Abstract
The assumption that genes encoding tyrosine kinase receptors could play a role in human cancers has been confirmed by the identification of oncogenic mutations in the kinase domain of RET and KIT. Recently, homologous residues were found mutated in MET, in papillary renal carcinomas (PRCs). The link coupling these genetic lesions to cellular transformation is still unclear. METPRC mutations result in increased kinase activity and—in some instances, i.e., M1250T substitution—in changes in substrate specificity. A direct correlation occurs between the transforming potential of METPRC mutants and their ability to constitutively associate with signal transducers through two phosphorylated tyrosines (Y1349VHVNATY1356VNV) located in the receptor tail. Substitution of these “docking tyrosines” with phenylalanines leaves unaffected the altered properties of the kinase but abrogates transformation and invasiveness in vitro. Uncoupling the receptor from signal transducers with a tyrosine-phosphorylated peptide derivative (YpVNV) inhibits invasive growth induced by METPRC mutants. These data indicate that constitutive receptor coupling to downstream signal transducers is a key mechanism in neoplastic transformation driven by mutated MET and suggest a therapeutic strategy to target neoplastic diseases associated with this oncogene.
Germ-line missense mutations involving the RET protooncogene are found in patients affected by multiple endocrine neoplasia types 2A and 2B (MEN2A and MEN2B) or familial medullary thyroid carcinomas (1, 2). Somatic mutations of KIT are found in mastocytomas and in gastrointestinal tumors (3–5). Germ-line and sporadic mutations of MET have been recently described in papillary renal carcinomas (PRCs) (6, 7). Interestingly, the mutations found within the kinase domain of RET (M918T) and KIT (D816V) are located in codons that are homologous to those mutated in MET (M1250T, D1228N, and D1228H)‡. These mutations invariably result in up-regulation of the tyrosine kinase activity of the encoded receptors (9–12). The molecular mechanism coupling this biochemical alteration to the oncogenic properties of the mutated proteins is unclear. In RET and KIT, mutations of specific residues located in the kinase domain alter substrate specificity, thus affecting signaling fidelity (9, 13–15). Mutated RET and KIT receptors also display a different array of autophosphorylation sites, an altered ability to interact with signal transducers, and modified down-regulation mechanisms (10, 13, 16–19). It is still unclear which of these alterations is relevant for the oncogenic properties of the mutants.
MET receptor variants have been identified harboring the mutations previously found in the kinase domain of RET and KIT. This provides the opportunity to elucidate the molecular mechanisms responsible for neoplastic transformation driven by activated tyrosine kinase receptors.
The MET receptor binds at high affinity to the hepatocyte growth factor/scatter factor (20, 21). Signaling via this ligand–receptor pair promotes a unique biological program leading to “invasive growth.” This phenotype results from the integration of distinct biological activities including cell proliferation, motility, extracellular matrix invasion, and protection from apoptosis (22). MET-mediated invasive growth requires concomitant activation of multiple signaling pathways (23). This is achieved by receptor coupling to a number of effectors, among which are the Grb2/SOS complex, the p85 regulatory subunit of phosphatidylinositol 3-kinase, Stat-3, and the multiadaptor protein Gab1 (24–30). The interaction occurs on phosphorylation of a unique docking site containing the tandemly repeated sequence Y1349VHVNATY1356VNV (26).
METHODS
Reagents, Peptides, Antibodies, and Cell Culture.
All reagents used were from Fluka and Sigma. Reagents for SDS/PAGE were from Bio-Rad. N-acetylated and C-amidated synthetic peptides YpVNVYp and FVNV were synthesized by fluorenylmethoxycarbonyl (Fmoc) chemistry and purified by HPLC (>99%). For in vivo electroporation, the phosphate group was replaced by the nonhydrolyzable methylenphosphonate group (30). The peptide containing the MET docking sites fused to an antennapedia sequence RQIKIWFQNRRMKWKKIGEHY1349VHVNATY1356VNVKCVA, the optimal peptide substrates for the epidermal growth factor receptor (AEEEEYFELVAKKKK), the Abl cytosolic kinase (EAIYAAPFAKKK), or the Src kinase (AEEEIYGEFEAKKK) were obtained from Genosys (U.K.). Anti-phosphotyrosine, anti-p130Cas, and anti-Gab1 antibodies were purchased from Upstate Biotechnology. Anti-MET antibodies were obtained as described (34). COS7 and BOSC-23 were from the American Type Culture Collection. NIH 3T3 cells (a kind gift of J. H. Pierce, National Cancer Institute, Bethesda, MD) were found to produce 10–30 scatter units/ml. Cultures of mammalian cells were maintained in DMEM supplemented with 10% or 5% heat-inactivated calf serum (Colorado Serum, Denver) in a humidified atmosphere of 5% CO2/air.
