Abstract
The mature T cell receptor (TCR) repertoire is shaped by positive- and negative-selection events taking place during T cell development. These events are regulated by interactions between the TCR and major histocompatibility complex molecules presenting self-peptides. It has been shown that many antagonist peptides are efficient at mediating positive selection. In this study we analyzed the effects of a transgene encoding an antagonist peptide (influenza NP34) that is presented by H-2Db in a Tap-1-independent fashion in mice expressing the influenza NP68-specific TCR F5. We find that the transgenic peptide does not mediate positive or negative selection in F5+Tap-1−/− mice, but inhibits maturation of CD8+ single positive thymocytes in F5+Tap-1+ mice without inducing signs of negative selection. We conclude that antagonism of antigen recognition occurs not only at the level of mature T cells but also in T cell development.
Keywords: thymus/positive selection
Thymocytes are subjected to positive- and negative-selection events during their development (1). Positive selection involves the differentiation of immature thymocytes with a T cell receptor (TCR) specificity restricted to self-major histocompatibility complex (MHC) into mature, functional T cells. Negative selection is the elimination of thymocytes with a TCR specificity that would result in self-reactivity in mature T cells. Both selection events are mediated by interactions between the TCR and self-peptide/MHC ligands (2–4). Negative selection occurs when the interaction between the TCRs and peptide/MHC ligands is strong, and positive selection takes place when the interaction is weak (5, 6). The strength of the interaction is influenced both by the affinity of the TCR for peptide/MHC ligand and the abundance of the ligand and, thus, by the avidity of the peptide/MHC–TCR interactions.
Several recent studies have focused on the relationships between the affinity of peptide/MHC–TCR interactions, their kinetics, and the effect on the signals transduced by the TCR (7–10). These studies have shown that the “on rate” of the ligand–receptor interaction is relatively constant, while the “off rate” varies widely depending on the specific combination of the ligand and the receptor. Thus, the affinity of the peptide/MHC–TCR interaction is determined primarily by the dissociation constant; a low-affinity interaction is caused primarily by a high dissociation rate, and a high-affinity interaction is caused by a low dissociation rate.
It was observed that peptide/MHC–TCR interactions with low dissociation rates often are efficient in mediating T cell activation, while interactions with high dissociation rates can antagonize T cell activation. Peptide/MHC–TCR interactions with a dissociation rate above a certain threshold have no influence on T cell activation. It is not entirely clear how short-lived peptide/MHC–TCR interactions antagonize the effects of long-lived interactions. It is possible that short-lived peptide/MHC–TCR interactions perturb the formation of stable TCR aggregates and/or mediate incomplete signaling events, causing depletion of downstream signaling molecules (7, 11, 12).
Many antagonistic peptides and peptides with antagonistic/partial agonistic properties seem to be efficient at inducing positive selection of double positive (DP) thymocytes (5), although there is no strict correlation between antagonism and positive selection (8, 13, 14). However, peptide/MHC–TCR interactions with dissociation rates above a certain threshold are incapable of mediating positive selection, while those with low dissociation rates tend to result in the production of nonfunctional single positive (SP) thymocytes or negative selection (8, 15, 16).
To date, the effects of low-affinity peptide/MHC–TCR interactions on positive and negative selection have been studied mostly in vitro. In this study we used mice transgenic for an influenza NP68 (nucleoprotein 366–374)-specific TCR (F5) (17). To analyze the effect of an antagonistic peptide on T cell development in vivo, we introduced a transgene encoding the antagonistic variant peptide NP34 fused to the adenovirus E19/3K signal sequence. The peptide was transported to the endoplasmic reticulum independently of Tap-1 and presented by H-2Db. We analyzed the effects of the transgene in mice with F5+Rag-1−/−Tap-1−/− and F5+Rag-1−/−Tap-1+ backgrounds. The peptide had no effect on T cell development in F5+Rag-1−/−Tap-1−/− mice, but inhibited the development of CD8+ T cells in F5+Rag-1−/−Tap-1+ mice without signs of negative selection.
MATERIALS AND METHODS
Construction of pC-NP34 Transgene and Generation of Transgenic Mice.
To generate mice that expressed the NP34 peptide independently of Tap-1, transgenes were constructed that encoded the adenovirus E3/19K signal sequence fused to the NP34 peptide, under the control of the chicken β-actin promoter and a cytomegalovirus-enhancer region. The construct was based on pCAGGS (a kind gift of J. Miyazaki, University of Tokyo, Tokyo). Two primers were synthesized encoding the 5′ and 3′ ends of the minigene (sense: CAT GCA TGG AAT TCA TAG GAT CCC CAC CAT GAG GTA CAT GAT TTT AGG CTT GCT GGC CCT TGC GGC AGT CTG CAG C; NP34 antisense: AGT TCT CAA GAA TTC TGA TCA CAT TGT TTC CAT GTT TTC GTT GCT TGC TGC CGC GCT GCA GAC TGC CGC AAG GGC) that contained a 21-nt overlap, thus serving as each other’s template for PCR. PCR was performed using the pfu polymerase (Stratagene). The product was digested with EcoRI, gel-purified, and ligated into the cloning site of pCAGGS. An additional alanine was introduced in the minigene between the E3/19K signal sequence and the peptide, since it is unknown where the signal peptide is cleaved off exactly. As N-terminal cleavage of additional amino acids can take place in the endoplasmic reticulum, the correct peptide would be produced if the peptide was cleaved at the N-terminal side of the extra alanine (18, 19). If the cleavage took place more to the C-terminal side than expected, the extra alanine avoids the production of an octameric peptide.
