Abstract
Natural killer (NK) cell cytotoxicity is regulated in large part by the expression of NK cell receptors able to bind class I major histocompatibility complex glycoproteins. The receptors associated with recognition of HLA-C allospecificities are the two-domain Ig-like molecules, p50 and p58 proteins, with highly homologous extracellular domains but differing in that they have either an activating or inhibitory function, respectively, depending on the transmembrane domain and cytoplasmic tails that they possess. We have compared the binding to HLA-Cw7 of an inhibitory p58 molecule, NKAT2, the highly homologous activating p50 molecule, clone 49, and a second activating p50 molecule, clone 39, which has homologies to both NKAT1 and NKAT2. NKAT2 binds to HLA-Cw7 with very rapid association and dissociation rates. However, the p50 receptors bind only very weakly, if at all, to HLA-C. The molecular basis of this difference is analyzed, and the functional significance of these observations is discussed.
Natural killer (NK) cells express a large repertoire of receptors able to bind to class I major histocompatibility complex (MHC) molecules. These include the CD94 and NKG2 lectin-like molecules (1–3) and the NK cell inhibitory receptors (NKIR), a family of proteins belonging to the Ig superfamily (4). The CD94 and NKG2 proteins associate to form a heterodimer able to bind the nonclassical class I MHC molecule HLA-E (5–7) whereas NKIR recognize classical class I MHC molecules (reviewed in ref. 8). The Ig-superfamily receptors can be classified according to the number of extracellular domains, the presence of a charged amino acid in the transmembrane domain, and the size of the intracellular tail, which implies the presence or absence of signal transduction motifs. A repertoire of two and three Ig-like domain receptors (p58/p50, p70) is expressed at the surface of NK cells from a single individual (9). The NK receptors associated with recognition of HLA-C contain two Ig domains (p58/p50) (10, 11) whereas those recognizing HLA-A and -B have three Ig domains (p70) (10, 12–14). The two-domain p58 NKIR molecules can be divided further on the basis of sequence into subgroups that correlate with recognition of one of the two group allospecificities of HLA-C (15, 16); 13 dimorphic residues in the extracellular region define the difference between the specificity 1 (e.g., NKAT1/clone 42) receptors that recognize HLA-Cw6 (and related alleles) and the specificity 2 (NKAT2/clone 49) receptors that recognize HLA-Cw7 (and related alleles). Moreover, these receptors exhibit a further level of diversity in that they exist in two forms, p58 and p50, highly homologous in their extracellular domains but differing in the transmembrane and cytoplasmic regions (17).
These different types of two-domain Ig-like receptors are functionally distinct. Receptors with a long cytoplasmic tail (p58 molecules, NKIR) contain immunoreceptor tyrosine-based inhibition motifs and are inhibitory receptors able to turn off NK- and T-cell activation after ligation by class I MHC proteins (refs. 18 and 19 and reviewed in ref. 8). In contrast, the p50 molecules (NKAR) have a truncated cytoplasmic tail that lacks the immunoreceptor tyrosine-based inhibition motifs and have a charged amino acid in the transmembrane region through which they associate noncovalently with a signaling molecule, DAP12, that contains an immunoreceptor tyrosine-based activating motif (20, 21). This complex is able to deliver an activating signal to NK and T cells (21–23).
NK cells as well as a small subset of T cell clones may express multiple, functionally independent, HLA-specific NKIR (16, 23–25). Thus, it is difficult, by using cellular assays, to define the class I HLA binding specificity of any given NKIR molecule. Moreover, the high degree of homology in the extracellular portion of these receptors means that the available NKIR-reactive mAbs bind both long and short-tailed receptors (17). Direct binding of p58 receptors to HLA-C has been demonstrated (10, 26–28), but to date there is no direct evidence of the binding of a p50 molecule to any HLA-C protein. The evidence for their binding to HLA-C is, therefore, indirect, being mainly mAb blockading the activating receptor in cellular assays (22, 23, 29–31). It has been reported that the action of an inhibitory receptor is always dominant over that of an activating receptor (22, 23). The question, therefore, arises as to how the binding to HLA-C of p50 and p58 receptors compare. In this paper, we have analyzed directly the binding of p50 and p58 NK receptors to HLA-C. While this paper was in preparation, another study, using a different methodology, described the differential binding to HLA-C of the NKIR NKAT2 and the NKAR clone 49 and the role of residue 45 in this phenomenon (32).
