Abstract
The new antigen receptor (NAR) gene in the nurse shark diversifies extensively by somatic hypermutation. It is not known, however, whether NAR somatic hypermutation generates the primary repertoire (like in the sheep) or rather is used in antigen-driven immune responses. To address this issue, the sequences of NAR transmembrane (Tm) and secretory (Sec) forms, presumed to represent the primary and secondary repertoires, respectively, were examined from the peripheral blood lymphocytes of three adult nurse sharks. More than 40% of the Sec clones but fewer than 11% of Tm clones contained five mutations or more. Furthermore, more than 75% of the Tm clones had few or no mutations. Mutations in the Sec clones occurred mostly in the complementarity-determining regions (CDR) with a significant bias toward replacement substitutions in CDR1; in Tm clones there was no significant bias toward replacements and only a low level of targeting to the CDRs. Unlike the Tm clones where the replacement mutational pattern was similar to that seen for synonymous changes, Sec replacements displayed a distinct pattern of mutations. The types of mutations in NAR were similar to those found in mouse Ig genes rather than to the unusual pattern reported for shark and Xenopus Ig. Finally, an oligoclonal family of Sec clones revealed a striking trend toward acquisition of glutamic/aspartic acid, suggesting some degree of selection. These data strongly suggest that hypermutation of NAR does not generate the repertoire, but instead is involved in antigen-driven immune responses.
Like the hallmark molecules of adaptive immunity (T cell receptors, Ig, and major histocompatibility complex) and the Ig gene rearrangement machinery, somatic hypermutation of Ig genes is present in the oldest group of extant jawed vertebrates, the cartilaginous fish (1–3). Similar to mutation and selection in adaptive evolution, somatic hypermutation in mice and humans is an antigen-driven process that randomly introduces mutations to Ig genes, and B cells whose receptors have affinity-enhancing mutations are selected in the germinal centers (GC) of secondary lymphoid tissues (reviewed in ref. 4). In sheep and cows, however, somatic hypermutation generates the primary repertoire in a foreign antigen-independent process during B-cell development in the Ileal Peyer’s Patches (5, 6). Generally, species that use somatic hypermutation and/or gene conversion to generate the repertoire use only one or a few variable (V) genes, whereas those using somatic hypermutation only in secondary responses express many V families (5–10). In the amphibian Xenopus, somatic hypermutation is not used to generate the repertoire (11), and the few examples where it is known to generate the repertoire occur in mammals. Such data suggest that somatic mutation first evolved for secondary immune responses and later was recruited to generate the repertoire in some species; studies of somatic hypermutation in the cartilaginous fish address this issue.
In the nurse shark, Ginglymostoma cirratum, a new class of antigen receptor gene (NAR) was discovered in our laboratory. NAR is a covalently linked dimer, each monomer consisting of one V domain and five constant (C) domains. NAR is found in both transmembrane (Tm) and secretory (Sec) forms, but it differs from bona fide Ig in that it does not associate with Ig light chains (2, 12). Further, the NAR V region is more similar to Ig light chain/T-cell receptor V than to Ig heavy chain V (2). The NAR V gene diversifies extensively by somatic hypermutation, unlike shark Ig genes, where the mutation frequency is low, and the mechanism seems unconventional (1). It is not known, however, whether somatic hypermutation of NAR is used to generate the repertoire or rather is a consequence of antigen-dependent reactions. Some features of NAR and of humoral responses in cartilaginous fish suggest that somatic mutation of NAR generates the repertoire: (i) like in sheep there is low germ-line diversity; fewer than five NAR loci are encoded in the nurse shark genome, and only two highly similar loci seem to be expressed (2, 12); (ii) GC formation has never been reported in an ectotherm, thus mutated clones may not be optimally selected; and (iii) there is no affinity maturation of shark IgM responses (13–15). Therefore, we hypothesized that NAR hypermutation is an antigen-independent mechanism used to generate a primary repertoire. However, its high mutation frequency may suggest that NAR is responsible for the true, adaptive immune responses in the cartilaginous fish. To test the hypothesis, we analyzed mutations in cDNA generated from NAR transcripts of the Tm and Sec forms. We reasoned that if NAR hypermutation is antigen dependent and involved in secondary immune responses, then a high proportion of Sec transcripts should be highly mutated (if, as expected, they are made in much larger quantities than Tm transcripts in activated memory cells that secrete NAR) whereas few highly mutated Tm transcripts should be detected. If conversely, NAR hypermutation diversifies the primary repertoire during development like in the sheep, then both Sec and Tm clones should be highly mutated. This approach is analogous to studies of IgM and IgG in mice and humans, where the co-occurrence of hypermutation and isotype switch in GC results in a larger proportion of IgG transcripts bearing mutations (16). However, because there is no class switch from IgM to NAR within a single shark lymphocyte (NAR transcripts never bear IgM V regions) we could not study mutations in different isotypes. Finally, we analyzed characteristics of mutations in NAR clones to determine whether the pattern is similar to that seen in IgM of sharks or IgG of mammals.
MATERIALS AND METHODS
DNA and RNA Isolation.