The minigene was excised by SalI and PstI digestion, gel-purified, and microinjected into fertilized F5+Tap-1−/−Rag-1+/− eggs obtained from Tap-1−/− females mated with F5+Tap-1−/−Rag-1−/− males. After overnight incubation, the eggs that matured into the two-cell stage were transferred to day 0.5 postcoitum pseudopregnant BDF1 females. Three weeks after birth, genomic DNA was prepared from tail tissue, and the presence of the transgene was analyzed by Southern blotting using the 0.9-kb XbaI-PstI fragment from the vector used for generating the transgenic mice
Mice and PCR Typing.
Tap-1−/− (20) and Rag-1−/− (21) mice were produced in this laboratory. F5 TCR transgenic mice (17) were kindly provided by D. Kioussis (The National Institute of Medical Research, London). Rag-1−/−H-2u mice were created as described (22). They were bred and maintained in a pathogen-free environment at the Massachusetts Institute of Technology. Three weeks after birth genomic DNA was prepared from tail tissue, and the presence of the F5 TCR, Tap-1, Rag-1, and NP34 transgene was analyzed by PCR. The primers used were F5: 5′-ACA AAT GCT GGT GTC ATC CA, 3′-GGG TGG AGT CAC ATT TCT CA; Tap-1: 5′-AGG CTC AGC GTG CCA CTA AT, 3′-ATT GAA GTT CCT GCG CCT CC; Rag-1: 5′-GAT CGA GCT GAA GGC AGA TG, 3′-GTC TCT TCC TCT TGA GTC CC; and Neo: 5′-CTT GGG TGG AGA GGC TAT TC, 3′-AGG TGA GAT GAC AGG AGG TC. Mice were analyzed between 8 and 10 weeks after birth.
Northern Blot Analysis.
Mice were anesthetized by using avertin and perfused by using PBS/1 mM EDTA. Organs were removed and placed on ice immediately. Whole RNA was prepared by using Tri-Reagent (Molecular Research Center, Cincinnati). Fifteen micrograms of RNA per lane was electrophoresed in a 1% agarose formaldehyde denaturing gel and transferred to Zeta-Probe GT membranes (Bio-Rad). Filters were hybridized with a radiolabeled 0.7-kb EcoRI-PstI fragment from the vector used for generating the transgenic mice, and washed blots were assessed by autoradiography.
Cytotoxicity Assay.
B6 mice were immunized i.p. with 100 μg NP-34 peptide in incomplete Freund’s adjuvant. After 9 days, the spleens were removed and single cell suspensions were prepared and stimulated in vitro with irradiated Tap-1−/− splenocytes loaded with 100 ng/ml NP-34. After 1 week, the cytotoxic T cells were restimulated in the presence of 15 units/ml interleukin 2. Five days later, the cytotoxic T cells were used in a chromium release assay as described (23). Con A-induced splenic lymphoblasts from Tap-1−/− or Tap-1−/−NP34+ mice were used as targets and prepared as described previously (23).
Flow Cytometry.
Three-color stainings were performed using fluorescein isothiocyanate or phycoerythrin-labeled anti-CD4, anti-Vβ11, or anti-CD69 antibodies (PharMingen) and Tri-Color-labeled anti-CD8 antibodies (Caltag, South San Francisco, CA). The cells were analyzed on a FACScan (Becton Dickinson) using cellquest SOFTWARE (BECTON DICKINSON).
TUNEL Assay.
Thymuses were embedded in OCT compound (Miles) on dry ice. Four-micrometer sections were prepared and stored at −80°C. The TUNEL assay was performed as described (24). In short, sections were fixed in formalin followed by ethanol/acetic acid (2:1). Endogenous peroxidase activity was blocked with 0.5% H2O2. Washed sections were incubated for 2 hr at 37°C with biotinylated dUTP (Boehringer Mannheim) and TdT (Promega). After the reaction was blocked, the sections were washed, incubated with avidin-biotinylated peroxidase (Dako), washed again, and developed with 3-amino-9-ethylcarbazole (Aldrich). Twenty different fields were counted per thymus (×40 objective).
RESULTS
Transgenic Mice.
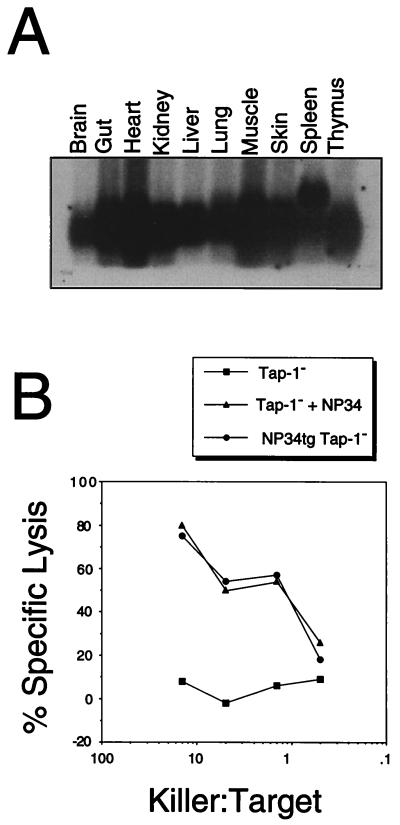
Two lines of mice carrying the pC-NP34 construct were obtained. Line 45 had two copies of the transgene, while line 61 had one copy. Northern blot analysis revealed that the peptide-encoding transgene was not expressed in transgenic line 61. The expression pattern of the transgene in different tissues of line 45 is shown in Fig. 1A. Total RNA was obtained from a Rag-1−/−Tap-1−/− mouse that had been perfused before sacrificing it to avoid the contribution of blood cells. The additional signal from a larger RNA in spleen probably was derived from the endogenous β-globin gene expressed in reticulocytes, probably a major source of total spleen RNA in Rag-1−/− mice. The transgene was expressed highest in heart, gut, and muscle and lowest in the brain. The level of expression in the thymus was intermediate compared with that in other tissues. We did not directly test the possibility that the signal obtained from thymus was derived from lymphoid cells rather than from thymic epithelium. However, the promoter used was a ubiquitous promoter and mediated high expression of the transgene in epithelial tissues such as skin, lung, and intestine. Therefore, it is likely that the transgene was expressed in thymic epithelium.
