Abstract
The chromosomal DNA of the bacteria Streptomyces ambofaciens DSM40697 is an 8-Mb linear molecule that ends in terminal inverted repeats (TIRs) of 210 kb. The sequences of the TIRs are highly variable between the different linear replicons of Streptomyces (plasmids or chromosomes). Two spontaneous mutant strains harboring TIRs of 480 and 850 kb were isolated. The TIR polymorphism seen is a result of the deletion of one chromosomal end and its replacement by 480 or 850 kb of sequence identical to the end of the undeleted chromosomal arm. Analysis of the wild-type sequences involved in these rearrangements revealed that a recombination event took place between the two copies of a duplicated DNA sequence. Each copy was mapped to one chromosomal arm, outside of the TIR, and encoded a putative alternative sigma factor. The two ORFs, designated hasR and hasL, were found to be 99% similar at the nucleotide level. The sequence of the chimeric regions generated by the recombination showed that the chromosomal structure of the mutant strains resulted from homologous recombination events between the two copies. We suggest that this mechanism of chromosomal arm replacement contributes to the rapid evolutionary diversification of the sequences of the TIR in Streptomyces.
The chromosomal DNA as well as numerous plasmids are linear in the genus Streptomyces (1–5). They belong to the class of genetic elements called invertrons, which are characterized by terminal inverted repeats (TIRs) and a protein covalently attached to their 5′ ends (6). The length of the TIR is highly variable, from 44 bp (SLP2 of S. lividans 66) to 180 kb (pPZG101 of S. rimosus) for the plasmids (7, 8) and from 24 kb (S. griseus) to 550 kb (S. rimosus) for chromosomes (3, 5).
Comparisons of the TIR sequences between different Streptomyces linear replicons revealed similarities only between the last few tens of bases. This area of similarity represents a very small fraction of the whole length of the repeated sequences (7, 9–11) and is rich in palindromic sequences that are involved in the priming of the 5′-terminal DNA segment of the lagging strand (12–14).
The discrepancy in the size of the TIRs is accompanied by a high level of sequence polymorphism between the terminal duplications of the different replicons. Therefore, despite a very close phylogenetic relationship (15, 16) and a highly conserved gene order among the S. coelicolor A3(2), S. lividans 66, and S. ambofaciens DSM40697 chromosomes (4, 17), there is no significant cross-hybridization of their TIR (18, 19). These data showed that the sequences at the ends of the chromosome are subject to a different evolution mechanism compared with the rest of the chromosome.
The chromosomal ends of Streptomyces are subject to a particularly high degree of genetic instability. Large-scale deletions and DNA sequence amplifications are intimately associated with a high frequency (>10−3) of spontaneous mutations. The deletable region corresponds to the chromosomal ends, and the deletions affect either one or both chromosomal arms. The amplifications most often are adjacent to a deletion termini (recently reviewed in refs. 20 and 21).
Here, we describe a new type of chromosomal rearrangement that results from a homologous recombination event and leads to a homogenization of the terminal sequences. This event, together with the reports of sequence exchanges between linear plasmids and chromosomes in Streptomyces (1, 7, 8), suggested to us a mechanism to account for the rapid evolution of the TIRs.
MATERIALS AND METHODS
Bacterial Strains and Culture Conditions.
Escherichia coli Sure (Stratagene) was cultured in Luria–Bertani medium and used as host strain for genomic libraries construction. E. coli JM101 and its derivative E. coli JM109 (Stratagene) were used as a recipient for electroporation and propagation of the M13 mp18 derivatives. Both were grown in 2xYT liquid medium (22) and poured as a top layer in Soft-Agar (SA) onto Hard-Agar (HA) plates (22).
S. ambofaciens was cultured in YEME medium (23) for pulsed-field gel electrophoresis (PFGE) DNA preparation or in Hickey–Tresner medium for classical DNA extraction.
DNA Manipulations, Reagents, and Enzymes.
Extraction of total DNA from S. ambofaciens for gene libraries preparation, extraction of high-molecular-weight genomic DNA, endonuclease cleaving in agarose blocks, and separation of the DNA fragments by PFGE were as reported previously (4). Restriction enzymes and molecular biology reagents were purchased from New England Biolabs and Boehringer Mannheim.
Restriction fragments were purified from agarose gels by using the Geneclean procedure (Bio 101).
DNA was labeled with digoxigenin-labeled dUTP, and hybrids were detected by using the Dig DNA labeling and detection kit (Boehringer Mannheim).
Gene libraries of mutant strains of S. ambofaciens were constructed from a BamHI partial digest of the total DNA in Supercos1 (24), according to the recommendation of the supplier of the Gigapack III gold encapsidation kit (Stratagene). The wild-type (wt) library was constructed previously (25).