Adult nurse sharks (1–2 years of age) were caught off the coast of Key West, FL and kept in open-air tanks at the University of Miami hatchery facility for at least 2 months before sampling. Peripheral blood lymphocytes from each shark were obtained as described (15). RNA was isolated with Trizol Reagent (Life Technologies) following the manufacturer’s protocol. Genomic DNA was isolated from red blood cells and incubated overnight with Proteinase K (50 μg) in TENS solution (50 mM Tris/100 mM EDTA/100 mM NaCl/1% SDS) followed by two rounds of phenol/chloroform extraction (1:1) and precipitation with 100% ethanol (2:1).
First-Strand Synthesis and PCR Amplification of NAR.
Total RNA (10 μg) was used to synthesize cDNA according to the protocol in the reverse transcription–PCR kit (Stratagene) with one of two primers: Tailr, which complements the Sec tail region of NAR (5′-TAGTACGACCTGAAACATTAAC-3′) or Tm, which complements the Tm region of NAR (5′-TACAAATGTGGTGTACAGCAT-3′) (12). PCR amplification of cDNA (25–30 cycles, annealing at 56–58°C) was carried out with Taq DNA polymerase (Gibco) by using the following forward primers: Leaderf (complements a region in the leader), 5′-CAGTCCTTTTAGCCTTATTACC-3′; or Frf (complements a region in framework 1), 5′-TGATCGAGTGGACCAAACACC-3′. The reverse primer was C1NARr (complements a region in the B-strand of the first C domain), 5′-ATAGTATCCGCTGATTAGACA-3′. Genomic DNA was amplified under similar conditions (25–30 cycles, 56°C) with the same forward primers as used for cDNA amplification, but with the following reverse primers: JT1r (complements the J segment of NAR type 1), 5′-GTCACGAMAGTGCCMKCTCCGC-3′; or JT2r (for NAR type 2 J segment), 5′-GTCCCGACAGTGCCASYKCCGT-3′, where M = A/C, K = G/T, S = G/C, and Y = C/T. To establish a background error rate that accounts for both the reverse transcriptase and the Taq polymerase, cDNA for the shark J chain (L. A. Steiner and M.F.F., unpublished work) was synthesized with primer Jchainr2, 5′-TGGATCTACTGTTGTTTCAAA-3′, and amplified (30 cycles, 56°C) with the forward primer, Jchainf, 5′-GATACGTTGGGGGTACTTGCA-3′, and the reverse primer, Jchainr, 5′-TAATCCAGTAGCTTTGTAGGA-3′.
Subcloning and DNA Sequencing.
PCR products of the expected size were excised from a 1.5% agarose gel, and the DNA was purified with the Gene Clean II Kit (Bio 101). The product was subcloned into the PCR2.1 vector, and both the ligation and the transformation into competent cells was done according to the protocol of the Original TA Cloning Kit (Invitrogen). A small sample of the transformed cells (0.2 ml) was added to prewarmed 5-bromo-4-chloro-3-indolyl β-d-galactoside-coated (40 mg/ml) Luria–Bertani (LB)-agar plates (17) and incubated for 14–16 hr at 37°C. Individual colonies were plugged from the plates and grown for 15 hr in 6 ml of LB (17) with 50 μg/ml of ampicillin at 37°C. Plasmid DNA was purified from the cultures with the Bio Robot 9600 (Quiagen). Approximately 1 μg of plasmid from each clone was sequenced in both directions with Sequenase (United States Biochemical).
Statistical Analysis.
Sec clones from each of the three sharks (n = 31, 20, 23) and Tm clones (n = 22, 21, 20) were analyzed for mutations. Standard nonparametric tests were used to test for (i) differences in the distribution of mutations among clones from the Sec and the Tm pools (Kolmogorov-Smirnov); (ii) differences in frequencies or average difference between samples (G test, t test for paired comparisons); and (iii) differences between observed and expected frequencies (χ2 analysis). The statistical package mega (18) was used to establish the relationship among members of an oligoclonal family, and for the analysis of proportion of synonymous and nonsynonymous changes per synonymous (Ps) and nonsynonymous (Pn) site, respectively, for different regions of the gene. Because each group of sequences was compared with a germ-line sequence, only the P values for the comparison between each sequence and its germ-line counterpart were used. The difference between Pn and Ps for each sequence was determined and the differences tested for higher than expected Pn values with the Wilcoxon Signed-Ranks Test. All statistical tests are described in Sokal and Rohlf (19). To determine clustering to the complementarity-determining regions (CDRs) in mouse passenger transgenes from previous studies, we analyzed nine mutated sequences from the VkOx1 transgenes (hybridomas: NQT15/2, 3, 4, 10, and 23; ref. 20), and 11 unique anti-nitrophenyl transgenes (21) of the Lk3 strain and 21 sequences from Peyer’s Patches of Lk[p5]bG-derived transgenes (22). The sequence analysis package Intelligenetics was used for alignment and sequence comparisons and for searches in various databases.
RESULTS
Mutation Frequencies in NAR Sec and Tm Transcripts.
