Abstract
Epstein-Barr virus (EBV) infects various cell types in a wide spectrum of benign and malignant diseases. Laboratory tests for EBV have improved and are increasingly used in diagnosis, prognosis, prediction, and prevention of diseases ranging from infectious mononucleosis to selected subtypes of lymphoma, sarcoma, and carcinoma. Indeed, the presence of EBV is among the most effective tumor markers supporting clinical management of cancer patients. In biopsies, localization of EBER transcripts by in situ hybridization remains the gold standard for identifying latent infection. Other RNA- and protein-based assays detect lytic viral replication and can distinguish carcinoma-derived from lymphocyte-derived EBV in saliva or nasopharyngeal brushings. Analysis of blood using EBV viral load and serology reflects disease status and risk of progression. This review summarizes prior research in the context of basic virologic principles to provide a rational strategy for applying and interpreting EBV tests in various clinical settings. Such assays have been incorporated into standard clinical practice in selected settings such as diagnosis of primary infection and management of patients with immune dysfunction or nasopharyngeal carcinoma. As novel therapies are developed that target virus-infected cells or overcome the adverse effects of infection, laboratory testing becomes even more critical for determining when intervention is appropriate and the extent to which it has succeeded.
Epstein-Barr virus (EBV) causes infectious mononucleosis and is also associated with a wide variety of malignancies including Hodgkin and non-Hodgkin lymphomas, post-transplant lymphoproliferative disorder (PTLD), nasopharyngeal carcinoma (NPC), and gastric carcinoma. The prevalence of EBV-related cancers, estimated to affect up to 1% of humans worldwide, warrants increased focus on laboratory assays to detect and characterize the infection. Within a given neoplasm, consistent presence of EBV implies that the virus might contribute to pathogenesis or maintenance of the clonal process. Furthermore, the physical location of viral DNA within every malignant cell of a given tumor implies that the virus is a biomarker that can be used to evaluate the extent of tumor spread and to monitor disease burden in response to therapy. Even before disease is clinically evident, high-risk individuals may benefit from screening tests that predict impending progression so that preemptive measures may be taken. Finally, improvements in EBV-directed therapy highlight the importance of laboratory detection and the potential for targeting viral gene products or their downstream pathways driving cell proliferation, inhibiting apoptosis, or evading immune response. While much research remains for understanding the full promise of EBV testing in patient care, it is clear that EBV is a broadly useful tumor marker.
Lifelong Infection in Healthy Carriers
EBV infects nearly all humans by the time they reach adulthood, after which the viral genome is retained for life in a small fraction of B lymphocytes. Healthy carriers have approximately 1 to 50 infected cells per million leukocytes,1 consistent with an average EBV viral load in whole blood of about 7 copies (range, 1 to 30 copies) of EBV DNA per million leukocytes.2 Any biopsy tissue may contain B lymphocytes and therefore may harbor amplifiable EBV DNA. Cell-free body fluids such as serum or plasma contain negligible amounts of EBV DNA, suggesting that EBV is detectable only in association with reactivated infection or EBV-related disease.2,3 To maximize the utility of EBV as a marker for disease, it is important to evaluate EBV in a quantitative rather than a qualitative fashion and to localize EBV to particular cell types when lesional tissue is available for histopathological analysis.
Oral mucosa is a major site of replication of EBV and shedding of infectious virions.4 Remotely infected healthy carriers have salivary EBV DNA levels varying from undetectable to over 1000 copies/ml, with periodic salivary shedding accounting for nearly universal infection of humans before adulthood.4 Stress and immunodeficiency are postulated to trigger EBV reactivation and increased oral shedding.
Cell Types Infected by EBV
EBV is capable of infecting B lymphocytes, squamous epithelial cells, glandular epithelial cells, myoepithelial cells, smooth muscle cells, T cells, NK cells, plasma cells, and follicular dendritic cells. This wide spectrum of susceptible cell types was determined because of pathological lesions in which EBV is localized to these cells, whereas healthy carriers seem to harbor EBV almost exclusively in B lymphocytes. The importance of B cells in the life cycle of EBV is emphasized by the inability of infection to take hold in children with Bruton's agammaglobulinemia, a rare genetic disorder in which B cells are absent.5 EBV can productively infect epithelial cells as evidenced by a tongue lesion called oral hairy leukoplakia in patients with human immunodeficiency virus and also by rare infection of healthy epithelial cells.6 It remains unclear whether the virions that are intermittently shed in saliva of healthy carriers originate from mucosal B lymphocytes, plasma cells, or squamous epithelial cells.