Mutagenesis, Cloning, and Expression of the METPRC Mutants.
A recombinant PCR-based approach using the human MET cDNA as a template was applied to construct the METPRC mutants. When possible, silent mutations creating restriction sites were introduced to facilitate identification of recombinants. Mutations were verified by sequencing the region of interest. Wild-type and mutant METPRC cDNAs were subcloned into the pCEV29.1 plasmid, which is an expression vector driven by the Moloney murine leukemia virus long terminal repeat and containing a Neo resistance gene (35). Transfection of the cDNAs in COS7 and BOSC-23 cells was performed with the DNA-calcium phosphate procedure (CellPhect transfection kit, Pharmacia).
Substrate Phosphorylation Assays.
Immunoprecipitation with anti-MET antibodies was performed from lysates of COS7 cells expressing wild-type or METPRC mutant receptors. Immunoprecipitates were incubated with kinase buffer [50 mM Hepes (pH 7.5), 150 mM NaCl, and 12.5 mM MgCl2] in the presence of 15 μM ATP including [γ-32P]ATP and the indicated substrates for 30 min at 4°C. When myelin basic protein was used as a substrate, samples were analyzed by 15% SDS/PAGE followed by autoradiography. Phosphorylation of myelin basic protein was estimated by densitometric scanning of the film with an LKB2202 Ultrascan laser densitometer. When peptides were phosphorylated, samples were spotted on P81 cation-exchange chromatography paper (Whatman) and scintillation was quantified after two 20-min washes with 1% orthophosphoric acid.
Association with Signal Transducers.
Cells expressing wild-type or METPRC mutants were lysed with EB buffer (100 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM sodium orthovanadate), immunoprecipitation was performed with the indicated antibodies. After SDS/PAGE, proteins were transferred to Hybond-enhanced chemiluminescence membranes (Amersham) by high-intensity wet blotting. Filters were probed with the appropriate antibodies and specific binding was detected by enhanced chemiluminescence system (Amersham). When indicated, the association experiments were performed in the presence of Met-derived peptides (50 μM) as described (29).
Focus-Forming and Soft Agar Growth Assays.
NIH 3T3 cells were transfected at a density of 1.5 × 105 cells per 100-mm plate by the calcium phosphate method, using 40 μg per plate carrier DNA (calf thymus high-molecular-weight DNA, Boehringer Mannheim). For detection of foci, cells were maintained in DMEM containing 5% calf serum (Colorado Serum, Denver), and foci were scored 3 weeks after transfection following fixation in 3% paraformaldehyde and Giemsa staining. For selection of stable transfectants, cells were maintained in DMEM containing 10% calf serum and 750 μg/ml G418. After selection, resistant clones were pooled and expanded. For analysis of colony formation in soft agar, different cell dilutions (105–106) in 4 ml of DMEM containing 10% calf serum and 0.5% Seaplaque agarose (FMC) were seeded in 6-mm plates containing a 1% agarose underlay. Cultures were fed every 4 days, and the formation of colonies was scored after 3 weeks.
Peptides, Electroporation, and Invasion Assays.