Cosmid DNA, and double-stranded or single-stranded bacteriophage M13 derivative DNA were isolated as reported (22).
DNA Sequencing and Sequence Analysis.
Restriction fragments from the sequenced regions were subcloned in the vector M13 mp18 (Sigma) (26). The dideoxy chain-termination sequencing method was performed on single-stranded or double-stranded DNA derived from the vector M13 mp18. The Sequenase 2.0 kit (United States Biochemical) was used with both dGTP or its analog, dITP, during the elongation step or the Taqtrack sequencing system (Promega) with the nucleotide analog 7-deaza-dGTP instead of dGTP in the reaction mixes. DNA fragments were radioactively labeled with [35S]dATP (NEN). The oligonucleotides used as primers were M13 forward, M13 reverse (Sigma), and an internal region of the ORFs 5′-CAGGGAGGCGGAGTTGT-3′. The blast 2.0 network service (27) and DNA or protein sequences from the EMBL, GenBank, PIR, and SWISSPROT databases were used to detect similarities between DNA sequences or potential protein encoded by these DNA sequences. The seqpup program was used to perform alignment of the DNA sequences corresponding to the hasR and hasL regions.
RESULTS
Chromosomal Arm Replacement in Strain NSA135HPP.
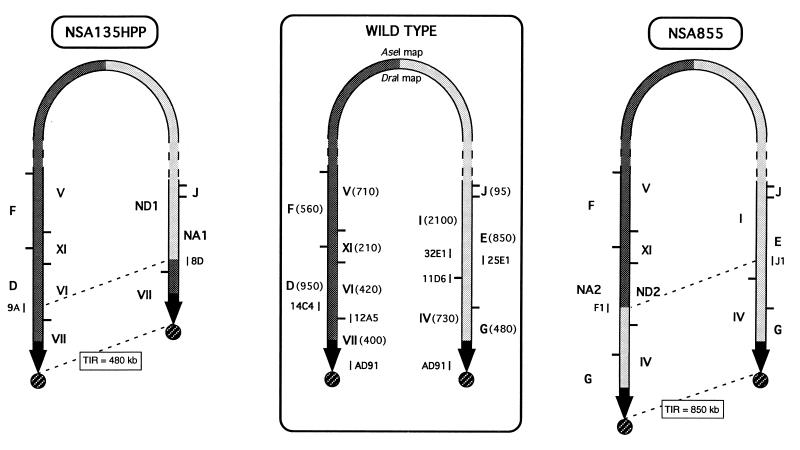
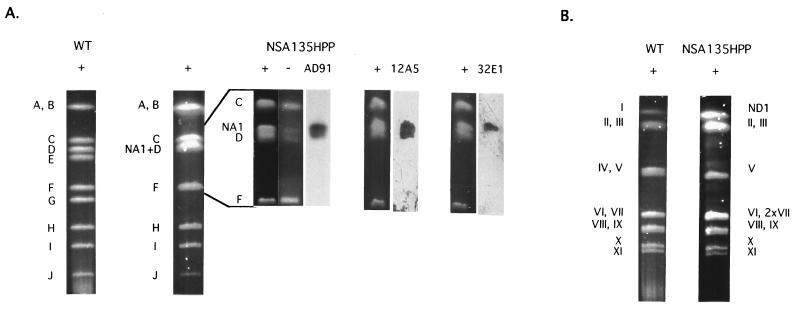
This strain, more properly, its ancestor, NSA135H (28), was isolated previously from genetic instability of S. ambofaciens as a mutant affected in colony morphology. Its chromosomal structure results from the deletion of 850 kb of the right chromosomal end (left and right are according to the map orientation on Fig. 1) and its replacement by 480 kb of sequences identical to the left end, the latter remaining unaffected by the rearrangement (Fig. 1). Thus, in the AseI PFGE pattern of the total DNA of this strain, fragments E (850 kb) and G (480 kb, right chromosomal end in the wt strain) were missing and a new AseI fragment of 960 kb (called NA1 for New AseI fragment 1 on Figs. 1 and 2A) was detected. This fragment, as well as the left-terminal AseI-D fragment of 950 kb, was covalently bound to a protein, as it did not enter a PFGE gel when the proteinase treatment was omitted of the DNA preparation (1, 4) (Fig. 2A). In addition, both fragments were revealed by cosmid AD91, flanking the ends of the TIR in the wt strain (4) (Figs. 1 and 2A). Therefore, the NA1 fragment corresponded to the right extremity of the linear chromosomal DNA of this strain (Fig. 1).
Figure 1.
Chromosomal arm replacement in strains NSA135HPP and NSA855. The AseI and DraI restriction maps of the unstable region of the wt strain DSM40697 are presented in the middle of the figure. The size of the restriction fragments, in kilobases, is given in brackets. Sequences from the left chromosomal arm are symbolized in dark gray while those from the right arm are in light gray. The 210-kb TIRs of the wt strain appear as black arrows and the terminal proteins appear as circles. The location of the cosmids is indicated by short, vertical bars. The new TIR size in the mutants is delimited by dotted lines.