Its presence in genital tract secretions of both genders, along with anecdotal reports of genital ulcers in infectious mononucleosis patients, implies that EBV could be sexually transmitted.7,8 Breast milk of nursing mothers may contain EBV, but this appears to be an uncommon route of vertical transmission.9
Clonality Assays Based on EBV Genomic Structure
A single virion successfully infects any given cell, as evidenced by the unique structure of the viral terminal repeat sequences in individual cell clones. The novelty of each virion relies on the number of tandem repeat sequences (up to 20) found at the ends of the 172,000-bp linear EBV genome. When a cell is infected, the double-stranded viral DNA circularizes to form an episome that may then replicate to produce 1 to 50 clonal copies of the EBV genome, and these clonal episomes are passed along to cellular progeny. If an infected cell undergoes malignant transformation, every neoplastic daughter cell inherits the same unique viral episomal structure, making the EBV genomic structure a marker for tumor clonality. On Southern blot analysis of the viral terminal repeat fragment, a monoclonal neoplasm displays as a single band, and the size of the band differs among various patients' tumors because of differing numbers of terminal repeats in the fused episome.10 Oral hairy leukoplakia and other lytic infections display a ladder of smaller bands corresponding to the polyclonal linear viral genomes.
Monoclonal EBV DNA is present in infected lymphomas and carcinomas.10,11 PTLDs are usually monoclonal, although occasional biclonal or oligoclonal examples imply multiple synchronous neoplastic transformations, and polyclonality characterizes early disease having a better prognosis.12 Although EBV DNA typically persists as an episome, in some instances it recombines with the human genome to create one or more chromosomal integrations of varying structure with respect to viral and host breakpoints.13 Whether integration contributes to neoplasia requires further study.
Viral Life Cycle Balances Latent and Lytic Infection
Infection of a B lymphocyte leads to two alternative outcomes supporting viral persistence and propagation.14 These outcomes reflect physiological processes underlying humoral immunity. EBV infection mimics antigen stimulation by triggering the host B cell to divide, producing daughter cells that become either memory B cells persisting long term or plasma cells supporting lytic viral replication with virion production. Throughout the remaining life of the host, EBV lies latent in some B cells by expressing few if any immunogenic viral proteins. When an infected cell is stimulated by its natural antigen, which would typically occur on a mucosal surface where foreign antigens abound, the B cell recapitulates the process described above by dividing to create more infected memory B cells and more terminally differentiated plasma cells producing thousands of virions that could reinfect the host or be shed from the mucosal surface to infect other human hosts.
Spectrum of EBV Gene Expression in Lesional Tissue
Latent infection is characterized by limited expression of viral proteins to avoid immune recognition and destruction. One tolerated protein is EBV nuclear antigen 1 (EBNA1) that functions to propagate the viral genome to daughter cells on cell division. EBNA1 is unable to elicit an effective cytotoxic immune response, partially explaining why EBV is never eliminated from the body.15 In immunocompromised hosts and in tissue culture systems in which immune surveillance is absent, infected B cells may express a broader range of viral proteins: LMP1, LMP2, and the EBV nuclear antigens (EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, and EBNA5). Inexplicably, Hodgkin lymphoma patients tolerate LMP2-expressing tumor cells even though their immune system seems to be competent in other regards. HLA type and human immunodeficiency virus status impact on risk of developing EBV-related Hodgkin lymphoma, emphasizing the importance of the immune system in controlling the outcome of infection.16 Prognosis relates to EBV status in a complex manner: children whose Hodgkin tumors are EBV-positive fare better, whereas older adults do better if their tumors are EBV-negative.17,18
Several patterns of latent viral gene expression have been described based on the spectrum of expressed proteins, transcripts, and noncoding RNAs like EBV-encoded RNAs (EBER1 and EBER2) and BamH1A rightward transcripts (BARTs) including microRNAs (Table 1).19,20,21 Even monoclonal monomorphous tumors may have heterogeneous viral gene expression among cells, so adequate sampling is required to fully appreciate the spectrum of expressed viral factors. EBERs are reliably expressed in virtually all latently infected cells in every benign and malignant lesion, which explains why EBER in situ hybridization is the assay of choice in clinical laboratories for defining a lesion as EBV-related.
Table 1.
Characteristic Patterns of EBV Gene Expression in Normal and Lesional Tissue
| Cell or tissue type | Typical EBV gene expression* |
|---|---|
| AIDS-related plasmablastic lymphoma | Type 0 latency (EBERs, BARTs) |
| Burkitt lymphoma | Type I latency (EBNA1, LMP2, EBERs, BARTs) |
| Hodgkin lymphoma | Type II latency (EBNA1, LMP1, LMP2, EBERs, BARTs) |
| AIDS-related Burkitt or primary effusion lymphoma | Type II |
| Peripheral T cell lymphoma | Type II |
| NK/T cell lymphoma, nasal type | Type II |
| Nasopharyngeal carcinoma | Type II plus BARF1 |
| Gastric adenocarcinoma | Type II plus BARF1 |
| Post-transplant lymphoproliferative disorder | Type III latency (EBNA1, −2, −3A, −3B, −3C; LMP1, LMP2, EBERs, BARTs) |
| AIDS-related immunoblastic or brain lymphoma | Type III |
| Infectious mononucleosis | Type III |
| Chronic active EBV infection | Type III |
| Lymphoblastoid cell lines in vitro | Type III |
| Oral hairy leukoplakia | Lytic infection (LMP1, LMP2, BZLF1, BMRF1, BHRF1, BCRF1, and other replication factors) |
| Remotely infected carriers | |
| Circulating B cells | Type 0 |
| Tonsil/mucosal B cells | Type II |
Viral gene expression may be focal or variable in a given lesion.