MATERIALS AND METHODS
Antibodies and Proteins.
mAb GL183 (33) was purchased from Immunotech (Marseille, FR). Soluble NKAT1, NKAT2, and HLA-C molecules were expressed in Escherichia coli and were refolded as described (28). The clone 49 and clone 39 cDNAs were obtained by PCR from NK lines prepared from donors J.L.S. and H.T.R., respectively. Constructs for expression of soluble clone 49 and clone 39 were prepared in pET22b, and the soluble proteins were expressed, refolded and purified as described (28). Peptides were purchased from the Biopolymers Laboratory at Harvard University and were checked for purity by HPLC.
Mutagenesis.
Mutagenesis was done by using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. Residue 35, glutamate (E) in clone 49, was mutated to glutamine (Q) by using the oligonucleotide 5′-GGTCAGATGTCAGGTTTCAGCACTTCCTTCTGCAC-3′ and its complement. Residue 45, tyrosine (Y) in clone 49, was mutated to phenylalanine (F) by using the oligonucleotide 5′-CACAGAGAGGGGAAGTTTAAGGACACTTTGCACC-3′ and its complement. That only the desired mutations were present in each mutant was checked by dideoxynucleotide chain termination sequencing of the entire extracellular domain.
Surface Plasmon Resonance Analysis.
Surface plasmon resonance analysis was carried out on a BIAcore 2000 (BIAcore, Piscataway, NJ) as described (28).
Molecular Modeling.
Molecular modeling was done by using the program rasmol 2.6 (49). The surface electrostatic potential maps were generated by using the program grasp (Columbia University, New York) on a Silicon Graphics (Mountain View, CA) workstation. Atomic coordinates of the extracellular region of NKAT1 (clone 42) were obtained from Qing Fan (Harvard University, Cambridge, MA) (34).
RESULTS
Recombinant proteins were expressed in E. coli as inclusion bodies and were refolded as described (28). HLA-Cw7 heavy chain was refolded by using the peptides RYRPGTVAL and KYFDEHYEY (35) and an amino acid substitution of the latter at position 8 (KYFDEHYLY). Several types of NK receptors were prepared: the inhibitory p58 molecules NKAT1 (clone 42) and NKAT2 (clone 6) and the activating p50 molecules NKAT5 (clone 49) and NKAT8 (clone 39).
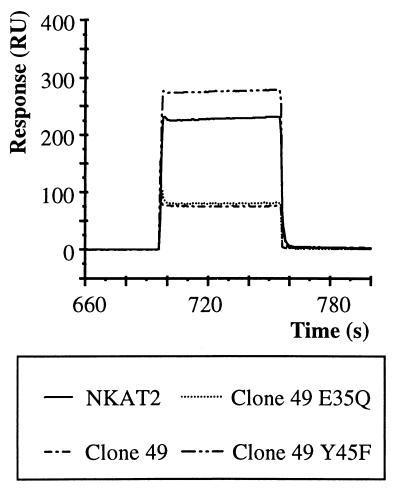
The p50 molecule clone 49 (NKAT5) is homologous to NKAT2 but has a short cytoplasmic tail (39 amino acids as compared with 84 or 76 in NKAT1 and NKAT2, respectively); the extracellular portions of these molecules differ at only three amino acids (positions 35, 45, and 216). These residues are not the dimorphic positions where the two NK specificities, NK1 and NK2, differ; that is, clone 49 possesses the same 13 dimorphic residues as NKAT2 and the other group 2 NK receptors. This high degree of homology between these proteins means that both are recognized by mAb GL183 [which recognizes group 2 NK receptors (33)]. Fig. 1 shows the BIAcore profile of GL183 binding both NKAT2 and clone 49 but not NKAT1. The binding of GL183 to both NKAT2 and clone 49 showed similar kinetics. GL183 is able to revert HLA-Cw7-mediated inhibition of NK2-specific, but not NK1-specific, NK cells (33). GL183 also reduces the cytolytic activity of T cell clones that express and have been activated through NKAR belonging to specificity 2 (23).