For electroporation, adherent cells were cultured onto 3.2-cm2 indium–tin oxide conductive glass surfaces and electroporated in situ with PBS containing the phosphorylated or nonphosphorylated peptides (100 μM). The electric pulse was generated by the Epizap apparatus (Ask Science Products, Ontario, Canada), as described (30). More than 90% of cells were permeabilized as tested by the uptake of Lucifer yellow, and viability was greater than 80%, as judged by trypan blue exclusion. For Matrigel (Becton Dickinson)invasion assays, cells were seeded on fibronectin-coated conductive slides and grown to confluence. After electroporation with the indicated peptides, the cell monolayer was wounded and covered with Matrigel containing laminin, collagen type IV, proteoglycans, and growth factors. Cells were cultured for 8–12 hr in serum-free medium. The invasive phenotype was evaluated by counting the cells that invaded the wounded area (≈0.5 mm2). Invasive growth in collagen was assessed in three-dimensional collagen gels (30). Single cells were suspended (2,000 cells per ml) on ice in gelling solution containing 8 parts type I collagen (2 mg/ml stock solution G collagen, Seromed, Milan), 1 part 10× DMEM, and 1 part Hepes (0.5 M, pH 7.4). The cold mixture was placed into microtiter plate wells and allowed to polymerize for 15 min at 37°C before adding 0.1 ml of DMEM supplemented with 20% fetal calf serum. Cells were cultured for 5 days and examined under a phase-contrast microscope.
RESULTS AND DISCUSSION
It was recently shown that MET mutants, found in PRCs, transform fibroblasts in vitro and promote tumorigenesis in nude mice (12). All PRC mutations are located within the receptor tyrosine kinase domain (Fig. 1). Molecular modeling indicated that some of the mutations (D1228N, D1228H, Y1230C, Y1230H, and M1250T) alter residues located in the activation loop (22). The activation loop plays a key role in converting inactive receptor tyrosine kinases into their active form and has been implicated in autoinhibition of substrate binding (31). We therefore analyzed the effects of the mutations on the catalytic properties of the MET kinase by measuring either phosphorylation of an exogenous substrate (Fig. 2A) or phosphorylation of the receptor tail (Fig. 2B) using a peptide mimicking the docking sites (Y1349VHVNATY1356VNV). Interestingly, the mutations affecting the activation loop are associated with the highest catalytic activity and with increased phosphorylation of the receptor tail. As expected (12), a correlation was found between the catalytic properties and the transforming ability, which was most marked for M1250T, the mutation occurring in RET in the MEN2B syndrome (2, 10).
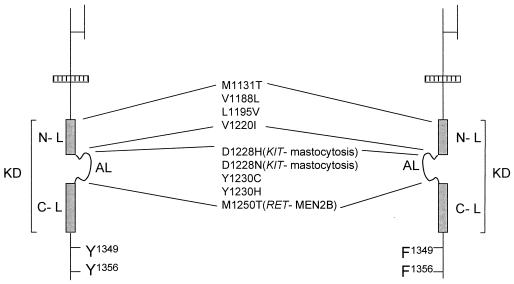
Figure 1.
Map of MET mutations found in PRCs. Schematic representation of the functional domains of the MET tyrosine kinase receptor. Mutations found in PRCs are listed. The tyrosine kinase domain (KD) is represented by a gray box. Y1349 and Y1356, on phosphorylation, generate the docking sites for signal transducers. These tyrosines were converted into phenylalanines (F) to generate PRC mutants unable to bind signal transducers (Right). Mutations of homologous residues found in RET and KIT together with the associated neoplastic diseases are listed in parentheses. AL, activation loop; N-L and C-L, N- and C-terminal kinase lobes.
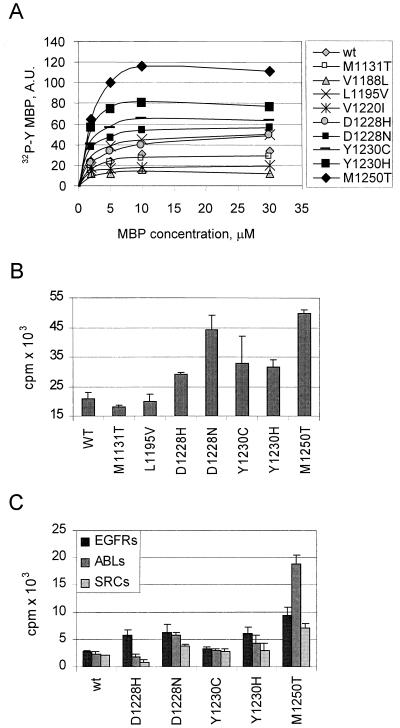
Figure 2.