Viral replication is associated with diminished EBERs and sequential expression of a cascade of lytic viral proteins beginning with BZLF1. Lytic proteins elicit strong humoral and cellular immune responses, explaining why viral infection is so well controlled in healthy carriers. Indeed, 5% of all circulating mononuclear cells are EBV-directed in healthy carriers.22
Serology
Serological tests confirm primary infection and document remote infection. The most widely used serological assay, the heterophile antibody test (colloquially called the “Monospot” test), was first introduced in 1932 well before EBV was identified as the causative agent of infectious mononucleosis. The original heterophile test was based on the discovery that serum or plasma from patients with infectious mononucleosis could agglutinate horse or sheep erythrocytes. Modern variants of the test detect serum-mediated agglutination of latex beads coated by bovine heterophile antigens. The heterophile test is used to facilitate rapid clinical decision-making in patients with infectious mononucleosis-like symptoms (fever, sore throat, lymphadenopathy, hepatosplenomegaly, malaise, headache). Complementary findings include an elevated white blood cell count and reactive lymphocytosis representing cytotoxic T cells responding to EBV infection.
A panel of sensitive and specific serological tests is applied in patients for whom the heterophile test is suspected to be false-negative (especially young children or older adults) and to document prior EBV infection (Figure 1). Indirect fluorescent antibody tests are useful, but they are also labor-intensive and subjective because they involve applying patient serum to infected cells immobilized on glass slides followed by microscopic interpretation of patterns of antibody localization to complexes of viral proteins collectively referred to as early antigen (EA), viral capsid antigen (VCA), or EBNA.23 In recent years, more economical and objective tests for antibodies against recombinant EBV proteins have been introduced using enzyme-linked immunosorbent assay technology and related methods.23,24,25
Figure 1.
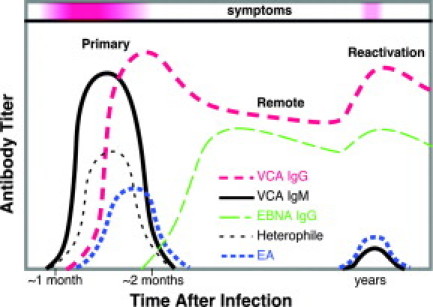
Serological titers distinguish primary infection from remote infection. IgG anti-VCA and IgM anti-VCA rise in concert with symptoms of primary infection and a positive heterophile test. After symptoms resolve, remote infection is characterized by EBNA and IgG anti-VCA without EA, although EA and IgM may reappear with or without symptoms on viral reactivation or EBV-related neoplasia.
EBV-related cancer is typically associated with high serological titers against EA and IgG VCA with low EBNA titer. Results should be interpreted with caution since similar patterns are possible in autoimmune disease and other reactive conditions. Furthermore, serology is not reliable when the immune system is dysfunctional, eg, acquired immunodeficiency syndrome (AIDS) or allogeneic transplant patients including solid organ and marrow recipients. Surprisingly, certain abnormal EBV serological patterns are a risk factor for developing lymphoma that is not EBV-infected, such as chronic lymphocytic leukemia, suggesting that atypical antibody response to EBV reflects a more general deficit in the immune system.26 A more direct role for the virus is postulated in Richter transformation of chronic lymphocytic leukemia in which EBV is localized to the higher grade lymphoma in 15% of cases.
There is one type of cancer for which EBV serology is critical: NPC patients often have high IgA titers against lytic EBV proteins in keeping with the origin of the cancer on the mucosal surface of the nasopharynx.27 A panel of serological and molecular tests on serum or plasma can screen for NPC in high-risk populations, assess prognosis, and monitor disease status over time.28,29,30
EBV Strain Types
There are two major strains of EBV (types A and B) as well as many minor strains that are being evaluated for their impact on disease association, oncogenicity, and drug resistance. Pilot data suggest that strain variation impacts aggressiveness for both lymphomas and carcinomas.31,32 Interestingly, healthy carriers often harbor multiple EBV strains, and strain type may differ by anatomical site and over time in a given individual.31 The variation appears to derive from infection by multiple strains more than acquired polymorphisms in a pre-existing strain.