Figure 1.
GL183 binds both NKAT2 and clone 49, but not NKAT1. GL183 was injected at a flow rate of 30 μl/min over a control surface and NKAT1 [484 resonance units (RU)], NKAT2 (879 RU), and clone 49 (582 RU) proteins, immobilized on a CM5 chip via amine coupling. This experiment is representative of several repeated by using different chips prepared with multiple batches of refolded proteins.
Because clone 49 is almost identical to NKAT2 in the extracellular portion, it has been postulated to interact with HLA-Cw7 and related alleles (9, 36) but cause activating events rather than inhibition. Fig. 2A shows two injections of a series of increasing concentrations of HLA-Cw7 over NKAT1, NKAT2, and clone 49. Although HLA-Cw7 clearly bound to the NKAT2 protein, it bound only very weakly, if at all, to clone 49 or to NKAT1. Scatchard plots (Fig. 2C) revealed that the binding of clone 49 to HLA-Cw7 appeared not to be saturable, suggesting either that it interacts with HLA-Cw7 with a very low affinity or that the binding is nonspecific.
Figure 2.
Clone 49 and clone 39 do not bind HLA-Cw7. (A) Increasing concentrations of HLA-Cw7 (11.4 and 22.7 μM) were injected over control, NKAT1 (148 RU), NKAT2 (437 RU), and clone 49 (631 RU). (B) Increasing concentrations of HLA-Cw7 (5.8 and 11.4 μM) were injected over control, NKAT2 (594 RU), clone 49 (737 RU), and clone 39 (884 RU). (C) Scatchard plots of the data shown in A. These data are representative of multiple experiments.
The fact that only three amino acids make the difference between binding and not binding was very striking. To analyze this more closely, point mutants of clone 49 were prepared at amino acids 35 and 45; the third difference between NKAT2 and clone 49 in the extracellular domains at residue 216 is in the stem region and not likely to be important. Position 45 is located in the putative binding region of NKIR for HLA-C and so was a good candidate to affect the interaction between clone 49 and HLA-Cw7. Indeed, as depicted in Fig. 3, the binding between clone 49 and HLA-Cw7 was restored when position 45 was mutated from Tyr to Phe (the residue present in NKAT2) but not when position 35 was changed from Glu to Gln. These data confirmed the importance of this region (residues 44–46) for the binding between the NK receptors and HLA-C (37).
Figure 3.
The mutation of clone 49 at position 45 restores binding to HLA-Cw7. HLA-Cw7 (5.8 μM) was injected over NKAT2 (456 RU), clone 49 (673 RU), and the two mutants of clone 49 Y45F (669 RU) and E35Q (1028 RU). These data are representative of multiple experiments.
Clone 39 is an unusual p50 molecule that, at the 13 dimorphic positions, has either those characteristic of the NK1 specificity (residues 33, 46, 102, 150, 151) or those characteristic of the NK2 specificity (residues 44, 50, 67, 68, 70, 89, 182, 190). It also has 15 unique residues different from any other two-domain Ig-like receptor and reacts with neither the NK1-specific (EB6) nor the NK2-specific (GL183) mAbs (data not shown). Clone 39 bound neither HLA-Cw6 nor HLA-Cw7 in BIAcore experiments (Fig. 2B and data not shown). Similarly, in native gel experiments, clone 39 and clone 49 bound neither HLA-Cw7 loaded with some of the peptides described below nor HLA-Cw6 loaded with two of its natural ligands (35) (data not shown).
The specificity of binding between the NK receptors and HLA-Cw7 was further analyzed by loading the HLA-C molecule with different peptides. Although position 45 appears to be a crucial residue for binding to HLA-Cw7, the failure of clone 49 to effectively bind HLA-Cw7 also could result from a negative influence of peptide. HLA-Cw7 was refolded with three different peptides: two natural ligand peptides RYRPGTVAL and KYFDEHYEY (35) and the latter peptide substituted at position 8 to remove the negatively charged glutamate residue (KYFDEHYLY), which points upwards and might interfere with binding to a NK receptor (38, 39). Of interest, HLA-Cw7 loaded with the natural ligand RYRPGTVAL bound the NKAT2 receptor, but not clone 39 or clone 49 (Fig. 4 and data not shown) whereas HLA-Cw7 reconstituted with either the other natural ligand or with its substitution at position 8 bound neither NKAT2 nor clone 49 (Fig. 4 and data not shown). These data confirm the importance of the peptide presented on the HLA-C molecule in the recognition by the NK receptors, but a more exhaustive analysis will be needed to define the molecular basis of this phenomenon.