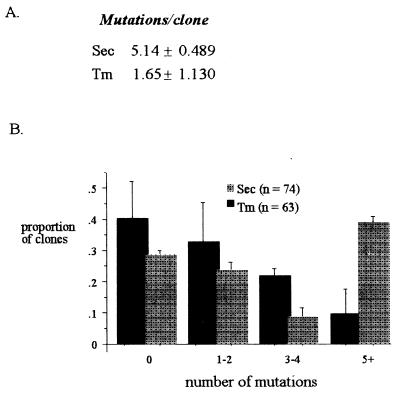
Two NAR genes constituted more than 90% of the clones obtained from all sharks in both Tm and Sec clones. Although the Tm sequences averaged 1.65 mutations/clone (overall frequency of 0.007/base), the average for Sec clones was 5.14 (0.022/base) (Fig. 1A). Furthermore, more than 40% of all Sec clones but fewer than 11% of the Tm clones had five mutations or more (Fig. 1B). Two unique Tm clones were highly mutated (>20), and because they were more than two SDs from the mean for Tm clones, they were excluded from the frequency analysis (but not Fig. 1B analysis). Even if included, however, the frequency in the Sec clones was higher in all sharks, the shark with the smallest difference still had 1.45 times more mutations in the Sec clones, and the overall frequency was still significantly higher in the Sec clones. There were no significant differences among the three sharks. The mutation frequencies observed for both Sec and Tm clones were significantly higher than the estimated background error from analysis of the J chain transcripts (0.0009/base). More than 25% and 40% of the Sec and the Tm clones, respectively, were unmutated, and they were considered in all frequency analyses.
Figure 1.
The frequency of mutations and their distribution among Tm and Sec clones. (A) Sec clones (n = 74) had a significantly higher number of mutations than the Tm clones (n = 62) (P < 0.05, one-tail t test for paired comparisons). Shared mutations in oligoclonal families were counted once. (B) The proportion of clones with a large number of mutations (>5/clones) was also highest in the Sec clones (P < 0.05, Kolmogorov-Smirnov Test).
Mutations in the Sec Clones But Not the Tm Clones Clustered to the CDRs.
Although the CDRs constitute 23% of the NAR V sequence analyzed, a majority of the mutations (59%) in the Sec clones in all three sharks clustered to the CDRs (Table 1). This high level of CDR clustering was not observed in the mutations from the Tm clones. In the oldest shark, 67% of the mutations from the Sec clones occurred in the CDRs, and only 24% were found in the CDR of Tm clones (Fig. 2 depicts this distribution for type 2 NAR sequences from this shark). Passenger Ig transgenes in mice that are not under antigenic selection do not display a high concentration of mutations in CDRs (CDRs constituted 20% of the sequence analyzed in those studies and 34% of the mutations occurred there; refs. 20–22). This low level of clustering to the CDRs in NAR Tm and mouse passenger transgenes probably is caused by the mutational hot spots found in the CDRs of antigen receptor genes in most vertebrates (23).
Table 1.
Mutations are targeted to the CDRs in the Sec clones and display evidence of positive selection in CDR1
| Region | % of analyzed sequence | % mutations* | R/S† | P̄n (SD)/P̄s (SD)* |
|---|---|---|---|---|
| Sec | ||||
| FR1 | 25% | 8.6% | 11/7 | 0.028 (0.03)/0.047 (0.05) |
| FR3 | 38% | 24% | 40/12 | 0.020 (0.02)/0.019 (0.02) |
| CDR1 | 13% | 34% | 61/10 | 0.097 (0.05)/0.073 (0.12)‡ |
| CDR2 | 10% | 24% | 43/8 | 0.109 (0.082)/0.126 (0.13) |
| CDRs total | 23% | 58.9% ± 7.9§ | ||
| Tm | ||||
| FR1 | 25% | 15% | 14/3 | 0.02 (0.012)/0.01 (0.033) |
| FR3 | 38% | 38% | 26/9 | 0.02 (0.012)/0.03 (0.029) |
| CDR1 | 13% | 25% | 18/7 | 0.06 (0.052)/0.09 (0.105) |
| CDR2 | 10% | 9.9% | 6/2 | 0.05 (0.024)/0.09 (0.123) |
| CDRs total | 23% | 34.7% ± 8.6 |
Mutated clones with fewer than 15 mutations (Sec, n = 66; Tm, n = 60).
Excludes tandem mutations where the type of change depends on which mutation occurred first. These were not excluded from the Pn, Ps analysis. R, replacement; s, silent.
P < 0.05 (Wilcoxon Signed-Ranks Test).
P < 0.05 (G test; Sec clones CDR mutation frequency was higher than Tm clones).