Figure 2.
(A) AseI restriction patterns of genomic DNA from S. ambofaciens DSM40697 (WT) and NSA135HPP. Running conditions were: 1% agarose gel, 200 V for 24 h with a ramped pulse time of from 20 to 150 sec for the whole patterns, and 0.9% agarose gel, 200 V for 40 h with a ramped pulse time of from 80 to 100 sec for the specific separation of the NA1 fragment (960 kb) and AseI-D (950 kb). Fragments were lettered according to the chromosome map (4). NA1 indicates the localization of the new AseI fragment; + and − represent pronase-treated and pronase-untreated samples of DNA, respectively. AD91, 12A5, and 32E1 indicate Southern blots of the pronase-treated lanes probed with the respective labeled cosmid DNA. (B) DraI restriction patterns of genomic DNA from S. ambofaciens DSM40697 (WT) and NSA135HPP. Running conditions were: 1% agarose gel, 200 V for 24 h with a ramped pulse time of from 20 to 150 sec. The high-molecular-weight fragments were slightly degraded in the wt genomic DNA. ND1 indicates the localization of the new DraI fragment in NSA135HPP; the faint additional bands in the pattern are a result of partial digests.
This fragment resulted from the fusion between the undeleted part of AseI-E and the duplicated part of AseI-D. This was shown by the hybridization of both cosmids 32E1 (from fragment AseI-E in the wt strain, Fig. 1) and 12A5 (between fragments DraI-VI and VII, Fig. 1) DNA onto NA1 (Fig. 2A).
As further evidence, the DraI restriction pattern of this strain was in agreement with the results presented above with AseI. Thus, DraI-IV fragment (right end in the wt chromosome) was the only missing fragment, and the intensity of the DNA band corresponding to the DraI-VII fragment (left chromosomal end) was compatible with a doublet (Fig. 2B). However, it was not possible to distinguish between the DraI-I fragment of 2,100 kb in the wt strain and the new DraI fragment (called ND1 for new DraI fragment 1) generated by this rearrangement, the size of which was estimated to be approximately 2,060 kb (Figs. 1 and 2B).
By hybridizing ordered overlapping cosmids corresponding to the AseI-E fragment onto BamHI patterns of total DNA of strain NSA135HPP, the deletion terminus was localized in cosmid 25E1 (Fig. 1). The extent of the deletion was estimated to be approximately 850 kb by adding together the sizes of the BamHI fragments of ordered overlapping cosmids from the unstable region of S. ambofaciens DSM40697.
The junction between the deletion terminus and the duplicated DNA was isolated in cosmid 8D (Fig. 1) from a gene library of the strain NSA135HPP. The region of the left chromosomal arm, corresponding to the boundary of the DNA segment duplicated on the right arm, similarly was isolated in cosmid 9A (Fig. 1). The BamHI map of these loci was constructed by ordering overlapping cosmids from the gene library and was compared with the map of the corresponding regions in the wt strain. The DNA rearrangement involved a single BamHI fragment on each chromosomal arm (14.2 kb from cosmid 25E1 and 17.5 kb from cosmid 14C4) to give a fusion BamHI fragment of 19.5 kb in cosmid 8D. No other change in the BamHI map was detected on either side of the junction nor on the undeleted arm (region of cosmid 9A).
Chromosomal Arm Replacement in Strain NSA855.
This strain was subcloned from the phenotypically heterogeneous progeny of the pigment-defective mutant NSA854, which arose from genetic instability of the wt strain (28).
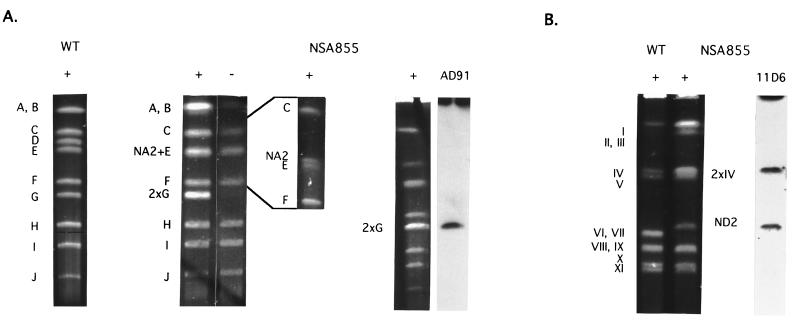
Conversely to the chromosomal structure of NSA135HPP, in strain NSA855, the left chromosomal arm is deleted for terminal 480 kb while 850 kb of the right end (corresponding to the deleted area in NSA135HPP) was duplicated and fused to the deletion terminus on the left arm (Fig. 1). Thus, the AseI restriction pattern showed that the AseI-D fragment (left end) was the only missing band while a new AseI fragment of approximately 860 kb, NA2, was detected (Figs. 1 and 3A). In addition, the intensity of the band corresponding to the AseI-G fragment (right end) was compatible with a doublet. This was also the only band of the pattern that remained trapped in the well in the proteinase-untreated assays and that hybridized with the wt terminal sequences (AD91 cosmid DNA, Fig. 3A). So, the AseI-G fragment was duplicated and lay at both chromosomal ends (Fig. 1).
