe.
RESULTS
Isolation of the TTI-Encoding cDNA.
Two degenerate overlapping oligonucleotides corresponding to N-terminal amino acid sequence of purified TTI (10) were hybridized to the tsetse salivary gland cDNA library, and four independent positive clones were isolated and characterized by DNA sequencing analysis. The four clones represented individual extension products of the TTI mRNA, as evidenced by their different 5′-extensions, distinct 3′-end poly(A) tails, and five A to T conversions noted in their 3′-untranslated segments (data not shown). Only one of the clones extended to contain the ATG start codon, but all were identical within their TTI-encoding sequences. The 5′ end of the TTI mRNA was confirmed by 5′-RACE analysis, which yielded one major extension product of approximately 150 bp that was sequenced from four independent clones (data not shown). Three of the inserts extended 26 bp and one extended 25 bp upstream of the initiation codon (ATG) that was previously identified in the cDNA. Southern analysis of the 5′-RACE PCR-amplification reaction product(s) showed this 150-bp fragment to be the only TTI-homologous product, indicating that it represents the start of the predominant TTI mRNA (data not shown). The complete 303-bp sequence of the TTI cDNA is shown in Fig. 1.
Figure 1.
The TTI-encoding cDNA and its protein sequence. The complete 303-bp nucleotide sequence of TTI and the corresponding protein sequence. ATG denotes the start of the 53-aa ORF and the underlined amino acids correspond to the putative signal–peptide sequence that was not present in the peptide purified and characterized from saliva. The ∗ represents the stop codon; the bold codons aaataaa in the 3′-untranslated segment represent the putative polyadenylation signal. The poly(A)19 tail is underlined.
Organization of the TTI-Encoding mRNA.
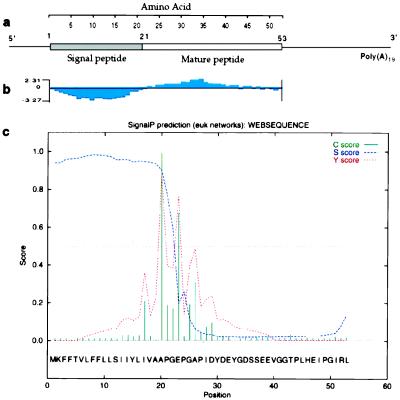
The full-length TTI cDNA contains a 26-bp untranslated segment at its 5′ end, followed by a 159-bp (53-aa) ORF and a 99-bp untranslated 3′-end sequence (schematically shown in Fig. 2a). The 5′ end of the ORF has a 63-bp sequence that corresponds to a 21-aa putative signal peptide not identified by N-terminal protein sequencing of native TTI (9). However, residues 22 to 42 of the predicted ORF are identical to the first 20 aa of TTI purified from tsetse salivary glands. The mature TTI encoded by the cDNA corresponds to a 32-aa peptide, with an acidic pI (3.71) and a predicted molecular mass of 3,370 Da, which is within 5% of that estimated for the purified native inhibitor (3,530 Da) by laser desorption mass spectrometry (10). The 3′-segment has an untranslated 99-bp sequence with a typical polyadenylation signal (A3TA3) 30 bp upstream of the poly(A) tail. A hydrophilicity (Kyte-Doolittle) plot of the 53-aa sequence was constructed by using protein analysis computer software (dnastar). This plot shows that the 20-aa residues at the 5′ end are significantly more hydrophobic than those corresponding to the mature TTI (Fig. 2b). The 53-aa TTI sequence was also analyzed by using the eukaryotic signal-peptide prediction program, psort (16), which strongly predicted cleavage between amino acid residues 19 and 20 (Fig. 2c).
Figure 2.