METPRC mutations affect the kinase properties. Wild-type or PRC-mutated MET receptors were transiently expressed in COS cells, immunoprecipitated, and used to phosphorylate different substrates, including (A) the exogenous substrate myelin basic protein (MBP, (B) the peptide containing the sequence Y1349VHVNATY1356VNV, corresponding to the signal transducer docking site located in the MET tail, and (C) the optimal peptide substrates for the epidermal growth factor transmembrane receptor (EGFRs: AEEEEYFELVAKKKK), the Abl cytosolic kinase (ABLs: EAIYAAPFAKKK), or the Src kinase (SRCs: AEEEIYGEFEAKKK). A shows the amount of 32P incorporated into MBP at different substrate concentrations under the conditions described in Methods. Symbols indicate the METPRC proteins harboring the mutation listed on the right. B and C show the extent of phosphorylation of the indicated peptide by METPRC mutants.
To assess whether the PRC mutations could alter the substrate specificity of MET, we measured the phosphorylation of peptides that are known to be optimal substrates for tyrosine kinases of either the receptor or the cytosolic type (14). Minor changes in substrate specificity, compared with wild-type MET, were observed in the Y1230H, D1228H, and D1228N mutants. A more marked shift in substrate specificity was observed with the M1250T mutant, which phosphorylated more efficiently peptides that are optimal substrates for cytosolic tyrosine kinases such as Abl (Fig. 2C). Thus, the substrate infidelity displayed by the RET receptor mutated in MEN2B is also observed in the corresponding METPRC mutant. It should be noted however, that even the M1250T mutant phosphorylates the tail peptide with higher efficiency (3-fold) than any other peptide substrate (Fig. 2 B vs. C). Altogether these experiments suggest that the transforming PRC mutations not only up-regulate the kinase activity, resulting in increased ability to phosphorylate the receptor tail, but in some cases may also alter the substrate specificity. We next assessed whether, in view of its altered substrate specificity, the M1250T mutant was able to phosphorylate cellular substrates that are normally phosphorylated by the Abl tyrosine kinase, among which are Crk and p130CAS (32). We found that in NIH 3T3 cells transfected with the M1250T mutant and other METPRC mutants, the level of tyrosine phosphorylation of p130CAS was slightly increased with respect to control or wild-type transfected cells, whereas the levels of Crk phosphorylation were unaffected (data not shown).
We next examined the effects of the PRC mutations on signal transduction. For this purpose, wild-type or PRC-mutated MET receptors were stably expressed in NIH 3T3 cells (Fig. 3 A and B). For each receptor mutant, the level of protein expression and of tyrosine phosphorylation was evaluated by immunoprecipitation experiments. The receptors harboring the Y1230C, Y1230H, D1228H, D1228N, and M1250T mutations were found to be tyrosine-phosphorylated in intact cells (Fig. 3A). Densitometric analysis showed that some of the METPRC mutants display an increased level of tyrosine phosphorylation with respect to wild-type MET. The increase varies from 2- to 3-fold for the M1131T, Y1230C, and Y1230H mutants to 5- to 7-fold for the V1220I, D1228H, D1228N, and M1250T mutants.
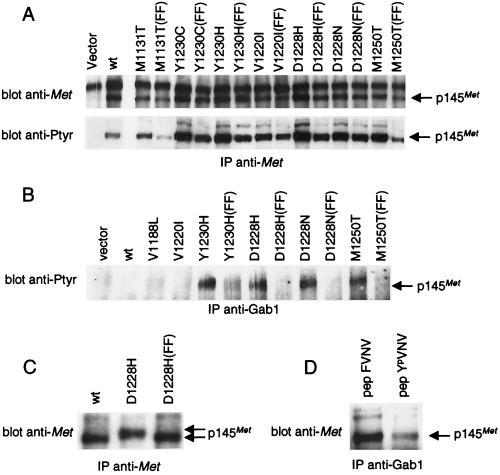
Figure 3.