Figure 3.
(A) AseI restriction patterns of genomic DNA from S. ambofaciens DSM40697 (WT) and NSA855. Running conditions were: 1% agarose gel, 200 V for 24 h with a ramped pulse time of from 20 to 130 sec for the first three lanes; 0.9% agarose gel, 200 V for 40 h with a ramped pulse time of from 80 to 100 sec for the specific separation of the NA2 fragment (860 kb) and AseI-E (850 kb); and, finally, 1% agarose gel, 200 V for 24 h with a ramped pulse time of from 40 to 160 sec for the last lane. Symbols +, −, and AD91 are as described in Fig. 2. The large fragments in the pronase-untreated lane are slightly degraded. (B) DraI restriction patterns of genomic DNA of S. ambofaciens DSM40697 (WT) and NSA855. Running conditions were: 1% agarose gel, 200 V for 23 h with a ramped pulse time of from 40 to 90 sec. 11D6 indicates Southern blots of the NSA855 pattern probed with labeled 11D6 cosmid DNA. The absence of a hybridization signal on the DraI-I fragment (2,100 kb) is explained by the fact that most of the very large fragments were not transferred to the membrane.
This structure was confirmed by the analysis of the DraI pattern. Fragments VI (420 kb) and VII (400 kb) were missing, and a new fragment of 450 kb was detected (ND2 in Fig. 3B). The terminal DraI-IV fragment showed double intensity compared with the wt pattern, as described above for the terminal AseI-G fragment. The hybridization with the DraI linking clone 11D6 (between fragments IV and I in the wt strain, Fig. 1) revealed the ND2 fragment (Fig. 3B), demonstrating that sequences from the right chromosomal arm were duplicated on the left chromosomal arm.
Using as probes ordered cosmids overlapping the AseI-D fragment, the deletion terminus was localized in cosmid 14C4 (Fig. 1). The junction of the rearrangement was isolated by using a gene library of strain NSA855 in a BamHI fragment of 11.9 kb cloned in cosmid F1 (Fig. 1). This fragment was homologous to both BamHI fragments of 17.5 kb of cosmid 14C4 and 14.2 kb of cosmid 25E1. Therefore, the chromosomal rearrangement characterized in strain NSA855 involved the same BamHI fragments as in NSA135HPP. No other difference was detected on either chromosomal arm when compared with the corresponding wt BamHI map.
A Duplicated Gene as a Substrate for Homologous Recombination.
To permit a more precise characterization of the rearrangements and to elucidate the mechanism of the chromosomal arm replacement, the nucleotide sequence of the duplication–deletion junctions was determined in both mutant strains and compared with the corresponding wt regions.
A total of 1,180 and 1,115 nt were sequenced from the wt regions surrounding the deletion termini on the right and left chromosomal arm, respectively. An ORF was identified on each arm by using the program frameplot 2.1 (29). The two DNA-coding sequences contained a G+C percentage of 69 and were 99% similar to each other. The deduced protein sequences also showed 99% identity (Fig. 4). Database searches using the blast 2.0 service (27) showed that they presented 64% of identity with the product of the gene crtS, a σ factor necessary for carotenoid biosynthesis in S. setonii (30). High similarities (47%) also were found with the products of the gene sigF, which controls the late steps of sporulation in S. coelicolor or S. aureofaciens (31). The similarities are particularly striking within the regions 2.4 (100% with crtS and 73% with sigF) and within the region 4.2 (62.5% with crtS and sigF), which are involved in the recognition of the −10 and −35 regions of promoters of the genes, respectively (32) (Fig. 4). So, these ORFs were designated homologous to alternative sigma factor (has) and R and L for right and left chromosomal arm, respectively.
Figure 4.
DNA nucleotide sequence and the deduced amino acid sequence of hasR. Nucleotide and amino acid differences in hasL are shown in bold. The conserved regions 2.4 (positions 342–408) and 4.2 (positions 727–822) are underlined. The translational stop codon is indicated by an asterisk.