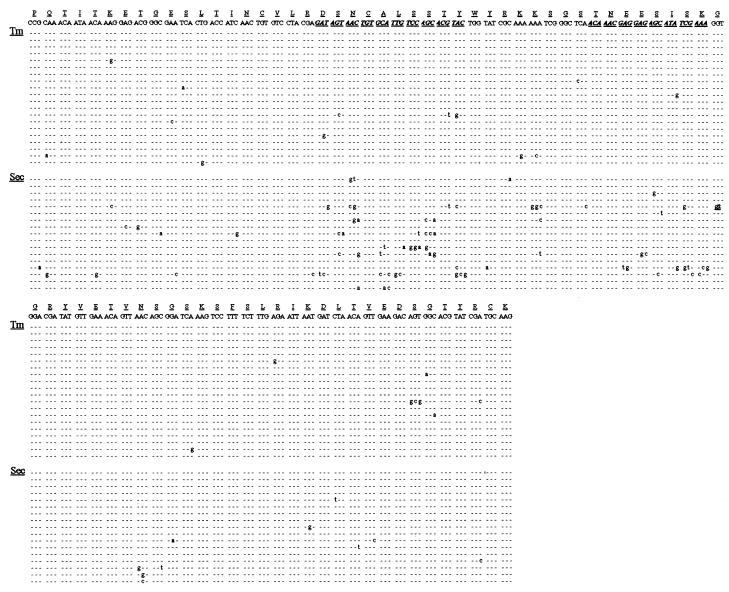
Figure 2.
Distribution of Tm and Sec mutations in a type 2 NAR gene of an individual shark. CDRs are in bold italics and underlined. A triplet insertion is underlined. The insertion precedes the triplet where indicated. The CDR3 of each sequence was unique.
The CDR1 of Sec Clones Had a Higher Proportion of Replacement Than Synonymous Changes, and the Pattern of Replacement Mutations Was Different in the Sec Clones (But Not Tm Clones) From That of Synonymous Changes.
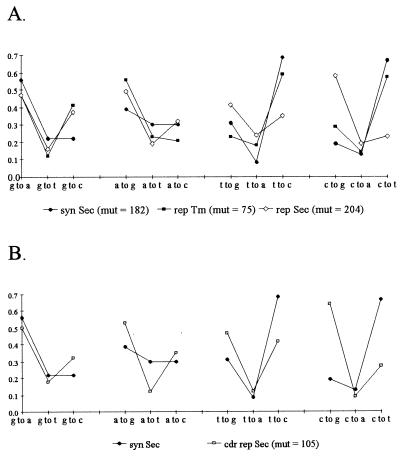
Among synonymous mutations in NAR there is a significant bias for transitions over transversions (even when considering the bias for transitions at the third base to generate synonymous changes as well as the high number of synonymous transitions from c to t in the AGC hotspot; M.D. and M.F.F., unpublished work). Because synonymous changes are presumably not under selection, this pattern should reflect the intrinsic properties of the mechanism, and it parallels the bias for transitions over transversions in mammalian Ig, heterologous sequences placed within Ig constructs, and JC intron sequences (24–26). Although the Tm clones displayed a pattern nearly identical to the synonymous changes, the Sec replacement mutations from cytosine and thymine or in the first two bases of the Sec CDRs revealed a distinct pattern, suggesting selection (Fig. 3 A and B). Interestingly, some of the uncommon synonymous changes remained at a relatively low frequency in all groups, perhaps reflecting constraints of the mechanism (i.e., low probability of a putative polymerase committing some errors). Also, the pattern of mutation from guanine and adenine was similar for all groups, but analysis of the data suggests that this results from saturation of mutations in the AGC/T hotspot (transitions in the third base would not generate replacements, thus this saturation would not impact replacements from cytosine or thymine).
Figure 3.
Deviation from the intrinsic mutation pattern of replacement substitutions in Sec (n = 66), but not Tm (n = 60), clones suggests selection. Replacement analysis excluded clones with more than 15 mutations (mut). (A) Synonymous changes include data from an additional 31 clones used in a previous study (2), but excludes c to t changes generated by synonymous changes to the AGC hotspot and to a TGC codon in framework region (FR) 2 of type 1 NAR that is under stringent selection (35). (B) Even when excluding the third base of each codon, the Sec CDR replacements differ from the intrinsic pattern of mutation. Conservatively, we excluded the few examples where a third base generated a replacement, because they tend to be generated by transversions. The overall G test comparison of all groups was significant (P < 0.05), which was mostly attributed to the comparison of the replacement Sec group vs. the synonymous group.
The CDR1 of the Sec clones displayed a significant bias toward replacements, suggesting antigen-driven selection (Table 1). Codons encoding Ig CDR1 have an intrinsic propensity for mutations to generate replacements (27), and thus one may expect a higher ratio in this region. However, the Pn and Ps analysis accounts for such a bias by estimating the baseline probability of replacement for each residue. Thus the significantly greater Pn than Ps values in CDR1 of the Sec clones reflect a true skewing for replacements.
A high replacement/silent ratio in the CDRs was seen in the sheep, where hypermutation generates the repertoire. In the sheep, however, the mutational mechanism targets guanines and adenines (a pattern not seen in NAR, see Fig. 5), and a significant proportion of CDR codons have guanine and/or adenine as the first two bases (5). Furthermore, mutations are highly targeted to the CDRs. Thus, given the lower targeting of mutations to CDRs in mouse passenger transgenes, it is likely that in the sheep somatic mutation is driven by some factor, such as self-antigen or superantigen. In addition, any intrinsic propensity to generate replacements in the CDRs of NAR caused by the mutational mechanism itself also would impact the Tm clones, which is clearly not the case (Table 1).