METPRC mutants are constitutively coupled to signal transducers via two tyrosines located in the tail of the receptor. (A) NIH 3T3 fibroblasts were transfected with either wild-type or PRC-mutated MET receptor mutants to generate stable cell lines. FF indicates MET-receptor variants in which tyrosines Y1349 and Y1356 were converted into phenylalanines. The amount of Met proteins and the level of phosphorylation was evaluated by immunoprecipitation with anti-Met antibodies followed by Western blotting with anti-Met or anti-phosphotyrosine (pTyr) antibodies, respectively. (B) Lysates of the same cells were immunoprecipitated with anti-Gab1 antibodies and analyzed by Western immunoblotting with anti-pTyr antibodies. The identity of the phosphorylated protein coimmunoprecipitated with Gab1 was confirmed by reprobing the same blot with an anti-Met antibody (data not shown). (C) Lysates of COS cells expressing either the wild-type (wt), the D1228H mutant, or its double phenylalanine counterpart D1228H (FF) were immunoprecipitated and analyzed by Western blotting with anti-Met antibodies. The gap between the arrows indicates the electrophoretic shift caused by differences in phosphorylation. (D) Lysates of NIH 3T3 cells expressing the D1228H mutant were incubated with the YpVNV phosphopeptide corresponding to the receptor’s docking site and immunoprecipitated with anti-Gab1 antibodies. The amount of associated receptor was determined by Western blot with anti-Met antibodies. The nonphosphorylated phenylalanine peptide analogue (FVNV) was used as a negative control. The arrows indicate the position of the 145-kDa MET receptor β-chain (p145Met).
We next analyzed, by coimmunoprecipitation experiments, the effect of the PRC mutations on receptor coupling to signal transducers. We found that receptors harboring the Y1230H, D1228H, D1228N, and M1250T mutations were constitutively associated with signal transducers (Fig. 3B). These include the multiadaptor protein Gab1 (Fig. 3B), the Grb2/SOS complex, and the p85 regulatory subunit of phosphatidylinositol 3-kinase (data not shown). Previous work has demonstrated that concomitant activation of these effectors is responsible for invasive growth (23, 33). The ability of METPRC mutants to associate in a constitutive fashion with signal transducers correlates directly with their transforming potential (Fig. 3B vs. Fig. 4A).
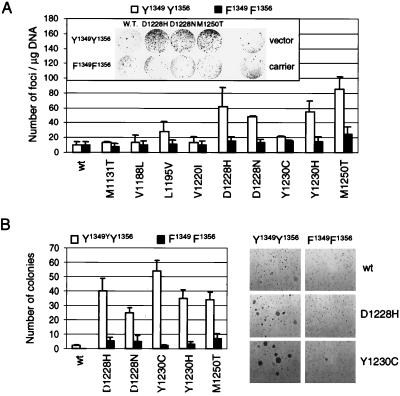
Figure 4.
Uncoupling METPRC mutants from signal transducers abrogates transformation and anchorage-independent growth. (A) The transforming potential of METPRC mutants (□, Y1349–Y1356) and of their signaling-inactive counterparts (■, F1349–F1356) was evaluated using the focus-forming assay. Values reported represent the average of three independent experiments (±SD). (Inset) micrograph showing a representative plate for each MET mutant after 3 weeks. Vector and carrier indicate cells transfected with empty vector or with calf-thymus DNA, respectively. (B) Anchorage-independent growth of METPRC mutant (□, Y1349–Y1356) and of their signaling-inactive counterparts (■, F1349–F1356) was assessed in soft agar. Values represent the average of two independent experiments (±SD). Micrographs show representative fields of colonies after 3 weeks of growth in soft agar.
To identify the critical mechanism(s) responsible for transformation (namely, the altered properties of the kinase versus constitutive coupling to physiological signal transducers), we uncoupled tyrosine kinase alterations from signal transduction by converting the two docking tyrosines of METPRC mutants into phenylalanines (Fig. 1). This Y → F conversion abrogated the association with signal transducers (Fig. 3B) but did not affect constitutive activation and autophosphorylation of the mutants (Fig. 3A). As previously shown (26), the lack of phosphorylation of the tail residues Y1349–Y1356 can be monitored by the faster electrophoretic mobility of the MET protein immunopurified from transfected COS cells (Fig. 3C). Interestingly, PRC mutants unable to bind their physiological signal transducers were dramatically impaired in their transforming potential in spite of the constitutive activation of the kinase (Fig. 4A). In addition, their ability to induce anchorage-independent growth (Fig. 4B) and to invade reconstituted basement membranes was also impaired (Fig. 5A). Previous work with cells transformed by a rearranged form of MET (TPR-MET) showed a direct correlation between the behavior in these in vitro assays and the invasive-metastatic phenotype in vivo (23).