Figure 4.
Influence of peptide on HLA-Cw7 binding to NKAT2. (A) Increasing concentrations of HLA-Cw7 loaded with peptide RYRPGTVAL were injected over control, NKAT1 (484 RU), NKAT2 (879 RU), and clone 49 (582 RU). (B) Increasing concentrations of HLA-Cw7 loaded with peptide KYFDEHYEY were injected over control, NKAT1, NKAT2, and clone 49. (C) Increasing concentrations of HLA-Cw7 loaded with peptide KYFDEHYLY were injected over control, NKAT1, NKAT2, and clone 49.These data are representative of multiple experiments.
DISCUSSION
In the work presented here, the binding of soluble p50 NKAR and p58 NKIR molecules to HLA-C has been compared. Two of these receptors, the p58 NKAT2 and the p50 clone 49, are highly homologous in their extracellular portions. The mAb GL183 bound both soluble proteins with similar affinity and with kinetic parameters typical of antibody–antigen interactions (Fig. 1). In contrast, NKAT2 bound HLA-Cw7 with extremely fast association and dissociation rate constants (Fig. 2A), as reported (28), but no binding was observed to HLA-Cw6.
However, binding of the p50 receptors clone 49 and clone 39 to HLA-C proteins could not be demonstrated convincingly. Some experiments suggested weak binding of HLA-Cw7 to clone 49, but this interaction was nonsaturable and possibly nonspecific (Fig. 2C). No interaction between clone 39 and either HLA-Cw6 or -Cw7 was detected. In contrast, indirect evidence previously obtained in cellular assays suggested that NKAR must interact with HLA-C proteins, as do NKIR (22, 23, 29, 31). Either p50 NKAR can bind HLA-C proteins with an affinity orders of magnitude weaker than that of p58 molecules or a third protein is required for their interaction.
Only a few experiments have been reported analyzing the binding of p50 molecules to HLA-C. The first observation of the function of p50 NKAR was a correlation between the expression of an EB6-reactive NKAR and enhanced cytolytic activity of the NK cell toward HLA-Cw4-expressing targets (22). Later, the p50 molecule EB6ActI, presumed to be responsible for this activation, was cloned from selected EB6-positive activating NK cells and was found to be homologous in its extracellular domains to NKAT1 (17). Strikingly, however, a soluble form of this receptor bound to HLA-Cw4 very weakly, if at all, in an assay measuring the binding of an EB6ActI–Ig fusion protein to HLA-C transfectants (30). The expression of p50 NKAR also has been reported on CD4+ T cell clones (23). In these experiments, a T cell clone, TANK-1, was isolated that expressed clone 39 and that showed enhanced proliferation (23), as well as increased interferon γ release (40), to superantigen presented on cells expressing HLA-Cw4 or -Cw7, but not HLA-Cw3 or -Cw6. HLA-Cw4 and -Cw6 belong to one allotype group of HLA-C proteins characterized by the presence of K80 whereas -Cw3 and -Cw7 belong to the other group that has N80 (41, 42). Because HLA-Cw4 and -Cw7 differ at residue 80, they should not interact with the same p50 protein (assuming a similar mode of binding to that which has been shown for p58 proteins). This puzzling result was thought to reflect the fact that clone 39 expressed at the dimorphic positions a mixture of the residues typical of NK1 and NK2 receptors (43, 44). However, as the field developed and more sequence information became available, a later analysis using additional PCR primers revealed that the TANK-1 clone may express mRNA for other p50 and p58 molecules (H.T.R., unpublished data), and, thus, the identity of the activating receptor(s) responsible for the unusual specificity of TANK-1 is unclear. A definitive demonstration of an interaction of clone 39 (or of additional receptors) with HLA-Cw4 and/or -Cw7 is, therefore, still required. Clone 39 also has been postulated to interact with HLA-Cw3, but, again, this suggestion is based on cellular assays and has not been confirmed in direct binding assays (31).