Strain variation may impact detectability of the virus by laboratory methods. Point mutation, deletion, or rearrangement of the EBV genome could contribute to false-negative probe hybridization results.33 Furthermore, integration into host chromosomal DNA may result in partial loss of viral gene sequences. Strain variation emphasizes the need to target a conserved viral gene segment (or preferably multiple segments) when designing a laboratory assay to detect EBV DNA. Pilot studies of AIDS lymphoma identified interfering sequence variants in the LMP1, LMP2, and BZLF1 genes, whereas BamH1W, EBER1, and EBNA1 regions were highly conserved.33 An advantage of targeting the reiterated BamH1W region is improved assay sensitivity at the expense of accuracy since BamH1W repeat number varies from 7 to 11 copies per EBV genome. However, accuracy may be less important than other performance characteristics for certain clinical applications such as detecting early stage NPC. Moreover, the number of viral genomes per cell varies across tumors, further emphasizing that EBV copy number is not synonymous with infected cell count.
In EBV-related cancer patients, the same EBV strain lies in the plasma as in the tumor, and the plasma EBV DNA is naked rather than encapsidated implying that it emanates from dying infected tumor cells rather than representing new virion production.34,35,36 By comparison, infectious mononucleosis patients have a mixture of encapsidated virions and naked EBV DNA in their plasma.36
EBER in Situ Hybridization Assay Localizes EBV in Biopsy Tissue
The most abundant viral transcripts in latently infected cells, EBER1 and EBER2, are non-polyadenylated and thus are not translated into protein; they function to inhibit interferon-mediated antiviral effects and apoptosis. These two transcripts, collectively called EBER, are expressed at such high levels (around a million copies per latently infected cell) that they are considered to be the best natural marker of latent infection. In situ hybridization targeting one or both EBERs is the gold standard assay for determining whether a biopsied tumor is EBV-related. Commercial systems for EBER in situ hybridization facilitate implementation in clinical laboratories (eg, Ventana [Tucson, AZ], Leica [Bannockburn, IL], Dako [Glostrup, Denmark], Invitrogen [Carlsbad, CA], Biogenex [San Ramon, CA]). As a control for adequate RNA preservation and for the hybridization process, a parallel assay should be done targeting endogenous RNA.37
Table 2 shows a list of EBV-associated diseases along with the proportion of cases harboring EBV within lesional cells. A biopsy assessed by EBER in situ hybridization is needed to confirm each diagnosis and its relation to EBV, with the exception of suspected infectious mononucleosis for which clinical findings and serology are usually diagnostic, whereas biopsy may be counterproductive because of histological overlap with Hodgkin and non-Hodgkin lymphoma. Another exceptional lesion is oral hairy leukoplakia in which EBER is down-regulated, whereas lytic proteins BZLF1 and BMRF1 are localized to ballooned cells in mid-layers of the hyperplastic stratified squamous epithelium.
Table 2.
EBV-Associated Diseases
| Disease | EBV-related (% cases) |
|---|---|
| Benign reactive infections | |
| Infectious mononucleosis | >99 |
| Oral hairy leukoplakia | >95 |
| EBV-related hemophagocytic syndrome | 100* |
| Chronic active EBV infection | 100* |
| Hodgkin lymphoma | |
| Hodgkin lymphoma, all subtypes | 40 |
| Hodgkin lymphoma, mixed cellularity | 70 |
| Hodgkin lymphoma, nodular sclerosis | 20 |
| Hodgkin lymphoma, lymph. predominant | <5% |
| Hodgkin lymphoma, lymphocyte depleted | 50 |
| Hodgkin lymphoma, AIDS-related | >95 |
| Carcinomas and soft tissue sarcomas | |
| Nasopharyngeal carcinoma, Asian | >95 |
| Nasopharyngeal carcinoma, USA | 75 |
| Lymphoepithelioma-like carcinoma | Most |
| Gastric adenocarcinoma | 7 |
| Smooth muscle tumor in AIDS/transplant | >95 |
| Follicular dendritic cell tumor, IP-like | Most |
| Non-Hodgkin lymphomas and related neoplasms | |
| Non-Hodgkin lymphoma, all subtypes | 5 |
| Non-Hodgkin lymphoma, diffuse large B cell subtype | 15 |
| Richter syndrome (transformed lymphoma) | 15 |
| Non-Hodgkin lymphoma, AIDS-related | 40 |
| Brain lymphoma, AIDS-related | 95 |
| Brain lymphoma, immunocompetent hosts | 5 |
| Post-transplant lymphoproliferative disorder | 95 |
| Burkitt lymphoma, African (endemic) | >95 |
| Burkitt lymphoma, North American | 20 |
| Burkitt lymphoma, AIDS-related | 30 |
| Lymphoma, primary immunodeficiency | Most |
| Pyothorax-associated lymphoma | 90 |
| Lymphomatoid granulomatosis (B cell lymphoma) | 90 |
| Plasmablastic lymphoma, AIDS-related | 60 |
| Primary effusion lymphoma, AIDS-related | 70 |
| Age-related EBV-associated B cell lymphoproliferation | 100* |
| Angioimmunoblastic T cell lymphoma (EBV+ B cells) | 80 |
| Peripheral T cell lymphoma, unspecified | 40 |
| Extranodal NK/T lymphoma, nasal type | >95 |
| NK leukemia | Most |
| γ δ T cell lymphoma, mucosal | Most |
| T cell lymphoma in chronic active EBV infection | Most |
By definition the disease is EBV-related.