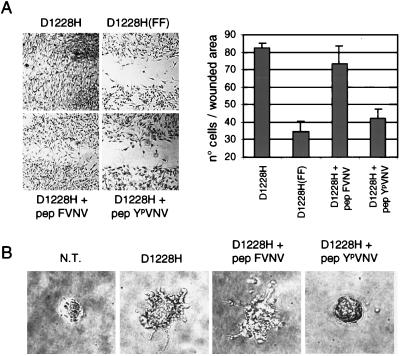
Figure 5.
Uncoupling METPRC mutants from signal transducers inhibits invasive growth. (A) The invasive potential of the D1228H mutants and of its signaling-inactive counterpart D1228H (FF) was evaluated by measuring the ability to invade reconstituted basal membranes made of laminin, collagen type IV, and proteoglycans (Matrigel). The YpVNV phosphopeptide was introduced into living cells by in situ electroporation; monolayers were then wounded and covered with Matrigel. The nonphosphorylated peptide FVNV was used as a negative control. The invasive phenotype was quantified by measuring the number of cells (±SD) found in the wounded area 12 hr later (see Methods). Micrographs show representative fields of cells plated on conductive slides used for in situ electroporation. (B) The invasive growth of the D1228H mutant was evaluated by measuring the sprouting and migration into a three-dimensional network of collagen type I. Human kidney cells (BOSC-23) either nontransfected (N.T.) or transiently expressing the D1228H receptor were seeded in collagen gels and grown for 5 days. The indicated peptides were introduced by in situ cell electroporation. Micrographs of representative fields were taken after 5 days.
These results indicate that constitutive coupling of signal transducers to the receptor tail is a key mechanism in METPRC-induced neoplastic transformation and suggest the possibility of interfering with this process by exogenous molecules. To this end, we tested whether a peptide mimicking the MET docking site could displace signal transducers (e.g., Gab1) in vitro. The phosphorylated synthetic peptide YpVNV, but not the unphosphorylated control FVNV, was found to be effective (Fig. 3D). Similar results were obtained with p85 and Grb2 (data not shown). To disrupt the interaction between METPRC and signal transducers in living cells, we synthesized derivatives of the same peptides, where the phosphate was replaced by a phosphonate group (resistant to phosphatases) and the N and C termini were modified to prevent proteolytic degradation (30). The peptides were introduced by in situ electroporation in cells transformed by METPRC mutants and the invasive phenotype evaluated in the above-mentioned in vitro assay. The tyrosine-phosphorylated peptide, but not its phenylalanine counterpart, prevented invasion of the reconstituted basement membrane by cells transformed by the D1228H mutant (Fig. 5A). The inhibition was as effective as the Y → F conversion in the receptor’s docking site. The same phosphorylated peptide specifically inhibited sprouting and migration of cells transformed by the D1228H mutant into a three-dimensional network of collagen (Fig. 5B). Similar inhibition in both assays was observed when other transforming METPRC mutants were tested.
These experiments show that the transforming and invasive potential of METPRC mutants relies on their constitutive coupling to downstream signal transducers. When this interaction is disrupted, the oncogenic properties are abrogated. Furthermore, these observations provide proof-of-concept for a signaling-based therapeutic strategy aimed at interfering with diseases associated with MET, or with tyrosine kinase receptors encoded by other oncogenes.
Acknowledgments
We thank Dr. Toru Miki for the pCEV29.1 plasmid, E. Wright and A. Cignetto for editing the manuscript, and Sarah Toniol for her enthusiastic help in performing the transducer-association experiments. The assistance of G. Petruccelli and R. Albano is gratefully acknowledged. This work was supported by research grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) and from the Giovanni Armenise-Harvard Foundation for Advanced Scientific Research.