The kinetics of interaction of p58 NKIR with HLA-C protein, which can be assessed from BIAcore data obtained at 25°C, revealed extremely rapid association and dissociation rate constants (28), which are also evident in Fig. 2A as compared, for example, to Fig. 1. The dissociation rate, koff, in the range of 2 s−1 (t1/2 = 0.43 s), and the equilibrium dissociation constant, Kd, of 10 μM both were determined from the BIAcore data. The association rate constant kon was too rapid for direct determination from the BIAcore data, but calculation from koff and Kd yielded values in the range of 2 × 105 M−1⋅s−1. These association and dissociation rate constants are among the fastest observed so far in the immune system and are comparable to the kinetics of interaction of adhesion receptors with their ligands. In kinetic terms, the lack of binding between p50 and HLA-C might be explained by either a dramatic decrease of the association rate (kon) resulting from the substitution of certain amino acids (e.g., Y45 in clone 49) or by a large increase of the dissociation rate, koff. An increase of the dissociation rate would make it faster than the association rate, and, therefore, binding would be undetectable. Both could change, but, in that case, the decrease in kon would have to be larger than the increase in koff. However, the methodology presently used is not adequate to measure these changes. These possibilities, which would involve a much lower affinity of the p50/HLA-C complex, might be explored by increasing the concentration of analytes or by lowering the temperature of the analysis.
For clone 49, the molecular basis of the defective binding was localized by mutagenesis to position 45, a Tyr residue located in the center of the putative HLA-binding site (refs. 30, 34, and 37, and see, specifically, ref. 32) that is a Phe on all other two-domain Ig-like receptors both p50 and p58 (44). The lack of interaction, on the other hand, of clone 39, which has Phe-45, cannot be explained on the same basis. Of interest, however, clone 39 possesses KFN at residues 44–46, a component of the putative binding site for HLA-C (Fig. 5). This sequence is a hybrid of the sequence found in NKIR1 (MFN) and that found in NKIR2 (KFK). Similarly, the other three reported NKAR have changes in one of the two sequences reported to be important in HLA-C binding (residues 44–46 and 67–70, SRMT or GPMM) (30, 37). NKAT7 and NKAT9 have changes in both regions: TFN at residues 44–46 and GRMR or GRMT at 67–70. EB6ActI and EB6ActII have changes only at residue 70: R or K instead of T. The change in EB6ActI, T70R, has been shown to affect binding of this molecule to HLA-Cw6 (30). All of these changes involve changes in charge of the sequences, with the exception of the F45Y change in clone 49.
Figure 5.
Sequence alignment of a region (residues 1–76) of domain 1 of several p50 and p58 NK receptors. Boxes indicate residues in the putative binding site for HLA-C. Dashes indicate identity with NKAT1.
The three-dimensional structure of the extracellular portion of another p58 molecule, NKAT1 (clone 42), has been solved (34). The very high degree of homology between the extracellular domains of the NKIR and NKAR two-domain receptors enabled molecular modeling of the NKAT2 and clone 49 proteins. Steric inhibition mediated by the presence of the hydroxyl group in the tyrosine residue present in clone 49, which points upwards (Fig. 6), could be responsible for the loss of the interaction of clone 49 with HLA-Cw7. As shown in Fig. 6, both regions important for binding to HLA-C (44–46 and 67–70) (30, 37) are located on top of the molecule relatively close to each other and are in the same domain. Because subtle amino acid changes in the HLA-C binding site can result in marked differences in binding affinity, it is not possible to confidently assign an HLA-binding specificity to a NKIR molecule solely on the basis of sequence comparison with other receptors. Lastly, another striking feature of the modeling is the presence of a unique polymorphism in clone 39 (residues 153–156) located at the extreme right in the models. Moreover, the surface potential of clone 39 is predicted to differ dramatically from that of NKAT2 or clone 49 (Fig. 6), particularly in the central regions between the 44–46 and 67–70 polymorphisms, which can affect interactions with other proteins (compare Fig. 6 C and F with Fig. 6I).
Figure 6.