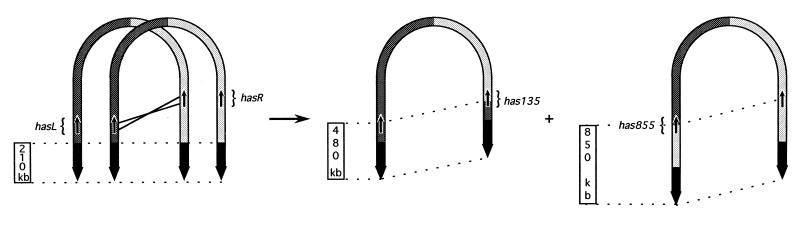
The ORFs, hasR and hasL, differ only by 1 nt in the 5′ and 5 nt in the 3′ coding regions (Fig. 4). These nucleotide differences correspond to nonconservative substitutions as 4 aa differed between the two putative products (Fig. 4). There are 5 potential translation initiation codons (ATG or GTG) at positions 1, 10, 46, 55, and 73. The localization of a potential ribosome-binding site (RBS), GGAGG, 10 bp upstream of the first putative ATG codon supports its functionality, as the distance from the initiation codon ranges from 5 to 12 bp in Streptomyces (33). However, the most downstream GTG codon (position 73) could also be used as the translation initiation codon as there is a sequence 10 bp upstream (GGA) that could also act as an RBS. The ORF terminated at a TGA codon at position 846, encoding a putative 281- or 257-residue polypeptide depending on the position of the start codon used. The transcriptional orientation of the duplicated genes hasR and hasL is divergent from the ends of the chromosome (Fig. 5).
Figure 5.
Sister chromosome exchange in S. ambofaciens DSM40697. The left and right chromosomal arms are shown in dark and light gray, respectively; the TIRs of the wt strain are shown as thick, black arrows. The transcriptional orientation is defined by the thin arrow. Both reciprocal products of the recombination correspond to the chromosomal structure of strains NSA135HPP (left product) and NSA855 (right product). The new TIRs are delimited by dotted lines.
No sequence similarity was detected between the 3′ regions of hasR and hasL (139 nt downstream from the stop codon TGA were compared). In the 5′ regions, 130 nt upstream from the ATG start codon were aligned. The similarities detected are limited to the area of the RBS.
In strain NSA135HPP, the chimeric region was sequenced (1,073 nt) and compared with the wt sequences. The hybrid ORF, called has135, is identical to hasL in the 5′ coding region (Gly-7, Fig. 4) and to hasR in the coding terminus region (Thr-279, Asp-280, and His-281, Fig. 4). The 127 nt from the 3′ region of has135 (i.e., the region proximal to the middle of the chromosome) also were identical to the corresponding region of hasR. However, the 100 nt in the 5′ region of has135 (i.e., on the side of the chromosomal ends) were completely identical to the 5′ region of hasL. This demonstrates that the chromosomal arm replacement in strain NSA135HPP resulted from homologous recombination between the identical parts of hasR and hasL.
The reciprocal situation was found in strain NSA855. A total of 1,312 nt were sequenced on the left chromosomal arm, at the deletion–duplication junction. The hybrid ORF characterized at this point (has855) was identical to hasR in the 5′ part of the gene and to hasL in the 3′ part. The 383 nt of the 3′ region downstream to has855 were identical to the 3′ part of hasL (Fig. 5). However, the 5′ region of has855 presented the same sequence as the corresponding region of hasR (87 nt compared).
DISCUSSION
The linear chromosomal DNA of the wt strain S. ambofaciens DSM40697 ends in TIRs of 210 kb (4). In the mutant strain NSA135HPP, a deletion of the terminal 850 kb was found on one chromosomal arm while 480 kb of sequence identical to the end of the other arm were duplicated and fused at the deletion terminus. The new TIR generated by this deletion–duplication event corresponded to the duplicated 480 kb of DNA since the undeleted arm has retained its wild-type structure (Fig. 1).
Conversely, in strain NSA855, the terminal 850 kb, corresponding to the deleted area in NSA135HPP, were duplicated and fused to the deletion terminus on the other arm (which was deleted of its terminal 480 kb corresponding to the translocated DNA in NSA135HPP). So, in strain NSA855, TIRs of 850 kb were generated by this event. The deletion contained sequences from the left chromosomal arm while the right one was not affected. This situation was unexpected because, usually, fragments from the left arm were only deleted when fragments AseI-G and E, from the right arm, were already deleted (28, 34).
The simplest hypothesis to account for these rearrangements is a sister chromosome exchange resulting from a single, nonallelic crossover between the two copies of the has gene (Fig. 5). The presence of numerous copies of the chromosome within the same article of the Streptomyces mycelium could favor interchromosomal recombination. Moreover, an additional crossing-over within the wt 210 kb TIR between these sister chromosomes would pass unnoticed and would generate the same products.