ABBREVIATIONS
- PRC
papillary renal carcinomas
- MEN
multiple endocrine neoplasia
Footnotes
The numbering of human MET residues is according to ref. 8.
References
- 1.Mulligan L M, Kwok J B, Healey C S, Elsdon M J, Eng C, Gardner E, Love D R, Mole S E, Moore J K, Papi L, et al. Nature (London) 1993;363:458–460. doi: 10.1038/363458a0. [DOI] [PubMed] [Google Scholar]
- 2.Hofstra R M, Landsvater R M, Ceccherini I, Stulp R P, Stelwagen T, Luo Y, Pasini B, Hoppener J W, van Amstel H K, Romeo G, et al. Nature (London) 1994;367:375–376. doi: 10.1038/367375a0. [DOI] [PubMed] [Google Scholar]
- 3.Nagata H, Worobec A S, Oh C K, Chowdhury B A, Tannenbaum S, Suzuki Y, Metcalfe D D. Proc Natl Acad Sci USA. 1995;92:10560–10564. doi: 10.1073/pnas.92.23.10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longley B J, Tyrrell L, Lu S Z, Ma Y S, Langley K, Ding T G, Duffy T, Jacobs P, Tang L H, Modlin I. Nat Genet. 1996;12:312–314. doi: 10.1038/ng0396-312. [DOI] [PubMed] [Google Scholar]
- 5.Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, et al. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt L, Duh F M, Chen F, Kishida T, Glenn G, Choyke P, Scherer S W, Zhuang Z, Lubensky I, Dean M, et al. Nat Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt L, Junker K, Weirich G, Glenn G, Choyke P, Lubensky I, Zhuang Z, Jeffers M, Vande Woude G, Neumann H, et al. Cancer Res. 1998;58:1719–1722. [PubMed] [Google Scholar]
- 8.Ponzetto C, Giordano S, Peverali F, Della Valle G, Abate M L, Vaula G, Comoglio P M. Oncogene. 1991;6:553–559. [PubMed] [Google Scholar]
- 9.Santoro M, Carlomagno F, Romano A, Bottaro D P, Dathan N A, Grieco M, Fusco A, Vecchio G, Matoskova B, Kraus M H, et al. Science. 1995;267:381–383. doi: 10.1126/science.7824936. [DOI] [PubMed] [Google Scholar]
- 10.Borrello M G, Smith D P, Pasini B, Bongarzone I, Greco A, Lorenzo M J, Arighi E, Miranda C, Eng C, Alberti L, et al. Oncogene. 1995;11:2419–2427. [PubMed] [Google Scholar]
- 11.Moriyama Y, Tsujimura T, Hashimoto K, Morimoto M, Kitayama H, Matsuzawa Y, Kitamura Y, Kanakura Y. J Biol Chem. 1996;271:3347–3350. doi: 10.1074/jbc.271.7.3347. [DOI] [PubMed] [Google Scholar]
- 12.Jeffers M, Schmidt L, Nakaigawa N, Webb C P, Weirich G, Kishida T, Zbar B, Vande Woude G F. Proc Natl Acad Sci USA. 1997;94:11445–11450. doi: 10.1073/pnas.94.21.11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piao X, Curtis J E, Minkin S, Minden M D, Bernstein A. Blood. 1994;83:476–481. [PubMed] [Google Scholar]
- 14.Zhou S, Carraway K L, III, Eck M J, Harrison S C, Feldman R A, Mohammadi M, Schlessinger J, Hubbard S R, Smith D P, Eng C, et al. Nature (London) 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 15.Bocciardi R, Mograbi B, Pasini B, Borrello M G, Pierotti M A, Bourget I, Fischer S, Romeo G, Rossi B. Oncogene. 1997;15:2257–2265. doi: 10.1038/sj.onc.1201413. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Vega Q C, Decker R A, Pandey A, Worby C A, Dixon J E. J Biol Chem. 1996;271:5309–5312. doi: 10.1074/jbc.271.10.5309. [DOI] [PubMed] [Google Scholar]