Utility of EBER in Situ Hybridization in Carcinomas
Nearly all undifferentiated NPCs are EBV-related, whereas a lesser proportion of keratinizing NPCs harbor EBV as demonstrated by EBER in situ hybridization. Around half of affected patients initially present with an enlarged cervical lymph node representing metastatic spread; identification of EBER-expressing carcinoma in a lateral or posterior retropharyngeal cervical node should trigger endoscopic examination of the nasopharynx in search of the primary site. In a left supraclavicular lymph node, EBER-positive undifferentiated carcinoma or adenocarcinoma should prompt consideration of a gastric primary, since about 7% of gastric carcinomas are infected.
Although some investigators claim to have identified EBER-negative cancers that harbor EBV by other analytic methods, these claims are controversial and are not universally accepted.38,39 DNA amplification results alone are inadequate to prove localization of the infection to malignant cells as opposed to reactive lymphocytes. Furthermore, RNA-based tests like EBER histochemistry are prone to false-negative results, emphasizing the need to interpret negative EBER stains in the context of a control assay to demonstrate that RNA is preserved and available for hybridization. In questionable cases, in situ hybridization to EBV DNA is the next most reliable histochemical test, and secondary support comes from Southern blot analysis of EBV clonality and other histochemical assays.
Utility of EBER in Situ Hybridization in Lymphomas
EBV-driven PTLD is a feared cause of morbidity and mortality in allogeneic transplant recipients. The disease is manifest when iatrogenic immunosuppression leads to diminished T cell immunity that predisposes to massive proliferation of EBV-infected B lymphocytes. EBER in situ hybridization is useful in the work-up of suspected PTLD as well as any lymphoproliferation arising in the setting of immunodeficiency, such as the following. 1) Immunosuppressive drugs (eg, methotrexate for rheumatoid arthritis) may lead to an EBV-driven lymphoproliferation that responds to withdrawal of the drug. 2) Impaired immunity in the elderly may cause “age-related EBV-associated B cell lymphoproliferative disorder.” 3) Immunodeficiency predisposes to lymphomatoid granulomatosis, a rare neoplasm of the lung or other extranodal sites in which infected neoplastic B cells are surrounded by far more abundant reactive T cells. 4) The longstanding chronic inflammation of tuberculosis predisposes to pleural-based pyothorax-associated B cell lymphoma that is EBV-related in 90% of cases. 5) AIDS patients are predisposed to develop B cell lymphoma (diffuse large B cell, immunoblastic, Burkitt, plasmablastic, Hodgkin, or primary effusion subtypes) with 50% of cases being EBV-related. In addition, brain lymphoma arising in an AIDS patient is nearly always EBV-related.40
Enlarged lymph nodes from infectious mononucleosis patients harbor variable numbers of EBER-expressing small to large lymphocytes. In contrast, Hodgkin and non-Hodgkin lymphomas generally lack EBER-expressing small lymphocytes while the larger tumor cells may be uniformly EBER-positive, a feature that can be helpful in resolving a benign versus malignant differential diagnosis. A histological mimic of Hodgkin lymphoma, anaplastic large cell lymphoma, is virtually never EBER-positive, at least in Western nations. Nasal NK/T lymphoma is a unique form of lymphoma that is so tightly linked to EBV that failure to identify the virus should prompt re-evaluation of the differential diagnosis. Aggressive NK cell leukemia, which may represent systemic dissemination of nasal NK/T lymphoma, is also EBV-related.
Peripheral T cell lymphoma frequently harbors EBV. Surprisingly, dual stains show that the infection is often localized to B lymphocytes that lie in proximity to uninfected malignant T cells.41 An example is angioimmunoblastic T cell lymphoma (AILT) which, in 80% of cases, has EBER localized to B cells. These B cells may be monoclonal, although their small numbers make it difficult to demonstrate clonality in the context of the more prevalent uninfected T cell clone.42 Regardless, B cell expansion correlates with EBER positivity in AILT as well as in peripheral T cell lymphomas of unspecified type.43 Attygalle et al found that expansion of EBV-infected lymphocytes (defined as ≥5 EBER-expressing cells per section) was helpful in supporting a diagnosis of AILT and distinguishing it from peripheral T cell lymphomas of unspecified type that harbor far fewer EBER-positive cells.42 This finding is controversial since Tan et al reported that both AILT and peripheral T cell lymphomas of unspecified type frequently contain infected cells (defined as more than three EBER-expressing cells per section).43 Cases of peripheral T cell lymphomas of unspecified type that are EBV-related tend to have a worse prognosis.41 Interestingly, some AILT are coinfected with HHV6 or are infected by HHV6 alone, and the histopathological appearance of AILT depends on which of these two viruses are present.44
Ongoing research is refining the criteria for distinguishing chronic active EBV infection or EBV-associated hemophagocytic syndrome from T cell lymphoma and NK cell neoplasia. These conditions are most prevalent in southeast Asia and may begin when an as yet uncharacterized immunodeficiency permits EBV infection of NK or T cells that then predisposes to subsequent development of NK or T cell lymphoma. The various preneoplastic and neoplastic lesions may occur separately or in combination in a given patient. In any case, EBER in situ hybridization, especially when combined with a dual stain for CD3 T cells, is useful in localizing the infection.45 Still to be resolved is whether identification of EBER-positive T cells is always indicative of neoplasia or whether latent EBV can be found in normal T cells.