Comparison of the modeled structures of NKAT2 (A, B, and C), clone 49 (D, E, and F), and clone 39 (G, H, and I) with the coordinates for NKAT1 (34). Using Rasmol (49), the front view, where the cytoplasmic region of the molecule would be at the bottom of the figure (A, C, and E), and the top view, obtained after rotating the front view 50° on a horizontal (x) axis (B, D, and F), are depicted. Dimorphic positions between group 1 and group 2 receptors are colored in green. The residues that differ between clone 49 and NKAT2 are colored yellow. Unique residues present only in clone 39 are colored orange. The potential surfaces, calculated by using grasp (50), of NKAT2, clone 49, and clone 39 are shown in C, F, and I, in the top view.
The functional significance of the different binding affinities of p58 and p50 receptors for HLA-C seems clear. If an activating and an inhibitory receptor are competing for binding to the same HLA-C molecule, then the inhibitory receptor is more likely to bind; that is, inhibition will be dominant over activation, as has been observed (22). It is interesting to consider this conclusion in the light of what is now understood about the processes of signal transduction mediated by the activating and inhibitory receptors. The short-tailed activating p50 molecules associate noncovalently with the DAP12 protein (20, 21). This molecule contains an immunoreceptor tyrosine-based activation motif (45) and ligation of this receptor complex results in an activation cascade that includes the kinases Syk and Zap70 (21), which can in turn phosphorylate other proteins; that is, the initial signal is amplified during the process of signal transduction. In contrast, ligation of p58 inhibitory receptors results in recruitment of a protein tyrosine phosphatase SHP-1, which presumably blocks the activation response by dephosphorylating some substrate (reviewed in ref. 46). This is essentially a process that is not amplified. Thus, if the affinity of activating and inhibitory receptors for HLA-C were equal, then, at any given concentration of peptide/MHC complex, the action of the activating receptor would be dominant. However, the system has evolved to a situation exactly the opposite of this prediction. In this context, it is interesting to note that, recently, data have been obtained that strongly suggest that the inhibitory CD94/NKG2-A heterodimer may have a higher affinity for HLA-E than the activating CD94/NKG2-C heterodimer (47). Further, the costimulatory ligand CD80 (B7–1) has been reported to bind the inhibitory receptor CTLA-4 with a 10-fold higher affinity than the costimulating receptor CD28 (48). Observations such as these suggest a general theme that inhibitory receptors may bind their ligands with a higher affinity than activating molecules bind the same or related ligands. An important caveat to the above is the influence of peptide on the recognition of HLA-C by p50 and p58 receptors. By using natural ligands of HLA-Cw7, the nature of the bound peptide made an absolute difference to its binding to NKAT2. The molecular basis of this effect is not clear, but it was not simply the result of the presence of charged amino acids toward the C terminus of the peptide. Whatever the basis of the effect seen in Fig. 4, the result leaves open the possibility that efficient recognition by p50 activating receptors could require a repertoire of peptides distinct from those required by inhibitory receptors, in the some manner as different T cell receptors respond to distinct peptides. Examination of this question probably will require a detailed analysis of the influence of peptide on NKIR and NKAR binding to HLA-C proteins.
Acknowledgments
We thank Dr. Miguel Lopez-Botet for helpful discussions and sharing data before publication. We also thank Dr. Javier Lopez Jaramillo (Harvard University) for much help and advice with the molecular modeling of the receptors and Qing Fan (Harvard University) for supplying the coordinates of the NKAT1 (clone 42) structure. This work was supported by National Institutes of Health Grant CA-47554, and was presented in the thesis of M.V.-G. in partial fulfillment of the requirements for a Ph.D. degree, Universidad Autónoma de Madrid, June 1998.