However, as both strains were isolated independently, each of the two chromosomal structures results from independent crossovers. Each of these crossovers could correspond either to a half-exchange generating only one of the two reciprocal structures or to a full exchange, which would have formed both structures at the same time (35). In this case, for each crossover only one of the two chromosomal structures would have been isolated.
Alternatively, a mechanism involving repair of a double-strand break cannot be ruled out. A recombinogenic free end within one copy of the has gene could invade the homologue and initiate a replication fork. The homologue used as substrate could be located either on the same or on another copy of the chromosome. If this replication proceeds until the end of the chromosome, it will lead to the duplication of the unbroken arm, in a way similar to the “copy break” duplication described in yeast (36). However, this mechanism implies unscheduled replication over hundreds of kilobases (e.g., 850 kb in strain NSA855) and terminal protein recruitment at the 5′ end.
In any case, both mechanisms involve homologous recombination. The 817-bp region of perfect identity between hasL and hasR is sufficient to promote homologous recombination since it requires at least 200 bp of identity in Streptomyces (37). Until now, chromosomal instability in Streptomyces was thought to result from illegitimate recombination (38). Here we show the implication of homologous recombination in the formation of large chromosomal deletion.
The sequence similarity between hasR and hasL is quite striking. Gene families are not a prominent feature of bacterial genetics but other examples of nearly identical copies of a gene, on distinct chromosomal loci, have been documented. This includes tufA and tufB (39, 40) or gadA and gadB in E. coli (41). Comparison of hasR and hasL sequences showed that the 6 nt that were different were clustered in the 5′ and 3′ coding regions of the genes. This also was true for the differences between the pairs of genes tufA, tufB and gadA, gadB. In Salmonella typhimurium, sequence information are transferred between tufA and tufB by gene conversion (42). The authors proposed that a mechanism of sister chromosome reciprocal exchange and segregation of the products in different cells could account for the high nucleotide similarities between the two genes. The high level of identity between the has gene could also be a result of sequence transfer. The nonallelic recombination described here shows that recombination between the two copies is possible.
Chromosomal arm replacement also altered the position of the replication origin compared with its balanced central location in the wt chromosome (Fig. 5) (25, 43). This situation is reminiscent of the chromosomal organization of S. rimosus where the dnaA region is asymmetrically located on the chromosome (from 34 to 44% of the chromosome from one end) and the TIR of 550 kb were the longest previously reported (5).
In addition, exchanges of DNA ends between the linear plasmid and chromosome via a single illegitimate recombination event were characterized (8, 44). These recombination events led to the formation of a hybrid chromosome with two different ends. However, all Streptomyces linear replicons are characterized by identical sequences in inverted orientation at their termini (6). The TIRs are highly polymorphic among linear replicons of Streptomyces, indicative of a dynamic turnover to such a degree that the selected character would be the presence of the repeated structure itself rather than the sequences. Thus, the universal presence of TIR supports the existence of a biological function for such a structure in nature. One explanation could be that the TIRs promote a strong interaction between the two ends as hypothesized by the racket-frame model (6). In addition, while the chromosomal DNA was shown to be physically linear, the circularity of the genetic map in several Streptomyces species supports this model (45, 46). In natural isolates, hybrid chromosomes might be selected against because of the lacking interaction between DNA ends.
The chromosomal arm replacement described here is a mechanism leading to the creation of new TIRs. One can speculate that this mechanism could re-form TIRs from a hybrid chromosome. Such a mechanism would provide an explanation of the structure of the TIR in S. lividans 66, where only the terminal 16 kb of the 25-kb TIRs are identical to one end of SLP2 (1). An ancestral exchange of ends between SLP2 and the chromosome would have generated a hybrid chromosome with two different ends. Subsequently, the terminal sequences on both chromosomal arms would have become the same via chromosomal arm replacement. This would have involved a DNA segment of 25 kb, larger than the 16 kb originating from the plasmid.
This mechanism would imply the duplication of sequences originating from one chromosomal arm onto the other. The presence of the catA gene in each copy of the TIR of S. coelicolor M145 (47) strongly suggests a duplication of a DNA segment that could have arisen by the mechanism described above. Another example is seen in strain NSA855, where the polyketide synthase gene localized on the AseI-E fragment in the wt strain (48) is duplicated within the new TIR of 850 kb.
Finally, large chromosomal rearrangements have evolutionary implications on the organization of the bacterial chromosome (35). The plasticity of the DNA ends may be a general feature of the linear replicons. The exchange of telomeric regions also was described in the bacteria Borrelia and Rhodococcus (49, 50), and in eukaryotes, numerous examples suggest that the sequestering of the telomere regions would allow this genomic domain to be plastic relative to the rest of the chromosomes (51). In Streptomyces, the plasticity of the ends of the linear replicons, associated to the horizontal transfer of genetic information (conjugative plasmids and chromosome transfer), could be seen as a rapid evolutionary diversification system.
