Abstract
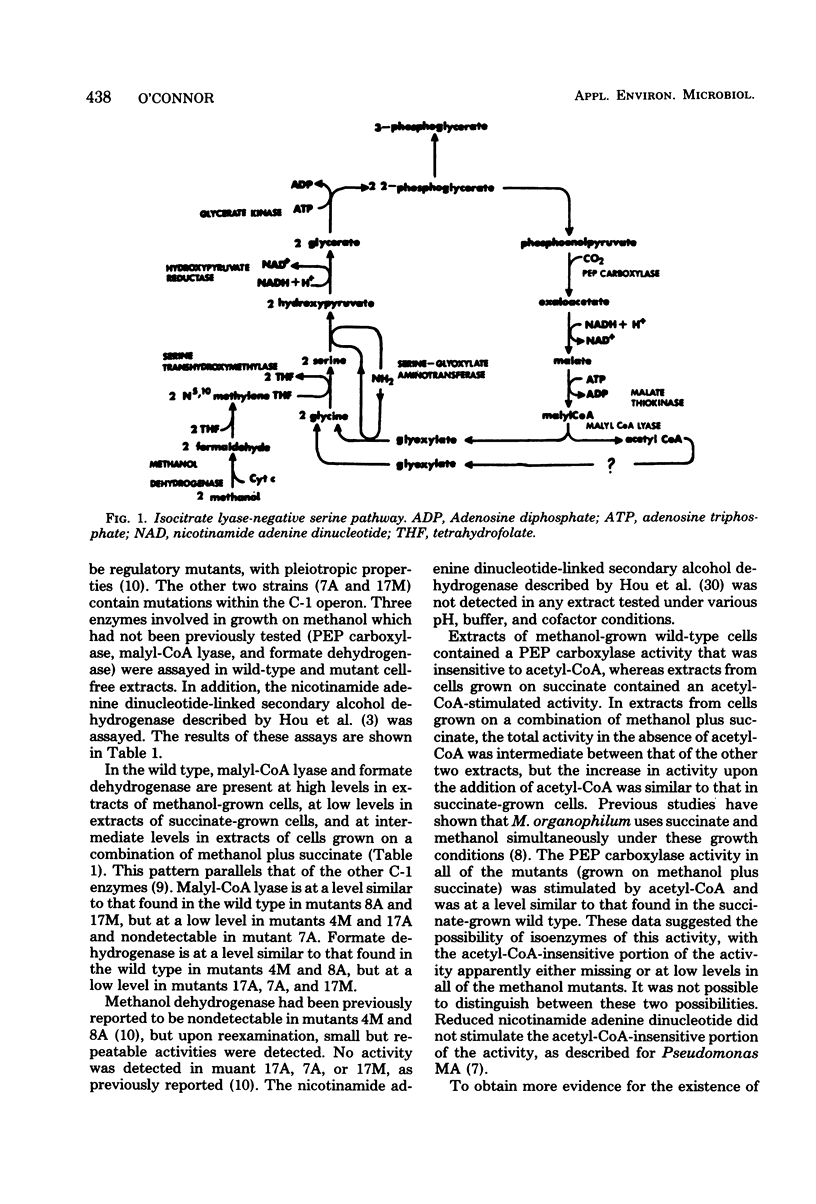
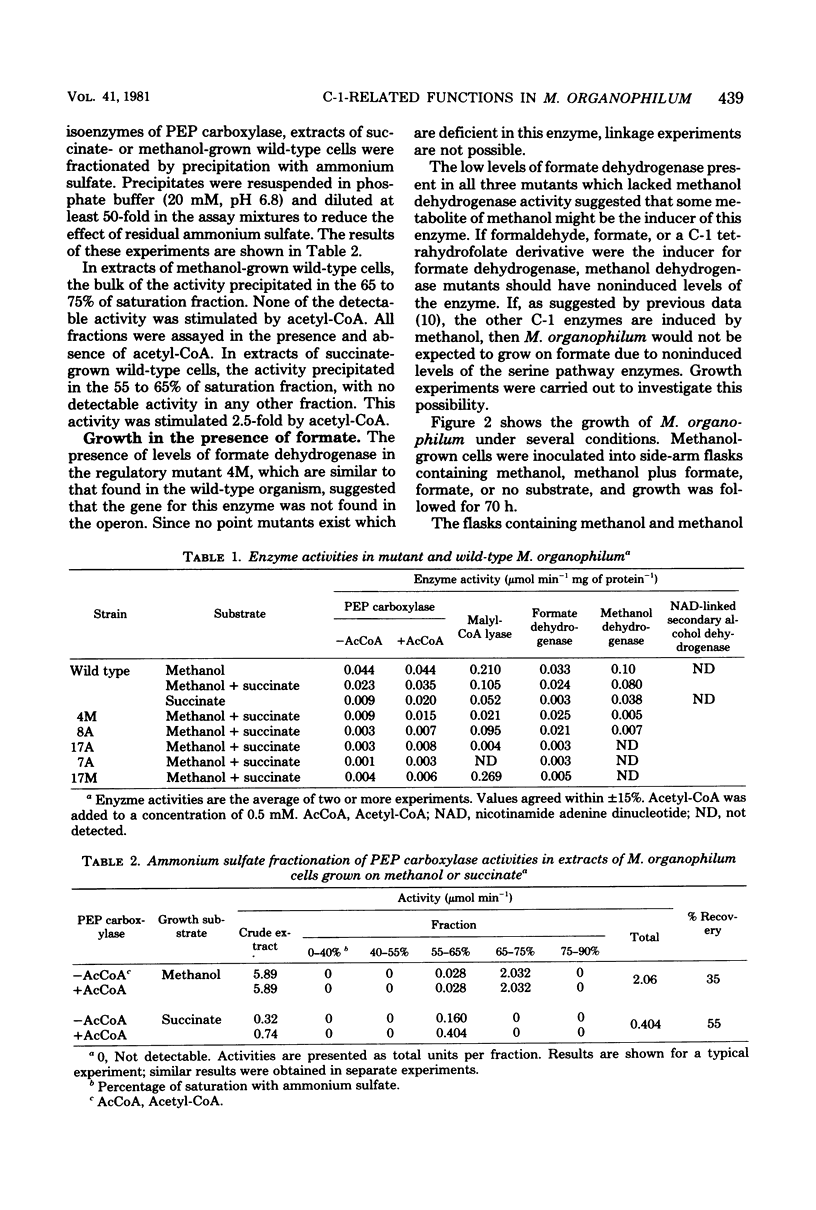
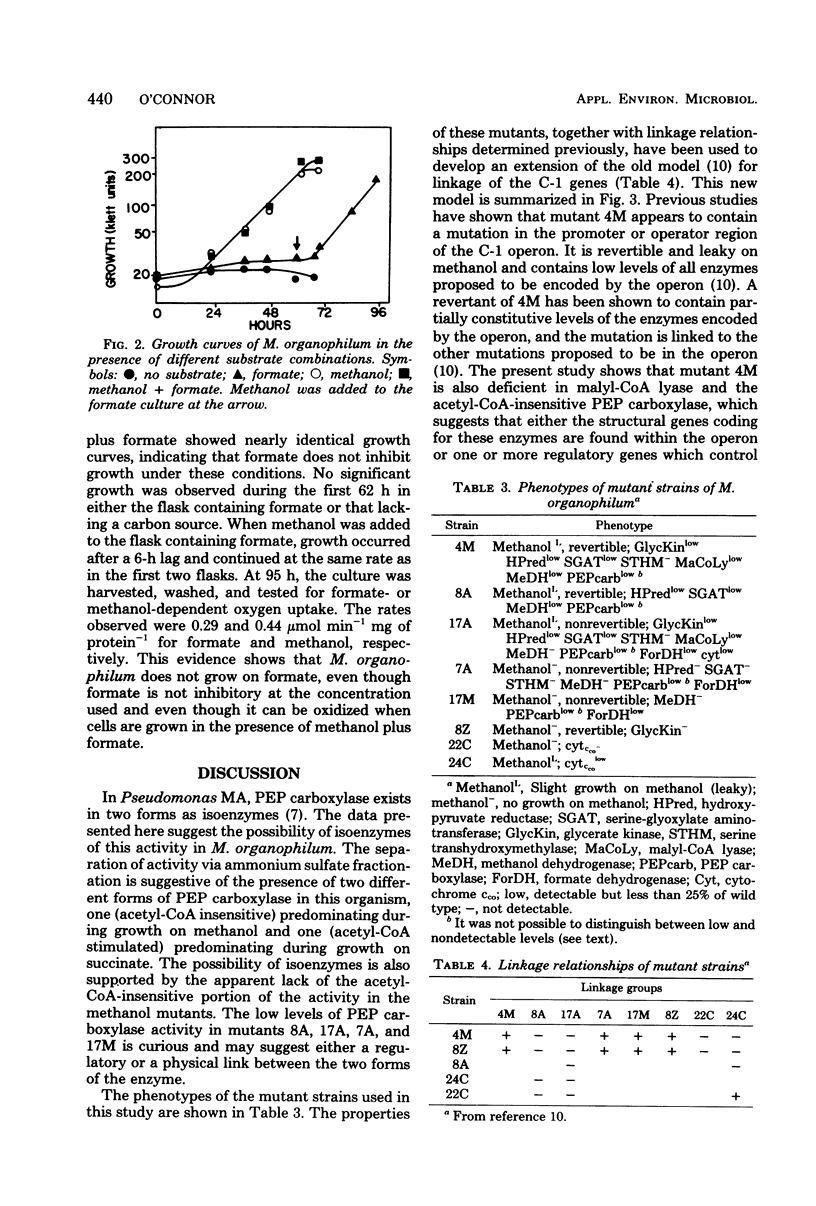
Evidence is presented which suggests that Methylobacterium organophilum contains isoenzymes of phosphoenolpyruvate carboxylase activity. Methanol-grown cells contained an acetyl coenzyme A (CoA)-insensitive activity which precipitated in a 65 to 75% of saturation ammonium sulfate fraction. Succinate-grown cells contained an acetyl-CoA-stimulated activity which precipitated in a 55 to 65% of saturation ammonium sulfate fraction. Mutants unable to grow on methanol appeared to lack acetyl-CoA-insensitive activity. This acetyl-CoA-insensitive phosphoenolpyruvate carboxylase, along with malyl-CoA lyase, is proposed to be encoded by the C-1 operon. The gene for formate dehydrogenase appeared to reside outside the operon and was not inducible by methanol. M. organophilum was unable to grow on formate, and evidence is presented suggesting that formate is unable to induce the enzymes which comprise the serine pathway for formaldehyde fixation. An expanded model for the C-1 operon is presented.