Figure 1.
(A) Northern blot analysis of NP34 transgene expression in various organs of a Rag-1−/−Tap-1−/−NP34+ mouse. (B) NP34 is presented in a Tap-1-independent manner. Con A blasts from Tap-1−/−NP34+ mice (•) were assayed for lysis by NP34-specific cytotoxic T cells. As negative and positive controls, Con A blasts from Tap-1−/− mice incubated with (▴) or without (■) 1 μg/ml synthetic NP34 peptide were used.
Presentation of the NP34 Peptide by MHC Class I.
Although Northern blot analysis showed ubiquitous expression of the NP34 transgene, we had to ascertain that the NP34 peptide actually was translated and transported to the endoplasmic reticulum in a Tap-1-independent manner and presented by H-2Db. To verify this, cytotoxic T cells recognizing the peptide were isolated from B6 mice immunized with the NP34 peptide and restimulated once in vitro. Subsequently, these cells were assayed for their ability to specifically lyse Con A-induced splenocyte blasts from Tap-1−/−NP34 transgenic mice, Con A blasts from Tap-1−/− mice, (negative control), or Con A blasts from Tap-1−/− mice incubated with 1 μg/ml NP34 peptide (positive control). As shown in Fig. 1B, NP34 transgenic cells were specifically lysed by the cytotoxic T cells in contrast to Tap-1−/− cells. This result confirmed that the NP34 peptide was presented by H-2Db on the surface of cells from NP34 transgenic mice in a Tap-1-independent manner.
Effect of NP34 Peptide on Selection of F5+ Thymocytes.
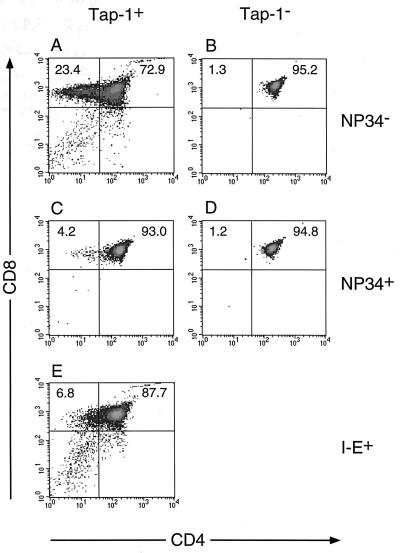
It has been observed that some antagonist peptides can mediate positive selection. Therefore, we examined whether F5+ thymocytes were selected in F5+Rag-1−/−Tap-1−/− mice by the transgenic NP34 peptide. We isolated thymocytes and cells from the spleen and lymph nodes of F5+Rag-1−/−Tap-1−/− and F5+Rag-1−/−Tap-1−/−NP34+ mice, stained them with antibodies against CD4, CD8, Vβ11, and CD69, and analyzed them by fluorescence-activated cell sorter (FACS). The presence of the NP34 transgene in F5+Rag-1−Tap-1−/− mice had no effect on the development of F5+ T cells. There was no change in the number of DP or CD8+ SP thymocytes (Fig. 2 B and D), and TCR expression was not up-regulated (Fig. 3 B and D). The percentage of CD69+ cells was unchanged (Fig. 4A). No peripheral CD8+ T cells were observed (not shown). These results contrasted significantly with results obtained from F5+Rag-1+Tap-1−/− mice expressing a transgene encoding the nominal peptide (NP68) under the control of the same promoter and enhancer sequences. In such mice, extensive negative selection occurred as could be concluded from the low number of thymocytes (2.6 ± 1.1 × 107, average of seven mice from four independent lines) and the increased percentage of CD69+ thymocytes (26.9 ± 1.2%) (data not shown). We observed a 6.5-fold increase of the percentage of DN thymocytes in these mice when compared with F5+Tap−/−Rag-1+ mice, indicating that negative selection took place predominantly during the early DP stage.
Figure 2.