Acknowledgments
This study was supported by the Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche. G.F. was the recipient of a grant from the Association pour la Recherche sur le Cancer. The sequencing advice from Dominique Schneider was greatly appreciated. We thank C. Hoyle for her help in the preparation of the manuscript.
ABBREVIATIONS
- has
homologous to alternative sigma factor
- PFGE
pulsed-field gel electrophoresis
- TIR
terminal inverted repeat
- wt
wild type
Footnotes
References
- 1.Lin Y-S, Kieser H M, Hopwood D A, Chen C W. Mol Microbiol. 1993;10:923–933. doi: 10.1111/j.1365-2958.1993.tb00964.x. [DOI] [PubMed] [Google Scholar]
- 2.Kinashi H. Actinomycetology. 1994;8:87–96. [Google Scholar]
- 3.Lezhava A, Mizukami T, Kajitani T, Kameoka D, Redenbach M, Shinkawa H, Nimi O, Kinashi H. J Bacteriol. 1995;177:6492–6498. doi: 10.1128/jb.177.22.6492-6498.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leblond P, Fischer G, Francou F X, Berger F, Guérineau M, Decaris B. Mol Microbiol. 1996;19:261–271. doi: 10.1046/j.1365-2958.1996.366894.x. [DOI] [PubMed] [Google Scholar]
- 5.Pandza K, Pfalzer G, Cullum J, Hranueli D. Microbiology. 1997;143:1493–1501. doi: 10.1099/00221287-143-5-1493. [DOI] [PubMed] [Google Scholar]
- 6.Sakaguchi K. Microbiol Rev. 1990;54:66–74. doi: 10.1128/mr.54.1.66-74.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C W, Yu T-W, Lin Y-S, Kieser H M, Hopwood D A. Mol Microbiol. 1993;7:925–932. doi: 10.1111/j.1365-2958.1993.tb01183.x. [DOI] [PubMed] [Google Scholar]
- 8.Pandza S, Biuković G, Paravić A, Dadbin A, Cullum J, Hranueli D. Mol Microbiol. 1998;28:1165–1176. doi: 10.1046/j.1365-2958.1998.00877.x. [DOI] [PubMed] [Google Scholar]
- 9.Wu X, Roy K L. J Bacteriol. 1993;175:37–52. doi: 10.1128/jb.175.1.37-52.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinashi H, Shimaji-Murayama M, Hanafusa T. Plasmid. 1991;26:123–130. doi: 10.1016/0147-619x(91)90052-x. [DOI] [PubMed] [Google Scholar]
- 11.Huang C H, Lin Y-S, Yang Y-A, Huang S-W, Chen C W. Mol Microbiol. 1998;28:905–916. doi: 10.1046/j.1365-2958.1998.00856.x. [DOI] [PubMed] [Google Scholar]
- 12.Chang P-C, Cohen S N. Science. 1994;265:952–954. doi: 10.1126/science.8052852. [DOI] [PubMed] [Google Scholar]
- 13.Chen C W. Trends Genet. 1996;12:192–196. doi: 10.1016/0168-9525(96)30014-0. [DOI] [PubMed] [Google Scholar]
- 14.Qin Z, Cohen S N. Mol Microbiol. 1998;28:893–903. doi: 10.1046/j.1365-2958.1998.00838.x. [DOI] [PubMed] [Google Scholar]
- 15.Stackebrandt E, Liesack W, Witt D. Gene. 1992;115:255–260. doi: 10.1016/0378-1119(92)90567-9. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi T, Sawada H, Tanaka F, Matsuda I. Int J Syst Bacteriol. 1996;46:476–479. doi: 10.1099/00207713-46-2-476. [DOI] [PubMed] [Google Scholar]
- 17.Leblond P, Redenbach M, Cullum J. J Bacteriol. 1993;175:3422–3429. doi: 10.1128/jb.175.11.3422-3429.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C W, Lin Y-S, Yang Y-L, Lin W-Y, Chang H-W, Kieser H M, Hopwood D A. Actinomycetol. 1994;8:103–112. [Google Scholar]
- 19.Fischer G, Kyriacou A, Decaris B, Leblond P. Biochimie. 1997;79:555–558. doi: 10.1016/s0300-9084(97)82003-2. [DOI] [PubMed] [Google Scholar]
- 20.Leblond P, Decaris B. FEMS Microbiol Lett. 1994;123:225–232. doi: 10.1111/j.1574-6968.1994.tb07229.x. [DOI] [PubMed] [Google Scholar]
- 21.Volff J-N, Altenbuchner J. Mol Microbiol. 1998;27:239–246. doi: 10.1046/j.1365-2958.1998.00652.x. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic Manipulation of Streptomyces: A Laboratory Manual. Norwich, CT: The John Innes Foundation; 1985. [Google Scholar]