Structure of the putative TTI mRNA. (a) The 303-bp TTI cDNA has a 53-aa ORF composed of a 21-aa putative signal peptide sequence and a 32-aa mature TTI peptide, based on the N-terminal aa analysis of the previously purified TTI from the saliva. (b) The hydrophobicity (Kyte-Doolittle) plot of the 53-aa ORF sequence constructed by using the Protean option of dnastar software shows a hydrophilic region corresponding to the putative signal peptide segment. (c) The eukaryotic signal peptide prediction program psort predicted cleavage between amino acid residues 19 and 20.
Genomic Organization of TTI.
Southern analysis was performed to determine the genomic copy number of TTI-encoding sequences (Fig. 3a). Only one hybridizing DNA fragment was detected in genomic DNA digested with restriction enzymes EcoRI, HindIII, XhoI, and PstI, indicating that the TTI is encoded by a single copy gene (Fig. 3a, lanes 1–4, respectively). The EcoRI digestion (lane 1) was found to be partial, and subsequent longer digestion times also revealed only one fragment (data not shown). To determine whether TTI genomic sequences contained any intervening sequences, PCR amplification was performed by using the TTI cDNA insert and tsetse genomic DNA as template. Both amplifications generated an approximately 200-bp fragment, in addition to an 80-bp fragment that was found only with genomic DNA (Fig. 3b). The 200-bp genomic fragment was sequenced and found to be identical to the cDNA, while the smaller product constituted a primer dimer (data not shown). Thus, no intervening sequences were noted within the coding segment of TTI.
Figure 3.
Genomic analysis of TTI. (a) Southern analysis of G. morsitans morsitans genomic DNA cleaved with EcoRI, HindIII, XhoI, and PstI restriction enzymes (lanes 1–4, respectively) and then hybridized to a TTI probe. (b) PCR-amplification analysis of TTI by using genomic DNA (lane 1) and TTI-cDNA (lane 2) as template DNA.
Expression of Recombinant TTI in E. coli.
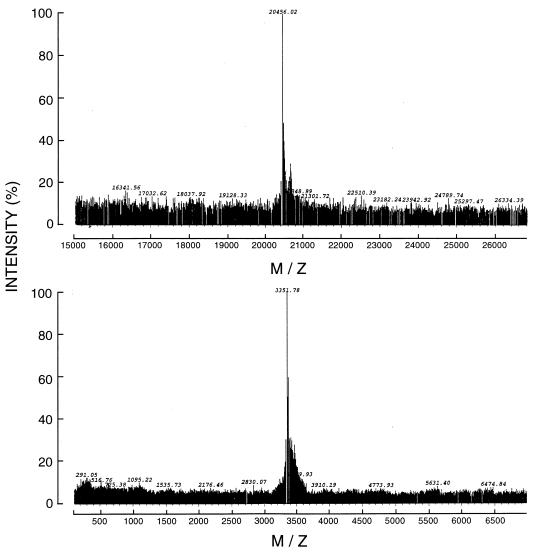
The cDNA fragment corresponding to the mature 32-aa TTI was cloned into the pET32 expression vector. Lysates of induced cells were found to inhibit thrombin activity as measured by a single-stage chromogenic assay, while noninduced transformed E. coli, as well as induced wild-type E. coli lysates, did not inhibit thrombin in vitro. The pET32–rTTI fusion protein was purified by using a combination of nickel resin affinity and RP-HPLC chromatographies. The purified recombinant protein was further characterized by MALDI-MS, which revealed a single ion species at a mass of 20,456 Da (Fig. 4), similar to the predicted mass of the pET32 vector fusion protein (approximately 17 kDa) plus TTI. The purity of this rTTI preparation was estimated based on MALDI-MS to be >95%. The 32-aa peptide comprising the mature TTI protein was also chemically synthesized and purified by RP-HPLC. The sTTI had a molecular mass of 3,351 Da, as determined by MALDI-MS (Fig. 4), with an estimated purity of >95%.
Figure 4.
MALDI-MS of rTTI and sTTI. Both proteins were purified by RP-HPLC before analysis. The rTTI (Upper) contains a 17-kDa N-terminal fusion peptide, while sTTI (Lower) corresponds to the 32-aa mature inhibitor.