- 17.Asai N, Iwashita T, Matsuyama M, Takahashi M. Mol Cell Biol. 1995;15:1613–1619. doi: 10.1128/mcb.15.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asai N, Murakami H, Iwashita T, Takahashi M. J Biol Chem. 1996;271:17644–17649. doi: 10.1074/jbc.271.30.17644. [DOI] [PubMed] [Google Scholar]
- 19.Pandit S D, Donis-Keller H, Iwamoto T, Tomich J M, Pike L J. J Biol Chem. 1996;271:5850–5858. doi: 10.1074/jbc.271.10.5850. [DOI] [PubMed] [Google Scholar]
- 20.Bottaro D P, Rubin J S, Faletto D L, Chan A M, Kmiecik T E, Vande Woude G F, Aaronson S A. Science. 1991;251:802–804. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- 21.Naldini L, Weidner K M, Vigna E, Gaudino G, Bardelli A, Ponzetto C, Narsimhan R P, Hartmann G, Zarnegar R, Michalopoulos G K, et al. EMBO J. 1991;10:2867–2878. doi: 10.1002/j.1460-2075.1991.tb07836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bardelli A, Pugliese L, Comoglio P M. Biochim Biophys Acta. 1997;1333:M41–M51. doi: 10.1016/s0304-419x(97)00026-7. [DOI] [PubMed] [Google Scholar]
- 23.Giordano S, Bardelli A, Zhen Z, Menard S, Ponzetto C, Comoglio P M. Proc Natl Acad Sci USA. 1997;94:13868–13872. doi: 10.1073/pnas.94.25.13868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graziani A, Gramaglia D, dalla Zonca P, Comoglio P M. J Biol Chem. 1993;268:9165–9168. [PubMed] [Google Scholar]
- 25.Ponzetto C, Bardelli A, Maina F, Longati P, Panayotou G, Dhand R, Waterfield M D, Comoglio P M. Mol Cell Biol. 1993;13:4600–4608. doi: 10.1128/mcb.13.8.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponzetto C, Bardelli A, Zhen Z, Maina F, dalla Zonca P, Giordano S, Graziani A, Panayotou G, Comoglio P M. Cell. 1994;77:261–271. doi: 10.1016/0092-8674(94)90318-2. [DOI] [PubMed] [Google Scholar]
- 27.Zhu H, Naujokas M A, Fixman E D, Torossian K, Park M. J Biol Chem. 1994;269:29943–29948. [PubMed] [Google Scholar]
- 28.Weidner K M, Di Cesare S, Sachs M, Brinkmann V, Behrens J, Birchmeier W. Nature (London) 1996;384:173–176. doi: 10.1038/384173a0. [DOI] [PubMed] [Google Scholar]
- 29.Bardelli A, Longati P, Gramaglia D, Stella M C, Comoglio P M. Oncogene. 1997;15:3103–3111. doi: 10.1038/sj.onc.1201561. [DOI] [PubMed] [Google Scholar]
- 30.Boccaccio C, Ando M, Tamagnone L, Bardelli A, Michieli P, Battistini C, Comoglio P M. Nature (London) 1998;391:285–288. doi: 10.1038/34657. [DOI] [PubMed] [Google Scholar]
- 31.Johnson L N, Noble M E, Owen D J. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 32.Salgia R, Pisick E, Sattler M, Li J L, Uemura N, Wong W K, Burky S A, Hirai H, Chen L B, Griffin J D. J Biol Chem. 1996;271:25198–25203. doi: 10.1074/jbc.271.41.25198. [DOI] [PubMed] [Google Scholar]
- 33.Weidner K, DiCesare S, Sachs M, Brinkmann V, Behrens J, Birchmeier W. Nature (London) 1996;384:173–176. doi: 10.1038/384173a0. [DOI] [PubMed] [Google Scholar]
- 34.Prat M, Crepaldi T, Gandino L, Giordano S, Longati P, Comoglio P. Mol Cell Biol. 1991;11:5954–5962. doi: 10.1128/mcb.11.12.5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michieli P, Li W, Lorenzi M V, Miki T, Zakut R, Givol D, Pierce J H. Oncogene. 1996;12:775–784. [PubMed] [Google Scholar]