- 24.Evans G A, Lewis K, Rothenberg B E. Gene. 1989;79:9–20. doi: 10.1016/0378-1119(89)90088-7. [DOI] [PubMed] [Google Scholar]
- 25.Berger F, Fischer G, Kyriacou A, Decaris B, Leblond P. FEMS Microbiol Lett. 1996;143:167–173. doi: 10.1111/j.1574-6968.1996.tb08476.x. [DOI] [PubMed] [Google Scholar]
- 26.Yanisch-Perron C, Viera J, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 27.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leblond P, Demuyter P, Simonet J-M, Decaris B. J Bacteriol. 1991;173:4229–4233. doi: 10.1128/jb.173.13.4229-4233.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bibb M J, Findlay P R, Johnson M W. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 30.Kato F, Hino T, Tanaka M, Koyama Y. Mol Gen Genet. 1995;247:387–390. doi: 10.1007/BF00293207. [DOI] [PubMed] [Google Scholar]
- 31.Potúc̆ková L, Kelemen G H, Findlay K C, Lonetto M A, Buttner M J, Kormanec J. Mol Microbiol. 1995;17:37–48. doi: 10.1111/j.1365-2958.1995.mmi_17010037.x. [DOI] [PubMed] [Google Scholar]
- 32.Lonetto M, Gribskov M, Gross C A. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strohl W R. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer G, Decaris B, Leblond P. J Bacteriol. 1997;179:4553–4558. doi: 10.1128/jb.179.14.4553-4558.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth J R, Benson N, Galitski T, Haack K, Lawrence J G, Miesel L. In: Escherichia coli and Salmonella typhimurium. Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2256–2276. [Google Scholar]
- 36.Morrow D M, Connelly C, Hieter P. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wohlleben W, Hartmann V, Hillemann D, Krey K, Muth G, Nussbaumer B, Pelzer S. Acta Microbiol Immunol Hung. 1994;41:381–389. [PubMed] [Google Scholar]
- 38.Birch A, Häusler A, Rüttener C, Hütter R. J Bacteriol. 1991;173:3531–3538. doi: 10.1128/jb.173.11.3531-3538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.An G, Friesen J D. Gene. 1980;12:33–39. doi: 10.1016/0378-1119(80)90013-x. [DOI] [PubMed] [Google Scholar]
- 40.Yokata T, Sugisaki H, Takanami M, Kaziro Y. Gene. 1980;12:25–31. doi: 10.1016/0378-1119(80)90012-8. [DOI] [PubMed] [Google Scholar]
- 41.Smith D K, Kassam T, Singh B, Elliott J F. J Bacteriol. 1992;174:5820–5826. doi: 10.1128/jb.174.18.5820-5826.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdulkarim F, Hughes D. J Mol Biol. 1996;260:506–522. doi: 10.1006/jmbi.1996.0418. [DOI] [PubMed] [Google Scholar]
- 43.Fischer G, Holl A C, Volff J-N, Vandewiele D, Decaris B, Leblond P. Res Microbiol. 1998;149:203–210. doi: 10.1016/s0923-2508(98)80080-6. [DOI] [PubMed] [Google Scholar]
- 44.Gravius B, Glocker D, Pigac J, Pandza K, Hranueli D, Cullum J. Microbiology. 1994;140:2271–2277. doi: 10.1099/13500872-140-9-2271. [DOI] [PubMed] [Google Scholar]
- 45.Pigac J, Alacevic M. Period Biol. 1979;81:575–582. [Google Scholar]
- 46.Hopwood D A. Genetics. 1966;54:1177–1184. doi: 10.1093/genetics/54.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Hopwood D A. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 48.Aigle B, Schneider D, Morilhat C, Vandewiele D, Dary A, Holl A C, Simonet J-M, Decaris B. Microbiology. 1996;142:2815–2824. doi: 10.1099/13500872-142-10-2815. [DOI] [PubMed] [Google Scholar]
- 49.Kalkus J, Dörrie C, Fischer D, Reh M, Schlegel H G. J Gen Microbiol. 1993;139:2055–2065. doi: 10.1099/00221287-139-9-2055. [DOI] [PubMed] [Google Scholar]
- 50.Casjens S, Murphy M, DeLange M, Sampson R, van Vugt R, Huang W H. Mol Microbiol. 1997;26:581–596. doi: 10.1046/j.1365-2958.1997.6051963.x. [DOI] [PubMed] [Google Scholar]
- 51.Pryde F E, Gorham H C, Louis E J. Curr Opin Genet Dev. 1997;7:822–828. doi: 10.1016/s0959-437x(97)80046-9. [DOI] [PubMed] [Google Scholar]