In Vitro Characterization of rTTI and sTTI.
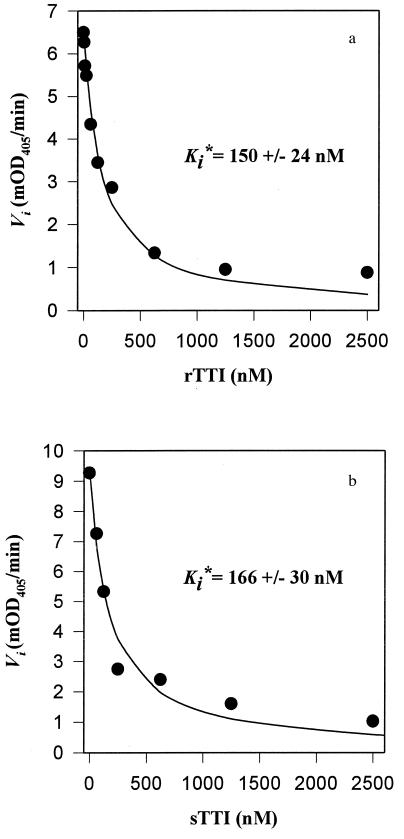
We characterized the inhibitory kinetics of rTTI by measuring the rate of thrombin-mediated hydrolysis of chromogenic substrate S2238 in the presence of increasing molar concentrations of rTTI. The data were fit to an equation that describes the kinetics of tight binding inhibitors (15), with a Ki* of 150 ± 24 nM (Fig. 5a). This value is significantly higher than that previously determined for native TTI (584 fM) under identical conditions (10). The Ki* for the sTTI peptide was 166 ± 30 nM, similar to that derived for the recombinant inhibitor (Fig. 5b).
Figure 5.
Inhibitory kinetics of rTTI and sTTI. The rate of thrombin (500 pM) hydrolysis of chromogenic substrate S2238 was determined in the presence of increasing concentrations of rTTI (a) and sTTI (b). The data from the above curves were fit to an equation describing the kinetics of tight-binding protease inhibitors (24) to derive the respective Ki* values.
Expression of TTI in Vivo.
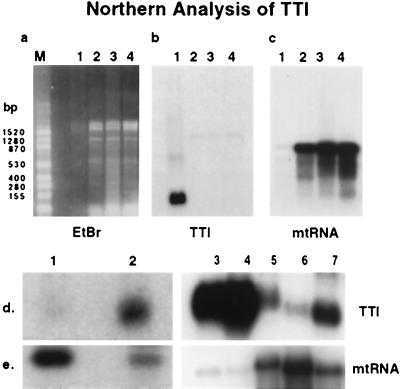
Total RNA from adult salivary glands, carcasses (without salivary glands and guts), pupal, and larval tissues, was analyzed by Northern analysis (Fig. 6a–c, lanes 1–4, respectively). An abundant transcript of approximately 250-bp was detected only in the adult salivary glands (Fig. 6b, lane 1) although both EtdBr staining (Fig. 6a) and mtRNA hybridization (Fig. 6c) indicated the presence of ample RNA in all preparations. Although newly emerged flies did not show significant levels of TTI expression in the gut (Fig. 6d, lane 1), we did find expression 6 hr following a bloodmeal (Fig. 6d, lane 2). Salivary gland expression of TTI was also up-regulated in response to bloodfeeding, with at least a 2-fold increase seen 12 hr after a bloodmeal (Fig. 6d, lane 3 vs. lane 4). When specific regions of the alimentary tract were evaluated, we found that the expression of TTI was approximately 5-fold greater in the anterior portion of the midgut than in the posterior half (Fig. 6d, lane 7 vs. lane 6). The amount of RNA detected in each sample was standardized according to the hybridization signal obtained with the mtRNA probe (Fig. 6e).
Figure 6.