Effects of the NP34 transgene on the development of CD8+ SP thymocytes. Cells from positively selecting F5+Rag-1−/−Tap-1+ (A), nonselecting F5+Rag-1−/−Tap-1−/− (B), and negatively selecting F5+Rag-1−/−Tap-1+I-E+ (E) thymuses were compared with cells from F5+Rag-1−/−Tap-1+NP34+ (C) and F5+Rag-1−/−Tap-1−/−NP34+ (D) thymuses. Cells were stained with anti-CD4 fluorescein isothiocyanate and anti-CD8 Tri-Color antibodies and analyzed by FACS. Density plots represent fluorescence intensities of CD4 and CD8. Indicated are mean percentages of CD8+ SP and CD4+CD8+ DP thymocytes from three to eight mice of each genotype. Density plots shown are from thymuses containing a similar percentage (less than 0.5% difference) of CD8+ SP and CD4+8+ thymocytes as the mean value.
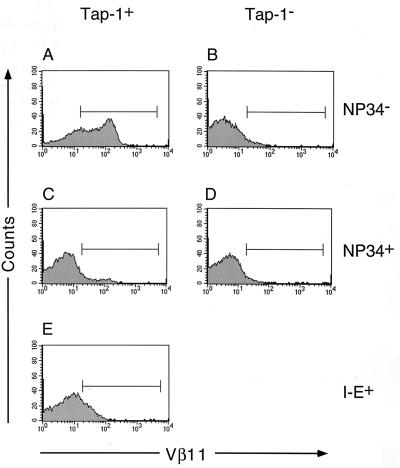
Figure 3.
Effects of the NP34 transgene on the expression of the F5 TCR on thymocytes. Cells from positively selecting F5+Rag-1−/−Tap-1+ (A), nonselecting F5+Rag-1−/−Tap-1−/− (B), and negatively selecting 5+Rag-1−/−Tap-1+I-E+ (E) thymuses were compared with cells from F5+Rag-1−/−Tap-1+NP34+ (C) and F5+Rag-1−/−Tap-1−/−NP34+ (D) thymuses. Cells were stained with anti-Vβ11 phycoerythrin antibodies and analyzed by FACS. Histograms represent the fluorescence intensity of Vβ11.
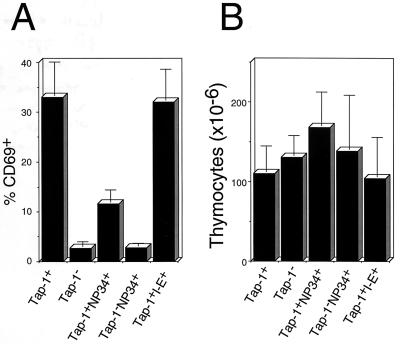
Figure 4.
(A) The NP34 transgene reduces the percentage of CD69+ thymocytes. Cells were isolated from F5+Rag-1−/−Tap-1+NP34+, F5+Rag-1−/−Tap-1−/−NP34+, positively selecting F5+Rag-1−/−Tap-1+, nonselecting F5+Rag-1−/−Tap-1−/−, and negatively selecting F5+Rag-1−/−Tap-1+I-E+ thymuses. Cells were stained with anti-CD69 phycoerythrin antibodies and analyzed by FACS. Bars represent the mean percentage of CD69+ thymocytes from three to eight mice of each genotype. SDs are indicated. (B) The NP34 transgene has no influence on the absolute numbers of thymocytes. Cells from F5+Rag-1−/−Tap-1+NP34+, F5+Rag-1−/−Tap-1−/−NP34+, positively selecting F5+Rag-1−/−Tap-1+, nonselecting F5+Rag-1−/−Tap-1−/−, and negatively selecting F5+Rag-1−/−Tap-1+I-E+ thymuses were isolated and counted. Bars represent the mean thymocyte numbers from three to eight mice of each genotype. SDs are indicated.
We then tested whether the NP34 transgene had any effect on the development of T cells in F5+Rag-1−/−Tap-1+ mice. As shown in Figs. 2A and 3A, F5+CD8+ thymocytes developed in F5+Rag-1−/−Tap-1+ mice, and 32% of all thymocytes expressed CD69 (Fig. 4A). Surprisingly, the presence of the NP34 transgene inhibited the development of CD8+ SP thymocytes (Fig. 2C) and peripheral CD8+ T cells (Table 1). Expression of CD69 was observed on only 11% of all thymocytes (Fig. 4A). The absolute number of thymocytes was not changed significantly (Fig. 4B) by the presence of the NP34 peptide. We did not observe up-regulation of the TCR on DP thymocytes of F5+Rag-1−/−Tap-1+NP34+ mice (Fig. 3 A and C). This does not seem to reflect down-regulation of TCR expression caused by the presence of NP34, because thymocytes that did mature to the CD8+ SP stage expressed the TCR at a level 90% of that found on thymocytes from F5+Rag-1−/−Tap-1+ mice.
Table 1.
Effects of NP34 or I-E on the number of F5+CD8+ SP T cells
| Genotype | Spleen | Lymph nodes |
|---|---|---|
| F5+Rag-1−/−Tap-1+ | 7.5 ± 2.8 × 106 | 3.6 ± 1.8 × 106 |
| F5+Rag-1−/−Tap-1+NP34+ | 1.4 ± 0.37 × 106 | 0.59 ± 0.47 × 106 |
| F5+Rag-1−/−Tap-1+I-E+ | 0.9 ± 0.45 × 106 | 0.14 ± 0.095 × 106 |
The NP34 transgene reduces the absolute numbers of CD8+ splenocytes and lymph node cells. Cells from F5+Rag-1−/−Tap-1+NP34+, positively selecting F5+Rag-1−/−Tap-1+, and negatively selecting F5+Rag-1−/−Tap-1+I-E+ lymphoid organs were isolated and counted. Cells were stained with anti-CD8 antibodies and analyzed by FACS. The absolute numbers of CD8+ T cells were calculated by multiplying the total cell number per organ with the percentage of CD8+ cells. Mean numbers of CD8+ T cells from six to eight mice of each genotype are shown. SDs are indicated.