It is interesting to note how often EBV-related lymphomas were historically categorized as reactive conditions. What was first described as Hodgkin's granuloma was later termed Hodgkin's disease and is now called Hodgkin lymphoma. What was once called lethal midline granuloma is now nasal NK/T lymphoma. Angioimmunoblastic lymphadenopathy with dysproteinemia is now AILT. Many follicular dendritic cell tumors were previously termed inflammatory pseudotumor. Lymphomatoid granulomatosis, now known to be an EBV-infected B cell lymphoma, should be more appropriately renamed. It is likely that EBV draws abundant reactive inflammatory cells into the region of an infected tumor, possibly contributing to misclassification of these lesions as benign. Further refinement of the World Health Organization criteria for classifying lymphoma and for distinguishing lymphoma from reactive conditions are predicted to depend increasingly on laboratory assays targeting EBV and other viruses such as HHV6, HHV8, HIV, HTLV1, and HCV that are cofactors in lymphomagenesis. Furthermore, advances in targeted therapy that eliminate virally infected cells or thwart their oncogenic effects will make it all of the more important to reliably detect and characterize viruses in lesional tissue.
Amplification Assays Targeting the EBV Transcriptome
The sequences of all 84 EBV genes and their variant transcripts are well described, as are the virally encoded microRNAs.20,21,46 Panels of real-time polymerase chain reactions (PCRs) characterizing the viral gene expression profile may identify novel biomarkers for subclasses of EBV-related disease.47,48 Individual real-time PCR or nucleic acid sequence-based amplification assays appear to help distinguish tumor from normal tissue even when the normal tissue harbors “background” EBV infection.49,50 For example, suspected NPC was confirmed by BARF1 nucleic acid sequence-based amplification in nasopharyngeal brushings since viral BARF1 is expressed only in carcinoma and not in normal nasopharynx nor in benign epithelial infection.49,50 Because BARF1 is specific to malignant epithelial cells, it should be explored as a biomarker for infected gastric carcinoma.50,51 DNA- and protein-based assays are complementary, with EBV DNA and BARF1 protein both elevated in nasopharyngeal brushings or saliva at high sensitivity and specificity for NPC.52,53 In a screening situation (eg, testing for local recurrence of NPC), high levels of EBV DNA, RNA, or protein in a brushing or in saliva should be followed by a biopsy, including “blind” biopsies if no lesion is seen by endoscopy. Rare malignant cells within a biopsy, especially in specimens with crush artifact, can sometimes be highlighted using EBER in situ hybridization.54
Immunohistochemical Assays Localize EBV Proteins in Biopsy Tissue
Western blot, flow cytometry, and enzyme-linked immunosorbent assay can potentially detect and measure selected viral proteins for which antibodies are available. However, the single most informative protein-based assay is immunohistochemistry, because it permits localization of protein in the context of histopathology, facilitating assessment of the medical significance of the infection. Localization is achievable in paraffin-embedded sections for latent and lytic viral factors including EBNA1, EBNA2, LMP1, LMP2, BHRF1, BZLF1, and BMRF1.45 Interpretation of results, including defining the spectrum of expressed genes and their localization to benign or malignant-appearing cells, complements EBER in situ hybridization for diagnosis of EBV-related disease.55
LMP1 immunohistochemistry is as informative as EBER in situ hybridization for defining whether a given Hodgkin lymphoma case is EBV-related as defined by localization of EBV to the neoplastic Reed-Sternberg/Hodgkin cells.37 Unfortunately, few other diseases so reliably express EBV-encoded proteins: Burkitt lymphoma, PTLD, and NPC may have diffuse, focal, or completely undetectable expression of viral proteins by immunostains. EBER in situ hybridization remains the most reliable histochemical assay to localize EBV in these cancer tissues.
Technical problems can foil interpretation of immunohistochemical results. For example, EBNA1 is thought to be expressed in virtually all latently infected tumors, yet EBNA1 immunohistochemistry is not sensitive enough to reliably substitute for EBER in situ hybridization. Furthermore, the 2B4 clone of EBNA1 antibody cross-reacts with human MAGEA4, potentially causing false-positive interpretations.56 Problems such as this emphasize the need to validate laboratory assays before they are used in clinical investigations.