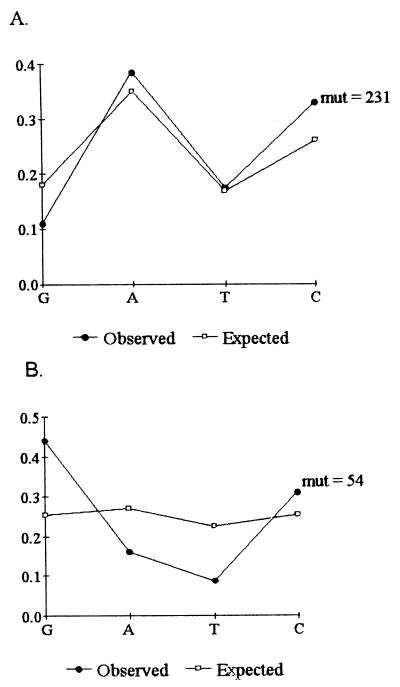
Figure 5.
Unlike shark IgM, NAR hypermutation does not target GC bases. (A) The proportion of synonymous mutations (mut) at each base in NAR is similar to expected based on the base composition at degenerate sites. (B) Mutation of shark IgM genes targets g:c bases, particularly guanines.
Three Oligoclonal Families of Sequences Were Detected Among the Sec Clones.
Hybridomas generated from GC foci during the course of an immune reaction often reveal families of sequences (28). These families share identical CDR3 (barring mutation) as well as other mutations that have been selected by antigen. The absence of GC in sharks prevented a similar analysis, but, unexpectedly, we detected three oligoclonal families among Sec clones from two sharks. Two of these families had two members with nearly identical CDR3 and shared mutations in other CDRs.
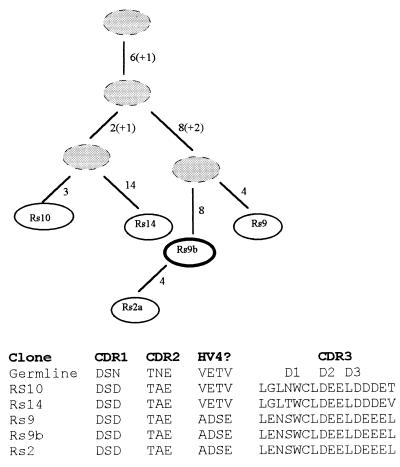
The third family had 11 members (five unique, Fig. 4). A striking feature of this family is the highly acidic nature of CDR3. Furthermore, a hierarchical analysis of this sequence reveals a progression toward acquisition of acidic residues; even when the codon of an acidic residue was mutated leading to a replacement, it was changed to the other acidic residue (glutamic acid ↔ aspartic acid). In a region of NAR likely to be homologous to hypervariable region 4 of T cell receptors (e.g., see ref. 29), three members of this family underwent a series of mutations (most were not in tandem) that generated four consecutive amino acid replacements. One of these was a change from glutamic acid to aspartic acid, and another from valine to glutamic acid. This latter change involved two t → a substitutions, mutations rarely seen among NAR synonymous changes (Fig. 3). Furthermore, all members of this family share a mutation in the CDR1 that replaces asparagine with aspartic acid. Apparently, this family is responding to a positively charged antigen.
Figure 4.
An oligoclonal family of Sec clones has been selected for changes to acidic residues. The numbers adjacent to lines connecting clones are the number of mutations shared (and unique mutations when ending the lineage) and numbers in parenthesis enumerate acquired acidic residues. Shaded circles represent theoretical intermediates. The bold circle reflects the most common member of the family. Hypervariable region 4 (HV4) is modeled to be in a similar position as in T cell receptors (29).
It is possible that these oligoclonal families are encoded by germ-line-joined NAR genes (germ-line-joined clusters are found in shark Ig genes, reviewed in ref. 30). This possibility, however, is unlikely for the following reasons: (i) prospective nonrearranged germ-line D segments were identified for all expressed genes; (ii) all of the genomic DNA PCR products were analyzed and revealed no complete germ-line-joined genes; (iii) this CDR3 was not detected in cDNA clones from any shark; and (iv) in a previous study, NAR clones from a genomic library revealed the same germ-line sequences that we obtained (12).
Unlike Xenopus Ig and, Strikingly, Shark IgM, NAR Hypermutation in the Nurse Shark Does Not Target GC Bases.
Among synonymous changes, a:t and g:c bases were mutated at frequencies proportional to the base composition at degenerate sites in the V region analyzed (Fig. 5A). This finding is in stark contrast to mutation of bona fide Ig in the shark and in Xenopus, where there is a strong bias to mutate g:c base pairs, particularly guanines (Fig. 5B; refs. 1, 11, and 31). There are further distinctions between NAR and shark IgM: limited data suggest that shark IgM mutation frequency is low and mutations do not cluster to the CDRs. NAR hypermutation in the Sec clones, in fact, is most similar to the pattern described for mammalian IgG.