Northern analysis of TTI. (a–c) Northern analysis of RNA purified from adult salivary gland (lane 1), carcasses (salivary gland and midgut removed, lane 2), pupae (lane 3), and larvae (lane 4). a shows the 2% agarose gel stained with ethidium bromide, indicating the presence of ample RNA in all lanes; b shows the hybridization with TTI–cDNA probe; c shows hybridization to mtRNA, again indicating the presence of RNA in the TTI-negative lanes. (d) Tissue-specific and bloodmeal-induced expression of TTI: RNA from teneral midguts (lane 1), from midguts 6 hr after bloodmeal (lane 2), from teneral salivary glands (lane 3), from salivary glands 12 hr after bloodmeal (lane 4), from total gut tissue 12 hr after bloodmeal (lane 5), from midguts 12 hr after bloodmeal (lane 6), and from anterior gut tissue 12 hr after bloodmeal (lane 7). The hybridization probe was TTI–cDNA. (e) The filter in d was hybridized to the mtRNA probe to quantitate the total amount of RNA in each preparation.
DISCUSSION
It has been known since the early part of this century that tsetse flies produce a potent anticoagulant. In 1924, Yorke and Macfie reported that a salivary gland emulsion from Glossina tachinoides profoundly inhibited the clotting of whole blood (6). Soon thereafter, Lester and Lloyd (7) confirmed that salivary glands from G. tachinoides, as well as G. morsitans, contained an anticoagulant and referred to this activity as an “anti-kinase.” However, it was not until 1966 that Hawkins (8) determined that all of the anticoagulant activity present in salivary extracts of Glossina austeni could be attributed to the inhibition of thrombin, the terminal protease in the mammalian coagulation cascade. A similar antithrombin activity was later identified in G. morsitans (9), and in 1996 we reported the purification and characterization of this low molecular weight anticoagulant, which we named the tsetse thrombin inhibitor (TTI) (10). In addition to its small size (3,530 Da), TTI is one of the most potent (Ki* = 584 fM) naturally occurring anticoagulants ever identified, with remarkable specificity for thrombin.
A full-length cDNA encoding TTI has now been isolated from a G. morsitans morsitans salivary gland specific library, and the 5′ end of the TTI mRNA was confirmed by using 5′ RACE analysis. The translated amino acid sequence contains a putative 21-aa secretory signal before the start of the mature protein. Interestingly, the eukaryotic signal peptide prediction software program psort predicts cleavage between aa residues 19–20. In contrast, direct amino acid sequencing of the purified TTI identified the first residue as glycine (position 21 of the translated TTI cDNA), raising the possibility that TTI might be expressed as a pre-pro-mature protein, and the dipeptide Ala-Pro is subsequently cleaved. A similar phenomenon has been observed for the cecropins, a family of arthropod-derived antibacterial peptides that contain a two aa propeptide (17), which is subsequently cleaved by a dipeptidyl amino peptidase. Based on Southern analysis, the 303-bp mRNA for TTI is encoded by a single-copy gene, with no identifiable intervening sequences. Of at least seven other tsetse salivary gland gene products currently being characterized by our laboratory, it appears that TTI is by far the most highly expressed transcript (unpublished observation).
Extensive database searches revealed no significant homology between TTI and any other previously identified naturally occurring anticoagulants, including rhodniin (18) and triabin (19), two thrombin inhibitors recently cloned from Rhodnius prolixus and Triatoma pallidipennis, respectively. The TTI sequence also shows no homology to hirudin (20, 21), the thrombin inhibitor isolated from the leech, Hirudo medicinalis, suggesting that G. morsitans has evolved a novel protease inhibitor designed to block mammalian coagulation.