ABBREVIATIONS
- NK
natural killer
- MHC
major histocompatibility complex
- NKIR
NK cell inhibitory receptors, NKAT, NK-associated transcript
- RU
resonance units
References
- 1.Lazetic S, Chang C, Houchins J P, Lanier L L, Phillips J H. J Immunol. 1996;157:4741–4745. [PubMed] [Google Scholar]
- 2.Brooks A G, Posch P E, Scorzelli C J, Borrego F, Coligan J E. J Exp Med. 1997;185:795–800. doi: 10.1084/jem.185.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carretero M, Cantoni C, Bellon T, Bottino C, Biassoni R, Rodriguez A, Perez-Villar J J, Moretta L, Moretta A, Lopez-Botet M. Eur J Immunol. 1997;27:563–567. doi: 10.1002/eji.1830270230. [DOI] [PubMed] [Google Scholar]
- 4.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari M C, Moretta L. Annu Rev Immunol. 1996;14:619–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 5.Borrego F, Ulbrecht M, Weiss E H, Coligan J E, Brooks A G. J Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braud V M, Allan D S, O’Callaghan C A, Soderstrom K, D’Andrea A, Ogg G S, Lazetic S, Young N T, Bell J I, Phillips J H, McMichael A J, et al. Nature (London) 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 7.Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, Geraghty D E. Proc Natl Acad Sci USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanier L L. Annu Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 9.Valiante N M, Uhrberg M, Shilling H G, Lienert-Weidenbach K, Arnett K L, D’Andrea A, Phillips J H, Lanier L L, Parham P. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 10.Wagtmann N, Rajagopalan S, Winter C C, Peruzzi M, Long E O. Immunity. 1995;3:801–809. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 11.Colonna M, Samaridis J. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 12.D’Andrea A, Chang C, Franz-Bacon K, McClanahan T, Phillips J H, Lanier L L. J Immunol. 1995;155:2306–2310. [PubMed] [Google Scholar]
- 13.Pende D, Biassoni R, Cantoni C, Verdiani S, Falco M, di Donato C, Accame L, Bottino C, Moretta A, Moretta L. J Exp Med. 1996;184:505–518. doi: 10.1084/jem.184.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dohring C, Scheidegger D, Samaridis J, Cella M, Colonna M. J Immunol. 1996;156:3098–3101. [PubMed] [Google Scholar]
- 15.Reyburn H, Mandelboim O, Valés-Gómez M, Sheu E G, Pazmany L, Davis D M, Strominger J L. Immunol Rev. 1997;155:119–125. doi: 10.1111/j.1600-065x.1997.tb00944.x. [DOI] [PubMed] [Google Scholar]
- 16.Valiante N M, Lienert K, Shilling H G, Smits B J, Parham P. Immunol Rev. 1997;155:155–164. doi: 10.1111/j.1600-065x.1997.tb00948.x. [DOI] [PubMed] [Google Scholar]
- 17.Biassoni R, Cantoni C, Falco M, Verdiani S, Bottino C, Vitale M, Conte R, Poggi A, Moretta A, Moretta L. J Exp Med. 1996;183:645–650. doi: 10.1084/jem.183.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moretta A, Vitale M, Bottino C, Orengo A M, Morelli L, Augugliaro R, Barbaresi M, Ciccone E, Moretta L. J Exp Med. 1993;178:597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips J H, Gumperz J E, Parham P, Lanier L L. Science. 1995;268:403–405. doi: 10.1126/science.7716542. [DOI] [PubMed] [Google Scholar]
- 20.Olcese L, Cambiaggi A, Semenzato G, Bottino C, Moretta A, Vivier E. J Immunol. 1997;158:5083–5086. [PubMed] [Google Scholar]
- 21.Lanier L L, Corliss B C, Wu J, Leong C, Phillips J H. Nature (London) 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 22.Moretta A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R, Bottino C, Moretta L. J Exp Med. 1995;182:875–884. doi: 10.1084/jem.182.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandelboim O, Davis D M, Reyburn H T, Valés-Gómez M, Sheu E G, Pazmany L, Strominger J L. Science. 1996;274:2097–2100. doi: 10.1126/science.274.5295.2097. [DOI] [PubMed] [Google Scholar]
- 24.Litwin V, Gumperz J, Parham P, Phillips J H, Lanier L L. J Exp Med. 1993;178:1321–1336. doi: 10.1084/jem.178.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanier L L, Gumperz J E, Parham P, Melero I, Lopez-Botet M, Phillips J H. J Immunol. 1995;154:3320–3327. [PubMed] [Google Scholar]