DISCUSSION
The results of this study suggest that, contrary to our original hypothesis, hypermutation of NAR genes does not generate their primary, antigen-reactive repertoire. Although an overall low level of mutation was found in Tm sequences, more than 35% of the clones had no mutations and nearly 40% had only one or two changes. In sheep, the one well-studied model in which hypermutation generates the primary diversity, all clones from young lambs are heavily mutated with extensive targeting to the CDRs (>80%), a result that is expected for creation of a diverse and heterogeneous repertoire. Our results with NAR demonstrate that low numbers of particular antigen receptor loci in the genome do not necessarily implicate somatic hypermutation (or gene conversion) in repertoire building.
How can one explain the few mutations detected in the majority of Tm clones that bear mutations? The pattern of both the Tm replacement and synonymous base changes is similar to that seen in synonymous sites of Sec clones (and indeed any set of synonymous sites examined; M.D. and M.F.F., unpublished work), and there is no significant targeting to the CDR. An idea that we favor is that the mutational mechanism may be down-regulated, but not entirely suppressed, during the early development of NAR+ cells. Consistent with this concept, conventional mouse IgM/IgD+ B cells found in peripheral blood sometimes have low levels of mutation (32).
Perhaps a more engaging theme, given the low number of NAR genes, is why the primary repertoire is not generated via hypermutation in addition to a second round of mutation when mature NAR+ cells respond to antigen. It is likely that sheep B cells undergo mutation in secondary lymphoid organs in response to antigen because chickens and rabbits, which generate their primary repertoire through gene conversion (and mutation in the rabbit), also experience mutation and conversion in mature B cells upon foreign antigen exposure (33, 34). Functional NAR genes are generated by way of rearrangement of three separate diversity (D) elements, among themselves and with V and J genes, resulting in a long and highly diverse CDR3. The majority of analyzed cDNA clones has undergone the predicted four rearrangement events (2, 35), which, in combination with the low number of NAR genes in the genome and a selection for preferred reading frames for the D elements, implies a great deal of cell wastage before peripheralization of mature NAR+ cells. Perhaps superimposing mutation upon this repertoire building through rearrangement would be too costly, especially in an ectothermic species whose cell generation time is long (36). In sum, our data suggest that the primary repertoire is entirely CDR3 based, with extensive mutational modification of mature cells.
We previously have proposed that IgM provides a first line of defense in cartilaginous fish and that NAR represents the true adaptive response (31). In this study, the high frequency of mutation, number of mutated clones, clustering of mutation to the CDRs, and even the lack of mutations in a significant portion of Sec clones are indeed all consistent with a selected, presumably antigen-driven mechanism. One possible scenario is that NAR hypermutation is antigen stimulated during a response but without selection for higher affinity. However, we found evidence for positive selection in the bias for replacements in CDR1 of the Sec clones (as seen in germ-line sequences of mammalian Ig; ref. 37) and the distinct patterns of CDR replacements in comparison to synonymous changes. In addition, the analysis of oligoclonal families reflects antigen-driven selection as one of the families had a clear tendency toward acquisition of charged amino acids. Thus, these data, taken together, indicate that antigen-driven mutation can occur in the absence of GC.
The NAR hypermutation pattern and frequency is most similar to that of mouse and human IgG and probably shares most of the components of the mammalian mechanism (M.D. and M.F.F., unpublished work). More strikingly, mutation in NAR is different from shark IgM and perhaps from all ectothermic vertebrates. Are there two different hypermutational mechanisms operative in ectothermic vertebrates? If true, is there a Xenopus counterpart to NAR hypermutation? In answer to the first question, it was proposed that the mechanism in shark and Xenopus Ig is the same as in mammals, but antigen-driven selection “disguises” the g:c bias in mammals (31). This hypothesis, however, does not account for the absence of a g:c bias in: (i) the synonymous mutations analyzed herein; (ii) the low-grade mutations found in the Tm clones; and (iii) intron sequences of mammalian Ig genes (38). We favor another explanation: the Ig hypermutational mechanism in frogs, sharks, and skates is the same as in NAR and mammalian IgG, but lacks some components—–perhaps recruitment of certain mismatch repair enzymes (31, 39–41). The third and least attractive possibility is that there are indeed different mechanisms, and one has been lost (or not found) in the amphibian lineage. This last possibility would imply remarkable convergence between the functionally related NAR and mammalian Ig loci. Once the molecular intricacies of the mutation mechanism are revealed, these questions can be addressed directly.