It has been described that superantigens can mediate negative selection of thymocytes at a late stage of their development, just before the thymocytes become single positive (25). To test the possibility that the NP34 transgene mediated a similar form of negative selection, we compared the phenotype of F5+Rag-1−/−Tap-1+NP34+ mice with that of mice expressing a superantigen for the Vβ11 chain of the F5 TCR. For this, we crossed F5+Rag-1−/−Tap-1+ mice to Rag-1−/−H-2u/u mice to obtain F5+Rag-1−/−Tap-1+H-2b/u mice. These mice express the I-E allele, which can present the mtv8/9 superantigens that delete Vβ11+ T cells (26). We did not directly assess the presence of the mtv superantigens in these mice, but because negative selection and strong TCR down-regulation on residual peripheral T cells occurred, we are confident that the superantigens were present. As shown in Fig. 2E and Table 1, the numbers of CD8+ thymocytes and T cells indeed were decreased. However, the absolute number of thymocytes was similar to that in control mice (Fig. 4B), indicating that negative selection took place at a late stage of T cell development. In contrast to F5+Rag-1−/−Tap-1+NP34+ mice, there was no decrease in the percentage of CD69+ thymocytes (Fig. 3B). In these mice, the low levels of TCR expression on CD4+CD8+ thymocytes likely is due to down-modulation upon interaction with the superantigen, because expression of the TCR on peripheral T cells was barely detectable (not shown).
Presence of Apoptotic Cells.
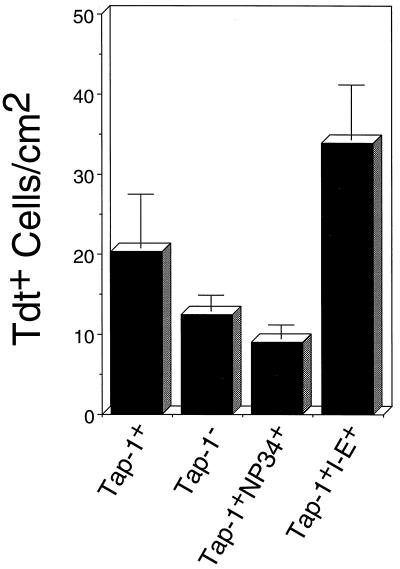
The FACS data showed that the phenotype of F5+Rag-1−/−Tap-1+NP34+ mice resembled the phenotype of nonselecting (F5+Rag-1−/−Tap-1−/−) mice more than that of negatively selecting (F5+Rag-1−/−Tap-1+H-2b/u) mice. To address the issue of apoptosis, we stained fresh frozen sections of thymuses from positively selecting (F5+Rag-1−/−Tap-1+), negatively selecting, nonselecting, and F5+Rag-1−/−Tap-1+ pC-NP34+ mice with the TUNEL method and counted the number of apoptotic (TdT+) cells. In Fig. 5, the number of apoptotic cells per square cm as counted with a ×40 objective is shown for each type of mice. Thymuses of negatively selecting mice contained more apoptotic cells than thymuses of positively selecting mice. Nonselecting mice showed decreased numbers of apoptotic thymocytes. The number of apoptotic cells in F5+Rag-1−/−Tap-1+NP34+ thymuses was comparable to that in nonselecting thymuses. It has been reported that apoptosis caused by superantigen-mediated negative selection occurs in the medulla, in contrast to apoptosis by neglect, which takes place in the cortex (27). However, we found that in all our mice, apoptosis occurred mainly in the cortex. We do not know the reasons for this apparent discrepancy. Several factors may have played a role, such as the altered thymic architecture caused by the Rag-1 deficiency or differences in the relative expression level or the tissue distribution of the superantigens involved. Despite this uncertainty, we maintain that the negative selection mediated by I-E is late as it is not accompanied by significant reduction of the number of DP thymocytes.
Figure 5.
Reduction of the numbers of apoptotic thymocytes by expression of the NP34 transgene. Sections were prepared from F5+Rag-1−/−Tap-1+NP34+, positively selecting F5+Rag-1−/−Tap-1+, nonselecting F5+Rag-1−/−Tap-1−/−, and negatively selecting F5+Rag-1−/−Tap-1+I-E+ thymuses and stained using the TUNEL assay. Bars represent mean values of the number of apoptotic cells per cm2 as examined with a ×40 objective in thymuses of three to four mice of each genotype. SDs are indicated.