- 26.Fan Q O R, Garboczi D N, Winter C C, Wagtmann N, Long E O, Wiley D C. Proc Natl Acad Sci USA. 1996;93:7178–7183. doi: 10.1073/pnas.93.14.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Chwae Y J, Kim M Y, Choi I H, Park J H, Kim S J. J Immunol. 1997;159:3875–3882. [PubMed] [Google Scholar]
- 28.Valés-Gómez, M., Reyburn, H. T., Mandelboim, M. & Strominger, J. L. (1998) Immunity, in press. [DOI] [PubMed]
- 29.Cambiaggi A, Orengo A M, Meazza R, Sforzini S, Tazzari P L, Lauria F, Raspadori D, Zambello R, Semenzato G, Moretta L, et al. Blood. 1996;87:2369–2375. [PubMed] [Google Scholar]
- 30.Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, Moretta L, Moretta A. Eur J Immunol. 1997;27:3095–3099. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 31.Campbell K S, Cella M, Carretero M, Lopez-Botet M, Colonna M. Eur J Immunol. 1998;28:599–609. doi: 10.1002/(SICI)1521-4141(199802)28:02<599::AID-IMMU599>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 32.Winter C C, Gumperz J E, Parham P, Long E O, Wagtmann N. J Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- 33.Moretta A, Tambussi G, Bottino C, Tripodi G, Merli A, Ciccone E, Pantaleo G, Moretta L. J Exp Med. 1990;171:695–714. doi: 10.1084/jem.171.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan Q R, Mosyak L, Winter C C, Wagtmann N, Long E O, Wiley D C. Nature (London) 1997;389:96–100. doi: 10.1038/38028. [DOI] [PubMed] [Google Scholar]
- 35.Falk K, Rotzschke O, Grahovac B, Schendel D, Stevanovic S, Gnau V, Jung G, Strominger J L, Rammensee H G. Proc Natl Acad Sci USA. 1993;90:12005–12009. doi: 10.1073/pnas.90.24.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uhrberg M, Valiante N M, Shum B P, Shilling H G, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier L L, Parham P. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 37.Winter C C, Long E O. J Immunol. 1997;158:4026–4028. [PubMed] [Google Scholar]
- 38.Rajagopalan S, Long E O. J Exp Med. 1997;185:1523–1528. doi: 10.1084/jem.185.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandelboim O, Wilson S B, Valés-Gómez M, Reyburn H T, Strominger J L. Proc Natl Acad Sci USA. 1997;94:4604–4609. doi: 10.1073/pnas.94.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandelboim O, Kent S, Davis D M, Wilson S B, Okazaki T, Jackson R, Hafler D, Strominger J L. Proc Natl Acad Sci USA. 1998;95:3798–3803. doi: 10.1073/pnas.95.7.3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colonna M, Spies T, Strominger J L, Ciccone E, Moretta A, Moretta L, Pende D, Viale O. Proc Natl Acad Sci USA. 1992;89:7983–7985. doi: 10.1073/pnas.89.17.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mandelboim O, Reyburn H T, Valés-Gómez M, Pazmany L, Colonna M, Borsellino G, Strominger J L. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati M S, Vitale M, Bottino C, Moretta L, Moretta A, Long E O. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 44.Steffens U, Vyas Y, Dupont B, Selvakumar A. Tissue Antigens. 1998;51:398–413. doi: 10.1111/j.1399-0039.1998.tb02981.x. [DOI] [PubMed] [Google Scholar]
- 45.Cambier J C. J Immunol. 1995;155:3281–3285. [PubMed] [Google Scholar]
- 46.Renard V, Cambiaggi A, Vely F, Blery M, Olcese L, Olivero S, Bouchet M, Vivier E. Immunol Rev. 1997;155:205–221. doi: 10.1111/j.1600-065x.1997.tb00953.x. [DOI] [PubMed] [Google Scholar]
- 47.Llano M, Lee N, Navarro F, Garcia P, Albar J P, Gerahty D E, Lopez-Botet M. Eur J Immunol. 1998;9:2854–2863. doi: 10.1002/(SICI)1521-4141(199809)28:09<2854::AID-IMMU2854>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 48.van der Merwe P A, Bodian D L, Daenke S, Linsley P, Davis S J. J Exp Med. 1997;185:393–403. doi: 10.1084/jem.185.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sayle R, Milner-White E J. Trends Biochem Sci. 1995;20:374–377. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 50.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]