EBV Viral Load Assays
Quantitative EBV DNA measurement is essential for differentiating the low-level infection of healthy carriers from the high levels characteristic of EBV-related disease. Patients with active infection or EBV-related cancer tend to have high levels of EBV DNA in the cell-free fraction of blood (plasma or serum), whereas in healthy carriers the virus is restricted to the intracellular compartment of the blood (Figure 2).
Figure 2.
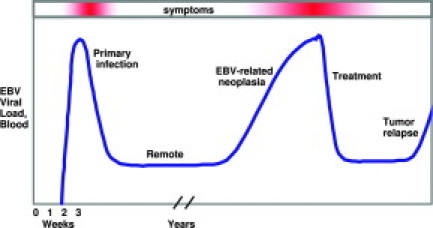
EBV viral load in whole blood reflects clinical status in patients with infectious mononucleosis, allogeneic transplant, and nasopharyngeal carcinoma. EBV DNA levels begin to rise within 2 weeks of primary infection and are already falling by the time the patient becomes symptomatic (due to interferon γ and other immune responses). Plasma or serum EBV DNA is undetectable in most remotely infected individuals; however, whole blood is low positive for the duration of life. If an EBV-related malignancy develops, levels may rise before clinical diagnosis, implying that high-risk patients benefit from routine monitoring. Successful therapy is marked by a decline to baseline, and rising levels may serve as a harbinger of relapse.
Real-time PCR is the principal technology used for modern EBV viral load measurement. It relies on amplification of a conserved sequence (typically ∼100 bp) using either an intercalating dye or a fluorescent probe (eg, TaqMan, MGB) to quantify the products against a series of standards representing serial dilutions of known EBV DNA content.33,57,58,59 Since the reaction vessel remains sealed, the risk of amplicon contamination is minimized. Published procedures target any of several viral sequences.60 Commercial primers and probes are available: Roche Molecular Diagnostics (Pleasanton, CA) targets LMP2, Qiagen (Valencia, CA) (Artus) targets BKRF1 (EBNA1), Nanogen (San Diego, CA) (Epoch) targets a major tegument protein (BNRF1), Argene (Varilhes, France) targets thymidine kinase (BXLF1), Bioactiva Diagnostica (Bad Homburg, Germany), and Amplimedical (Milan, Italy) targets BKRF1 (EBNA1).3,61,62,63,64
There is no consensus on the optimal methods for performing or reporting EBV viral load results, so it is the responsibility of each testing laboratory to validate their own procedures.65 There is concern that prolonged or improper storage of whole blood may allow EBV DNA to escape from the intracellular compartment, potentially causing false-positive EBV results in plasma or serum specimens that are subsequently separated from the cellular fraction. False-negative findings due to nucleases are also a concern. EBV DNA in plasma is known to be partially degraded, although this may reflect apoptotic pathways in the cell from which it emanated.35
Automated extraction instruments promote reproducibility of DNA isolation procedures. The efficiency of extraction and amplification (as judged by amplification of an endogenous or spiked control sequence) should be considered. Units of measurement vary from copies per milliliter, to copies per microgram of DNA, to copies per 100,000 leukocytes.65 Commercial sources of quantified EBV genomic DNA are now available that could potentially be used for assay calibration or for interlaboratory comparisons (eg, Advanced Biotechnologies Inc. [Columbia, MD], Accrometrix [Benicia, CA], Affigene [Bromma, Sweden], and ATCC's Namalwa Burkitt lymphoma cell line containing two integrated EBV genomes per cell [American Type Culture Collection, Manassas, VA]). In the future, international agreement on a reference standard and availability of FDA-approved kits will facilitate standardization of procedures across laboratories.
Despite wide variability in methods and the potential pitfalls of laboratory testing, multiple publications as well as proficiency survey data demonstrate that EBV viral load assays are sensitive, specific, precise, linear across a wide dynamic range, rapid, reasonably inexpensive, and useful in patient care. Prospective studies are needed to address methodologic concerns and also to refine the indications for testing and the use of test results in patient management.
EBV Viral Load in Infectious Mononucleosis
Circulating EBV DNA levels (in whole blood, plasma, serum, or memory B cells) spike within 2 weeks after primary EBV infection and then drop to low or undetectable over a period of several weeks to months.36,66,67,68,69,70 Interestingly, the kinetics vary from patient to patient and may be accompanied by a rebound after an initial decline.4 It may take a year or more to reach steady state,71 after which a healthy carrier's whole blood contains about 1 to 50 copies of EBV DNA per million white blood cells,1 whereas plasma or serum EBV DNA is nearly always undetectable.2,3,25,33,67
Laboratory confirmation of infectious mononucleosis should begin with the “Monospot” test followed, when necessary, by the more definitive serological assays described above. EBV DNA testing is reserved for atypical cases and for immunosuppressed patients.