DISCUSSION
In this study, we examined the influence of the antagonistic NP34 peptide on positive and negative selection of thymocytes carrying the NP68-specific TCR, F5. For this, we created mice that present the NP34 peptide on H-2Db in a Tap-1−/−-independent manner and bred them to F5+Rag-1−/−Tap-1−/− or F5+Rag-1−/−Tap-1+ background. We found that in F5+Rag-1−/−Tap-1−/− mice, the NP34 transgene did not induce positive or negative selection: there was no up-regulation of CD69 or TCR on CD4+8+ DP thymocytes (28). No CD8+ SP thymocytes or mature T cells developed in these mice. F5+ mice expressing a transgene encoding the nominal NP68 peptide showed a strong reduction of the number thymocytes, indicating that negative selection occurred. Surprisingly, in F5+Rag-1−/−Tap-1+ mice carrying the NP34 transgene, the development of CD8+ SP thymocytes was inhibited. The number of peripheral T cells was reduced accordingly. However, the thymuses of F5+Rag-1−/−Tap-1+NP34+ mice showed no obvious signs of negative selection such as reduced numbers of DP thymocytes or down-regulation of the coreceptors CD4 and CD8. This contrasted with the situation in F5+Tap-1−/− mice expressing the nominal NP68 peptide, in which the thymuses showed a 4- to 5-fold reduction in cellularity. Because negative selection mediated by superantigens was shown previously to occur without a reduction of the number of DP cells (25), it was possible that the NP34 transgene mediated a similar form of negative selection (29). Alternatively, the presence of the NP34 peptide could have interfered with positive selection of F5+ thymocytes.
To distinguish between these possibilities, we compared the thymuses of F5+Rag-1−/−Tap-1+NP34+ mice with thymuses from mice in which no positive or negative selection takes place (F5+Rag-1−/−Tap-1−/− mice) or in which negative selection takes place (F5+Rag-1−/−Tap-1+I-Eu+ mice). In F5+Rag-1−/−Tap-1+I-Eu+ mice, negative selection of F5+ cells is mediated by mtv superantigens presented by I-E (17). We compared numbers of apoptotic cells in F5+Rag-1−/−Tap-1+NP34+ mice with those in negatively selecting F5+Rag-1−/−Tap-1+I-E+ mice. We found more apoptotic thymocytes in F5+Rag-1−/−Tap-1+I-E+ mice than in F5+Rag-1−/−Tap-1+NP34+ mice. The numbers of apoptotic cells in the latter mice were comparable to F5+Rag-1−/−Tap-1−/− mice in which no selection occurs. This finding is in line with the observation that injection of 1 mg NP34 peptide does not induce apoptosis in F5+Rag-1−/−Tap-1+ mice (30). Furthermore, we observed that the percentage of thymocytes expressing CD69, an early activation marker that is up-regulated upon positive and negative selection events (28, 31, 32), is lower in F5+Rag-1−/−Tap-1+NP34+ mice than in F5+Rag-1−/−Tap-1+I-E+ mice. Thus, if negative selection is the cause of the low number of mature CD8+ T cells in F5+Rag-1−/−Tap-1+NP34+ mice, this form of negative selection must be accompanied by an efficient mechanism to dispose of cells showing the first signs of programmed cell death. This idea is hard to reconcile with the lack of reduced cell numbers in F5+Rag-1−/−Tap-1+NP34+ thymuses. Therefore, we favor the hypothesis that positive selection is inhibited by the NP34 transgene. This idea accounts for our observations that early events associated with positive selection, such as up-regulation of the TCR and CD69, are reduced in F5+Rag-1−/−Tap-1+NP34+ mice as compared with F5+Rag-1−/−Tap-1+ mice, and that the number of apoptotic cells in these mice is comparable to that in nonselecting thymuses.
How could the presence of the NP34 peptide inhibit positive selection? The NP34 peptide has been described to antagonize target recognition by mature F5+ cytotoxic T cells and inhibit negative selection of F5+ thymocytes mediated by the NP68 peptide (33). Therefore, it is not unlikely that the NP34 peptide is able to antagonize positive selection events as well. Data supporting such an idea have been obtained previously from experiments employing fetal thymic organ cultures from transgenic mice carrying a class II-restricted TCR recognizing the moth cytochrome c peptide (34). Although apoptosis was not assessed directly, the data did support the notion that negative selection was not responsible for the absence of the CD4+ thymocytes. The data presented here indicate that antagonism of positive selection is not limited to thymocytes carrying CD4-restricted TCRs or to in vitro culture conditions.
In view of the results showing that some antagonist peptides are efficient in mediating positive selection, it seems somewhat surprising that an antagonist peptide is capable of antagonizing positive selection. However, it has been shown that the interaction of antagonistic peptide/MHC ligands with the TCR-CD3 complex can result in partial phosphorylation of CD3ζ. This partially phosphorylated form of CD3ζ is incapable of recruiting the protein-tyrosine kinase zap-70 (35, 36). Furthermore, it has been shown that lck does not become activated by engagement of the TCR with antagonistic peptide/MHC ligands (37). As lck (38, 39) and zap-70 (40) are both important for positive selection, such processes may explain the inhibition of positive selection by an antagonist peptide.
Taken together, these results implicate that the signal mediating positive selection must be different from the signal resulting in antagonism. This does not necessarily contradict the finding that some antagonistic peptides are able to mediate positive selection. In fact, we do observe that the NP34 peptide can mediate positive selection of F5+ thymocytes in FTOC at relatively high concentrations (>10 μM) (C.N.L. and S.T., unpublished results). As positive selection was not induced in F5+Rag-1−/−Tap-1−/−NP34+ mice, it is likely that the concentration of peptide needed for mediating positive selection is higher than that resulting in antagonism of positive selection. This parallels the situation in mature T cells, to which certain peptides act as antagonists at lower concentrations, and as weak agonists at high concentrations (41).