We expressed rTTI as a fusion protein in E. coli, and the inhibitor was subsequently purified by using affinity chromatography and RP-HPLC. Although rTTI inhibits the catalytic activity of thrombin in vitro, it is substantially less active (Ki* = 150 ± 24 nM) than the native salivary protein (10). We also evaluated the in vitro inhibitory activity of the sTTI, in part to determine whether the presence of the large N-terminal fusion tag was responsible for the decreased activity of rTTI. Interestingly, sTTI is virtually identical to rTTI with regard to its kinetics of thrombin inibition, each with a Ki* approximately 2,000-fold greater than native TTI. These data suggest that posttranslational modifications or additional folding are required in order for TTI to become fully active, despite the presumed absence of intramolecular disulfide bonds or obvious glycosylation sites in the mature protein.
We report here evidence that expression of TTI in vivo occurs both in salivary glands and the midgut of the adult tsetse. To our knowledge, anticoagulant gene expression has never before been demonstrated in nonsalivary tissues of any hematophagous arthropod. Moreover, TTI expression in the gut appears to be induced in response to a bloodmeal, as detected by Northern analysis of guts dissected at 6 hr after feeding. These data suggest a previously uncharacterized bifunctional role for TTI in the biology of G. morsitans. First, injection of TTI with saliva into the skin would most likely facilitate feeding from lacerated vessels, presumably by inhibiting both local fibrin generation and thrombin-mediated platelet aggregation. Second, de novo secretion of TTI in the gut would potentially serve to keep the ingested blood in a liquid state, thereby aiding in its processing and digestion. Interestingly, this hypothesized role of TTI in the management of a bloodmeal corroborates the early observations of Lester and Lloyd (7), who showed that flies with dissected salivary glands fed without difficulty, but later died with clots found throughout the alimentary canal. This finding also raises questions about the biologic function of the trypsin-like midgut fibrinolysins previously isolated from tsetse (22), particularly with regard to their putative role in clot lysis.
Because of the extensive antigenic variation African trypanosomes display within their mammalian hosts, there are currently no effective vaccines available to prevent infection in humans or cattle. Moreover, the widespread use of antitrypanosomal drugs is expensive and has been associated with the development of parasite resistance (23). Therefore, vector control strategies, including reduction of tsetse populations within farming and residential communities, has become an important disease management strategy. Characterization of tsetse gene products such as TTI, particularly as they relate to insect survival and trypanosome acquisition, remains an important step in the development of new vector-based strategies for parasite control. It will be important to determine whether inhibition of TTI in vivo impairs the ability of tsetses to feed on their vertebrate hosts, to evaluate the potential of a TTI-based vaccine to reduce disease transmission. Furthermore, it remains to be seen whether TTI also plays an indirect role in trypanosome transmission, perhaps by facilitating the escape of the parasite from the ingested blood bolus within the gut of the fly. We hope to fully characterize the transcriptional regulatory elements for TTI, which are responsible for bloodmeal-induced expression and secretion in salivary glands and the gut. As insect transformation systems become more readily available, this may allow us ultimately to use the TTI promoter to express and secrete foreign gene products with antitrypanosomal activity in the two primary insect tissues where parasites reside.
Acknowledgments
We thank Danny L. Jue and Tamara K. Crews of the Biotechnology Core Facility Branch, National Centers for Infectious Diseases, Centers for Disease Control and Prevention, for production of the synthetic peptide used in this study. We also thank S.L. O’Neill for his critical reading of the manuscript, as well as Qiuying Cheng for helpful discussions of this work. This work was supported by National Institutes of Health Grants AI-34033 (S.A.), AI-01299 (M.C.), and HD-27757 (M.C.), and National Science Foundation Grant 9511806 (S.A.). S.L. is funded by the Li Foundation, and X.C. is funded by the McKnight Foundation.
ABBREVIATIONS
- TTI
tsetse thrombin inhibitor
- rTTI
recombinant TTI
- sTTI
synthetic TTI
- pTTI1
plasmid TTI
- RACE
rapid amplification of cDNA ends
- MALDI
matrix-assisted laser desorption ionization
- mtRNA
mitochondrial RNA
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF054616).