The seemingly inefficient mutation of shark Ig (low frequency, GC targeting, no clustering to CDR) points to NAR as the molecule responsible for antigen-driven responses in sharks, which would reconcile previous findings of poor affinity maturation of shark IgM and predicts affinity maturation of the NAR antigen-specific responses. The genomic arrangement of shark IgM (many clusters of V, D, and J genes for the heavy chain and of V and J for the light chain, with some clusters germ-line-joined; refs. 1, 30, 42, and 43) may hinder clonal expansion and antigen-driven mutation. The very low number of NAR loci and the absence of an associated light chain may have been selected to increase the probability of expression of a single receptor/lymphocyte, permitting effective expansion and antigen-driven mutation in the absence of germinal centers. The deficiency in GC does not necessarily indicate that ectotherms lack centralized areas within secondary lymphoid tissues where selection can occur; it would be surprising if they did not have some type of analogous structures. It is also possible, however, that NAR is specialized to respond to a specific class of antigens. Interestingly, the oligoclonal family we obtained is rich in negatively charged amino acids, as is the germ-line CDR2 of NAR genes (Fig. 2) and at least one of the D elements from most of the NAR genes examined.
Here we furnish evidence that hypermutation in adaptive humoral responses of cartilaginous fish can be antigen driven. The same conclusion was reached in studies of immunized Xenopus (31), although, as described, the mutational pattern was unusual and the frequency was low. Because representatives of almost all vertebrate classes now have been shown to use somatic hypermutation for antigen-driven responses, and its use for repertoire generation only has been shown in mammals, one can hypothesize that the mechanism evolved first for antigen-dependent reactions and later was recruited in some species to generate the repertoire. However, if adaptive immune responses driven by diverse antigen receptors arose in evolution before the advent of recombination activating gene-driven rearrangement mechanisms, mutation may well have been the generator of diversity. If the latter scenario is true, then mutation to generate the repertoire was lost in most species, perhaps because germ-line V sequences have evolved to interact with commonly encountered pathogens or for structural flexibility (44), explaining the persistence of some Vh families over hundreds of millions of years (45).
Acknowledgments
We are grateful to Dr. Michael Neuberger for providing mouse passenger transgene Ig sequences, Dr. Austin Hughes for helpful suggestions on Pn and Ps analysis, and Dr. Robert Blanden for comments on the manuscript. This work was supported by National Institutes of Health Grant RR06603. M.D. is a Burroughs Wellcome Fund Fellow of The Life Sciences Research Foundation.
ABBREVIATIONS
- NAR
new or nurse shark antigen receptor
- CDR
complementarity-determining region
- FR
framework region
- GC
germinal center
- Sec
secretory
- Tm
transmembrane
- Ps
per synonymous
- Pn
per nonsynonymous
Footnotes
References
- 1.Hinds-Frey K R, Nishikata H, Litman R T, Litman G W. J Exp Med. 1993;178:815–824. doi: 10.1084/jem.178.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenberg A S, Avila D, Hughes M, Hughes A, McKinney E C, Flajnik M F. Nature (London) 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 3.Diaz M, Flajnik M F. Immunol Rev. 1998;162:13–24. doi: 10.1111/j.1600-065x.1998.tb01425.x. [DOI] [PubMed] [Google Scholar]
- 4.Tsiagbe V K, Inghirami G, Thorbecke G J. Crit Rev Immunol. 1996;16:381–421. [PubMed] [Google Scholar]
- 5.Reynaud C-A, Garcia C, Hein W R, Weill J-C. Cell. 1995;80:115–125. doi: 10.1016/0092-8674(95)90456-5. [DOI] [PubMed] [Google Scholar]
- 6.Lopez O, Perez C, Wylie D. Immunol Rev. 1998;162:55–66. doi: 10.1111/j.1600-065x.1998.tb01429.x. [DOI] [PubMed] [Google Scholar]
- 7.Reynaud C-A, Dahan A, Anquez V, Weill J-C. Cell. 1989;59:171–183. doi: 10.1016/0092-8674(89)90879-9. [DOI] [PubMed] [Google Scholar]
- 8.Reynaud C-A, Anquez V, Dahan A, Weill J-C. Cell. 1985;40:283–291. doi: 10.1016/0092-8674(85)90142-4. [DOI] [PubMed] [Google Scholar]
- 9.Knight K, Crane M A. Adv Immunol. 1994;56:178–218. doi: 10.1016/s0065-2776(08)60452-6. [DOI] [PubMed] [Google Scholar]
- 10.Sitnikova T, Su C. Mol Biol Evol. 1998;15:617–625. doi: 10.1093/oxfordjournals.molbev.a025965. [DOI] [PubMed] [Google Scholar]
- 11.Wilson M, Hsu E, Marcuz A, Courtet M, Du Pasquier L, Steinberg C. EMBO J. 1992;11:4337–4347. doi: 10.1002/j.1460-2075.1992.tb05533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg A S. Ph.D thesis. Coral Gables, FL: University of Miami; 1994. [Google Scholar]