How is it possible that certain peptides have no agonistic effects on mature T cells whatsoever, but induce the activation events associated with positive selection of thymocytes (5, 8, 42)? We suggest that this is because of differences between accessory molecules expressed by immature and mature T cells. The coreceptors CD4 and CD8 stabilize already formed TCR/ligand interactions (43) probably by serial engagement and induction of conformational changes that stabilize the TCR/ligand complex (44). This may not be achieved if the TCR/ligand interaction is too short-lived (45). It is known that increased expression levels of coreceptors greatly augment the sensitivity of T cells toward low-affinity ligands (13, 45, 46). Therefore, it is possible that coreceptor engagement is accelerated with increased coreceptor to TCR ratios, thus shifting the threshold for activation events toward peptide/MHC ligands with higher off-rates. Thymocytes express higher levels of CD8 and lower levels of TCR than mature T cells. We assume that this is the reason why thymocytes may recognize ligands that cannot be recognized by mature T cells.
Acknowledgments
The authors thank Drs. W. Haas and A. Coutinho for the critical reading of the manuscript and Dr. J. Miyazaki for the gift of plasmid pCAGGS. C.N.L. received a postdoctoral fellowship from the Cancer Research Institute. This research was supported by National Institutes of Health Grant CA53874 and a gift from the Chiba Prefectural Government of Japan to S.T. and by National Institutes of Health Grants DK43551 and DK47677 to A.K.B.
ABBREVIATIONS
- SP
single positive
- DP
double positive
- TCR
T cell receptor
- FACS
fluorescence-activated cell sorter
- MHC
major histocompatibility complex
References
- 1.Kisielow P, Teh H S, Bluthmann H, von Boehmer H. Nature (London) 1988;335:730–733. doi: 10.1038/335730a0. [DOI] [PubMed] [Google Scholar]
- 2.Ashton-Rickardt P, van Kaer L, Schumacher T N, Ploegh H L, Tonegawa S. Cell. 1993;73:1041–1049. doi: 10.1016/0092-8674(93)90281-t. [DOI] [PubMed] [Google Scholar]
- 3.Hogquist K A, Gavin M A, Bevan M J. J Exp Med. 1993;177:1469–1473. doi: 10.1084/jem.177.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sebzda E, Wallace V A, Mayer J, Yeung R S, Mak T W, Ohashi P S. Science. 1994;263:1615–1618. doi: 10.1126/science.8128249. [DOI] [PubMed] [Google Scholar]
- 5.Hogquist K A, Jameson S C, Heath W R, Howard J L, Bevan M J, Carbone F R. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 6.Ashton-Rickardt P, Bandeira A, Delaney J R, van Kaer L, Pircher H P, Zinkernagel R M, Tonegawa S. Cell. 1994;76:651–663. doi: 10.1016/0092-8674(94)90505-3. [DOI] [PubMed] [Google Scholar]
- 7.Lyons D S, Lieberman S A, Hampl J, Boniface J J, Chien Y, Berg L J, Davis M M. Immunity. 1996;5:53–61. doi: 10.1016/s1074-7613(00)80309-x. [DOI] [PubMed] [Google Scholar]
- 8.Alam S M, Travers P J, Wung J L, Nasholds W, Redpath S, Jameson S C, Gascoigne N R. Nature (London) 1996;381:616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- 9.Matsui K, Boniface J J, Steffner P, Reay P A, Davis M M. Proc Natl Acad Sci USA. 1994;91:12862–12866. doi: 10.1073/pnas.91.26.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corr M, Slanetz A E, Boyd L F, Jelonek M T, Khilko S, al Ramadi B, Kim Y S, Maher S E, Bothwell A L, Margulies D H. Science. 1994;265:946–949. doi: 10.1126/science.8052850. [DOI] [PubMed] [Google Scholar]
- 11.Rabinowitz J D, Beeson C, Lyons D S, Davis M M, McConnell H M. Proc Natl Acad Sci USA. 1996;93:1401–1405. doi: 10.1073/pnas.93.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reich Z, Boniface J J, Lyons D S, Borochov N, Wachtel E J, Davis M M. Nature (London) 1996;387:617–620. doi: 10.1038/42500. [DOI] [PubMed] [Google Scholar]
- 13.Hogquist K A, Tomlinson A J, Kieper W C, McGargill M A, Hart M C, Naylor S, Jameson S C. Immunity. 1997;6:389–399. doi: 10.1016/s1074-7613(00)80282-4. [DOI] [PubMed] [Google Scholar]
- 14.Sebzda E, Kundig T M, Thomson C T, Aoki K, Mak S Y, Mayer J P, Zamborelli T, Nathenson S G, Ohashi P S. J Exp Med. 1996;183:1093–1104. doi: 10.1084/jem.183.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogquist K A, Jameson S C, Bevan M J. Immunity. 1995;3:79–86. doi: 10.1016/1074-7613(95)90160-4. [DOI] [PubMed] [Google Scholar]
- 16.Girao C, Hu Q, Sun J, Ashton-Rickardt P. J Immunol. 1997;159:4205–4211. [PubMed] [Google Scholar]