References
- 1.Lehane M J. Biology of Blood-Sucking Insects. London: HarperCollins; 1991. [Google Scholar]
- 2.Ribeiro J M C. Annu Rev Entomol. 1987;32:463–478. doi: 10.1146/annurev.en.32.010187.002335. [DOI] [PubMed] [Google Scholar]
- 3.Stark K R, James A A. Parasitol Today. 1996;12:430–437. doi: 10.1016/0169-4758(96)10064-8. [DOI] [PubMed] [Google Scholar]
- 4.Ribeiro J M C. Infect Agents Dis. 1995;4:143–152. [PubMed] [Google Scholar]
- 5.Ribeiro J M C. Exp Parasitol. 1989;69:104–106. doi: 10.1016/0014-4894(89)90177-x. [DOI] [PubMed] [Google Scholar]
- 6.Yorke W, Macfie J W S. Ann Trop Med Parasitol. 1924;18:103–108. [Google Scholar]
- 7.Lester H, Lloyd L. Bull Entomol Res. 1926;19:39–60. [Google Scholar]
- 8.Hawkins R I. Nature (London) 1966;212:738–739. [Google Scholar]
- 9.Parker K, Mant M. Thromb Haemostasis. 1979;42:743–751. [PubMed] [Google Scholar]
- 10.Cappello M, Bergum P W, Vlasuk G P, Furmidge B A, Pritchard D I, Aksoy S. Am J Trop Med Hyg. 1996;54:475–480. doi: 10.4269/ajtmh.1996.54.475. [DOI] [PubMed] [Google Scholar]
- 11.Gooding R, Jordan A. Can J Genet Cytol. 1986;28:1016–1021. [Google Scholar]
- 12.Moloo S K. Parasitology. 1971;63:507–512. doi: 10.1017/s0031182000080021. [DOI] [PubMed] [Google Scholar]
- 13.Chirgwin J, Przybyla A, MacDonald R, Rutter W. Biochemistry. 1979;18:5294. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 14.Barany G, Merrifield R B. In: The Peptides. Gross E, Meienhofer J, editors. New York: Academic; 1980. [Google Scholar]
- 15.Morrison J F, Walsh C T. Adv Enzymol Relat Areas Mol Biol. 1988;61:201–300. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 16.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boman H G. Eur J Biochem. 1991;201:23–31. doi: 10.1111/j.1432-1033.1991.tb16252.x. [DOI] [PubMed] [Google Scholar]
- 18.Tapparelli C, Matternich R, Ehrhardt C, Zurin M, Claeson G, Scully M F, Stone S R. J Biol Chem. 1993;268:4734–4741. [PubMed] [Google Scholar]
- 19.Noeske-Jungblut C, Haendler B, Donner P, Alagon A, Possani L, Schleuning W. J Biol Chem. 1995;270:28629–28634. doi: 10.1074/jbc.270.48.28629. [DOI] [PubMed] [Google Scholar]
- 20.Rydel T J, Ravichandran K G, Tulinsky A, Bode W, Huber R, Roitsch C, Fenton J W. Science. 1990;249:277–280. doi: 10.1126/science.2374926. [DOI] [PubMed] [Google Scholar]
- 21.Markwardt F. Thromb Haemostasis. 1991;66:141–152. [PubMed] [Google Scholar]
- 22.Endege W O, Lonsdale-Eccles J D, Olembo N K, Moloo K, ole-MoiYoi O K. Comp Biochem Physiol. 1989;92B:25–34. doi: 10.1016/0305-0491(89)90308-8. [DOI] [PubMed] [Google Scholar]
- 23.Bacchi C J. Parasitol Today. 1993;9:138–145. doi: 10.1016/0169-4758(93)90145-6. [DOI] [PubMed] [Google Scholar]
- 24.Williams K R, Samandar S M, Stone K L, Saylor M, Rush J. In: The Protein Protocols Handbook. Walker J M, editor. Totowa, NJ: Humana; 1996. pp. 541–555. [Google Scholar]