- 13.Clem L W, Leslie G A. Proc Natl Acad Sci USA. 1971;68:139–143. doi: 10.1073/pnas.68.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mäkelä O, Litman G W. Nature (London) 1980;287:639–640. doi: 10.1038/287639a0. [DOI] [PubMed] [Google Scholar]
- 15.Petty C L, McKinney E C. Eur J Immunol. 1983;13:133–138. doi: 10.1002/eji.1830130208. [DOI] [PubMed] [Google Scholar]
- 16.Siekevitz M, Kocks C, Rajewsky K, Dildrop P. Cell. 1987;48:757–770. doi: 10.1016/0092-8674(87)90073-0. [DOI] [PubMed] [Google Scholar]
- 17.Sambrock J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Kumar S, Tamura K, Nei M. mega: Molecular Evolutionary Genetics Analysis, Version 1.01. University Park: The Pennsylvania State University; 1993. [Google Scholar]
- 19.Sokal R P, Rohlf F J. Biometry. 2nd Ed. New York: Freeman; 1981. pp. 780–810. [Google Scholar]
- 20.Sharpe M J, Milstein C, Jarvis J M, Neuberger M S. EMBO J. 1991;10:2139–2145. doi: 10.1002/j.1460-2075.1991.tb07748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betz A G, Rada C, Pannell R, Milstein C, Neuberger M S. Proc Natl Acad Sci USA. 1993;90:2385–2388. doi: 10.1073/pnas.90.6.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betz A G, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger M S. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 23.Hsu E. Int Immunol. 1996;8:847–854. doi: 10.1093/intimm/8.6.847. [DOI] [PubMed] [Google Scholar]
- 24.Betz A G, Neuberger M S, Milstein C. Immunol Today. 1993;14:405–411. doi: 10.1016/0167-5699(93)90144-a. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Fernandez A, Gupta S K, Pannell R, Neuberger M S, Milstein C. Proc Natl Acad Sci USA. 1994;9:12614–12618. doi: 10.1073/pnas.91.26.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yelamos J, Klix N, Goyenechea B, Lozano F, Chui Y L, Gonzalez-Fernadez A, Pannell R, Neuberger M, Milstein C. Nature (London) 1995;376:225–229. doi: 10.1038/376225a0. [DOI] [PubMed] [Google Scholar]
- 27.Chang B, Casali P. Immunol Today. 1994;15:367–373. doi: 10.1016/0167-5699(94)90175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacob J, Kelsoe G, Rajewsky K, Weiss U. Nature (London) 1991;354:389–392. doi: 10.1038/354389a0. [DOI] [PubMed] [Google Scholar]
- 29.Garcia K C, Rejano M, Pease L R, Huang M, Peterson P A, Teyton L, Wilson I A. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 30.Warr G W. Dev Comp Immunol. 1995;19:1–12. doi: 10.1016/0145-305x(94)00052-h. [DOI] [PubMed] [Google Scholar]
- 31.Du Pasquier L, Wilson M, Greenberg A S, Flajnik M F. In: Somatic Diversification of Immune Responses. Kelsoe G, Flajnik M F, editors. Berlin: Springer; 1998. pp. 199–216. [Google Scholar]
- 32.Klein U, Kuppers R, Rajewsky K. Eur J Immunol. 1993;23:3272–3277. doi: 10.1002/eji.1830231232. [DOI] [PubMed] [Google Scholar]
- 33.Arakawa H, Kuma K, Yasuda M, Furusawa S, Ekino S, Yamagishi H. J Immunol. 1998;160:4232–4241. [PubMed] [Google Scholar]
- 34.Mage R S. Immunol Rev. 1998;162:49–54. doi: 10.1111/j.1600-065x.1998.tb01428.x. [DOI] [PubMed] [Google Scholar]
- 35.Roux K H, Greenberg A S, Greene L, Strelets L, Avila D, McKinney E C, Flajnik M F. Proc Natl Acad Sci USA. 1998;95:11804–11809. doi: 10.1073/pnas.95.20.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du Pasquier L. Nature (London) 1982;296:311–313. doi: 10.1038/296311a0. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka T, Nei M. Mol Biol Evol. 1989;6:447–459. doi: 10.1093/oxfordjournals.molbev.a040569. [DOI] [PubMed] [Google Scholar]
- 38.Lebecque S G, Gearhart P J. J Exp Med. 1990;172:1717–1727. doi: 10.1084/jem.172.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winter D, Phung Q H, Umar A, Baker S M, Tarone R E, Tanaka K, Liskay R M, Kunkel T A, Bohr V A, Gearhart P J. Proc Natl Acad Sci USA. 1998;95:6953–6958. doi: 10.1073/pnas.95.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phung Q H, Winter D B, Craston A, Tarone E R, Bohr V A, Fishel R, Gearhart P J. J Exp Med. 1998;187:1745–1751. doi: 10.1084/jem.187.11.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cascalho M, Wong J, Steinberg C, Wabl M. Science. 1998;279:1207–1210. doi: 10.1126/science.279.5354.1207. [DOI] [PubMed] [Google Scholar]
- 42.Hinds K R, Litman G W. Nature (London) 1986;320:546–549. doi: 10.1038/320546a0. [DOI] [PubMed] [Google Scholar]
- 43.Shamblott M J, Litman G W. EMBO J. 1989;8:3733–3739. doi: 10.1002/j.1460-2075.1989.tb08549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wedemayer G J, Patten P A, Wang L H, Schultz P G, Stevens R C. Science. 1997;276:1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- 45.Ota T, Nei M. Mol Biol Evol. 1994;11:469–482. doi: 10.1093/oxfordjournals.molbev.a040127. [DOI] [PubMed] [Google Scholar]