- 17.Mamalaki C, Elliott J, Norton T, Yannoutsos N, Townsend A R, Chandler P, Simpson E, Kioussis D. Dev Immunol. 1993;3:159–174. doi: 10.1155/1993/98015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snyder H L, Yewdell J W, Bennink J R. J Exp Med. 1994;180:2389–2394. doi: 10.1084/jem.180.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott T, Willis A, Cerundolo V, Townsend A. J Exp Med. 1995;181:1481–1491. doi: 10.1084/jem.181.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Kaer L, Ashton-Rickardt P, Ploegh H L, Tonegawa S. Cell. 1992;71:1205–1214. doi: 10.1016/s0092-8674(05)80068-6. [DOI] [PubMed] [Google Scholar]
- 21.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 22.Lafaille J J, Nagashima K, Katsuki M, Tonegawa S. Cell. 1994;78:399–408. doi: 10.1016/0092-8674(94)90419-7. [DOI] [PubMed] [Google Scholar]
- 23.Wunderlich J, Shearer G. In: Current Protocols in Immunology. Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Stober W, editors. New York: Wiley; 1992. pp. 3.11.14–13.11.17. [Google Scholar]
- 24.Clayton L K, Ghendler Y, Mizoguchi E, Patch R J, Ocain T D, Orth K, Bhan A K, Dixit V M, Reinherz E L. EMBO J. 1997;16:2282–2293. doi: 10.1093/emboj/16.9.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasquez N J, Kaye J, Hedrick S M. J Exp Med. 1992;175:1307–1316. doi: 10.1084/jem.175.5.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gollob K J, Palmer E. Proc Natl Acad Sci USA. 1992;89:5138–5141. doi: 10.1073/pnas.89.11.5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surh C D, Sprent J. Nature (London) 1994;372:100–103. [Google Scholar]
- 28.Kishimoto H, Surh C D, Sprent J. J Exp Med. 1995;181:649–655. doi: 10.1084/jem.181.2.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Page D M, Alexander J, Snoke K, Appella E, Sette A, Hedrick S M, Grey H M. Proc Natl Acad Sci USA. 1994;91:4057–4061. doi: 10.1073/pnas.91.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wack A, Ladyman H M, Williams O, Roderick K, Ritter M A, Kioussis D. Int Immunol. 1996;8:1537–1548. doi: 10.1093/intimm/8.10.1537. [DOI] [PubMed] [Google Scholar]
- 31.Swat W, Dessing M, von Boehmer H, Kisielow P. Eur J Immunol. 1993;23:739–746. doi: 10.1002/eji.1830230326. [DOI] [PubMed] [Google Scholar]
- 32.Brandle D, Muller S, Muller C, Hengartner H, Pircher H. Eur J Immunol. 1994;24:145–151. doi: 10.1002/eji.1830240122. [DOI] [PubMed] [Google Scholar]
- 33.Williams O, Tanaka Y, Bix M, Murdjeva M, Littman D R, Kioussis D. Eur J Immunol. 1996;26:532–538. doi: 10.1002/eji.1830260305. [DOI] [PubMed] [Google Scholar]
- 34.Spain L M, Jorgensen J L, Davis M M, Berg L J. J Immunol. 1994;152:1709–1717. [PubMed] [Google Scholar]
- 35.Sloan L J, Shaw A S, Rothbard J B, Allen P M. Cell. 1994;79:913–922. doi: 10.1016/0092-8674(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 36.Madrenas J, Wange R L, Wang J L, Isakov N, Samelson L E, Germain R N. Science. 1995;267:515–518. doi: 10.1126/science.7824949. [DOI] [PubMed] [Google Scholar]
- 37.Windhagen A, Scholz C, Hollsberg P, Fukaura H, Sette A, Hafler D A. Immunity. 1995;2:373–380. doi: 10.1016/1074-7613(95)90145-0. [DOI] [PubMed] [Google Scholar]
- 38.Hashimoto K, Sohn S J, Levin S D, Tada T, Perlmutter R M, Nakayama T. J Exp Med. 1996;184:931–943. doi: 10.1084/jem.184.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molina T J, Kishihara K, Siderovski D P, van Ewijk W, Narendran A, Timms E, Wakeham A, Paige C J, Hartmann K U, Veillette A, et al. Nature (London) 1992;357:161–164. doi: 10.1038/357161a0. [DOI] [PubMed] [Google Scholar]
- 40.Negishi I, Motoyama N, Nakayama K, Nakayama K, Senju S, Hatakeyama S, Zhang Q, Chan A C, Loh D Y. Nature (London) 1995;376:435–438. doi: 10.1038/376435a0. [DOI] [PubMed] [Google Scholar]
- 41.Evavold B D, Sloan-Lancaster K, Allen P M. Immunol Today. 1993;14:602–609. doi: 10.1016/0167-5699(93)90200-5. [DOI] [PubMed] [Google Scholar]
- 42.Pawlowski T J, Singleton M D, Loh D Y, Berg R, Staerz U D. Eur J Immunol. 1996;26:851–857. doi: 10.1002/eji.1830260419. [DOI] [PubMed] [Google Scholar]
- 43.O’Rourke A M, Rogers J, Mescher M F. Nature (London) 1990;346:187–189. doi: 10.1038/346187a0. [DOI] [PubMed] [Google Scholar]
- 44.Garcia K C, Scott C A, Brunmark A, Carbone F R, Peterson P A, Wilson I A, Teyton L. Nature (London) 1996;384:577–581. doi: 10.1038/384577a0. [DOI] [PubMed] [Google Scholar]
- 45.Hampl J, Chien Y H, Davis M M. Immunity. 1997;7:379–385. doi: 10.1016/s1074-7613(00)80359-3. [DOI] [PubMed] [Google Scholar]
- 46.Madrenas J, Chau L A, Smith J, Bluestone J A, Germain R. J Exp Med. 1997;185:219–229. doi: 10.1084/jem.185.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]