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Holloway B. W. Plasmids that mobilize bacterial chromosome. Plasmid. 1979 Jan;2(1):1–19. doi: 10.1016/0147-619x(79)90002-7. [DOI] [PubMed] [Google Scholar]
- Hou C. T., Patel R., Laskin A. I., Barnabe N., Marczak I. Microbial oxidation of gaseous hydrocarbons: production of methyl ketones from their corresponding secondary alcohols by methane- and methanol-grown microbes. Appl Environ Microbiol. 1979 Jul;38(1):135–142. doi: 10.1128/aem.38.1.135-142.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. A., Quayle J. R. Microbial growth on C-1 compounds. 6. Oxidation of methanol, formaldehyde and formate by methanol-grown Pseudomonas AM-1. Biochem J. 1964 Nov;93(2):281–290. doi: 10.1042/bj0930281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Newaz S. S., Hersh L. B. Reduced nicotinamide adenine dinucleotide-activated phosphoenolpyruvate carboxylase in Pseudomonas MA: potential regulation between carbon assimilation and energy production. J Bacteriol. 1975 Nov;124(2):825–833. doi: 10.1128/jb.124.2.825-833.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M. L., Hanson R. S. Serine transhydroxymethylase isoenzymes from a facultative methylotroph. J Bacteriol. 1975 Nov;124(2):985–996. doi: 10.1128/jb.124.2.985-996.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem A. R., Hacking A. J., Quayle J. R. Cleavage of malyl-Coenzyme A into acetyl-Coenzyme A and glyoxylate by Pseudomonas AM1 and other C1-unit-utilizing bacteria. Biochem J. 1973 Sep;136(1):89–96. doi: 10.1042/bj1360089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf H. J., Hanson R. S. Alcohol dehydrogenase from Methylobacterium organophilum. Appl Environ Microbiol. 1978 Jul;36(1):105–114. doi: 10.1128/aem.36.1.105-114.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]