EBV Viral Load in Chronic Active EBV Infection and Related Lymphoid Neoplasms
Chronic active EBV infection is a life-threatening disease that is more common in Asia than in western countries. It is characterized by persistent or recurrent infectious mononucleosis-like symptoms for at least 6 months along with atypical serology (high titers against several latent and lytic antigens with unexpected absence of EBNA antibody) and high EBV viral load in the absence of immunosuppression.72 Levels of EBV DNA in blood mononuclear cells correlate with disease severity.68 Interestingly, clonal EBV genomes are often found in histologically abnormal organs (liver, spleen, lymph nodes, marrow), even though multiple lineages of lymphocytes may be infected (B cells, helper and cytotoxic T cells, NK cells), leading to the hypothesis that neoplastic transformation of an infected lymphoid progenitor cell gave rise to cells of various lineages.73,74 Mutation in an immune-modulating gene such as perforin may be responsible for faulty control of EBV infection, resulting in chronic infection and subsequent clonal outgrowth of infected T and/or NK cells.75 Affected patients seem to lack LMP2-specific cytotoxic T cells, and replacement by infused EBV-specific T cells is being evaluated as a potential therapy.
EBV Viral Load in X-Linked Lymphoproliferative Disorder
An EBV-specific immunodeficiency, X-linked lymphoproliferative disorder, is caused by heritable mutation of SH2D1A rendering a child susceptible to fatal primary EBV infection or EBV-related lymphoma. Both innate (NK cell) and antigen-driven (T cell) immune responses appear to be deranged, resulting in failure to kill B lymphocytes appropriately and high EBV viral load. Clinical recognition of this rare syndrome is difficult but important given the divergent therapy required for sepsis, which has similar clinical features in young children.76
EBV Viral Load in Nasopharyngeal Carcinoma
Undifferentiated NPC is strongly associated with EBV in southeast Asia, where NPC is endemic, and to a lesser extent in western nations, where NPC is rare.77,78,79 Infection precedes malignant transformation as shown by clonal viral terminal repeat sequences as well as EBER expression in all neoplastic cells at the primary site and in distant metastases. Indeed, EBV infection serves as a marker of the neoplastic clone that can be capitalized on for diagnosis and management of affected patients.
The level of circulating EBV DNA in plasma or serum tracks with tumor stage at initial diagnosis and likewise serves as a marker of tumor burden during therapy.28,80,81,82,83,84 Early stage patients can be further subdivided into those at low versus high risk of relapse by evaluating their plasma EBV load.85 High pretreatment levels portend poor survival.59,80,82,85 In contrast, low or undetectable EBV DNA in plasma implies localized NPC, assuming that EBER in situ hybridization reveals that the primary carcinoma is EBV-related. High risk individuals from endemic areas (eg, adult family members of NPC patients from southeastern China) could potentially be screened for early NPC using EBV-directed laboratory methods (serology, Q-PCR or BARF1 gene products in nasopharyngeal brushings).52,86,87
Kinetic studies show that plasma EBV DNA becomes undetectable about 2 hours after surgical resection.88 When radiotherapy is used, the initial rise in plasma EBV DNA may reflect release of EBV DNA on tumor lysis, followed by declining EBV with a median half-life of 3.8 days.89 Patients with residual disease retained measurable EBV at 6 to 8 weeks, whereas those with undetectable plasma EBV maintained stable remission.82,89,90 Serial testing can predict relapse before it becomes symptomatic.28,81,82
EBV viral load is a better test of recurrence than is serology.28,59 This is not surprising given that serology provides indirect evidence of prior infection, whereas Q-PCR measures active disease. While distant relapse is usually marked by EBV DNA in plasma,91 local relapse is better assessed endoscopically using biopsy or perhaps less invasive tests like BARF1 RNA in nasopharyngeal brushings, BARF1 protein in saliva, or promoter hypermethylation that distinguishes carcinoma from “background lymphocyte” infection.49,52,53 Prospective clinical trials are needed to examine the utility of various laboratory approaches for diagnosing and managing NPC.
EBV Viral Load in Gastric Adenocarcinoma
EBV DNA is detectable within the malignant epithelial cells of about 10% of gastric adenocarcinomas, especially in those having a lymphoepithelioma-like appearance (ie, undifferentiated with abundant tumor-infiltrating lymphocytes) and in those arising in the stump after surgical gastrectromy.11,92 Surprisingly, little has been done to evaluate the role of EBV testing in diagnosis and monitoring of gastric cancer. A pilot study has indicated that serum EBV DNA levels are often elevated in patients whose cancer harbors EBV.93
EBV Viral Load in Transplant Recipients
Blood represents a convenient and noninvasive specimen type in which to evaluate EBV as a biomarker for PTLD. Affected patients have high levels of EBV DNA in both the cellular fraction of blood and in cell-free (serum or plasma) fractions.94 EBV DNA levels often rise before a mass lesion or symptoms become evident, implying that routine monitoring of high-risk patients may predict incipient PTLD.95,96,97,98 Early warning permits preemptive therapy to reverse disease progression.
Despite the high mortality rate of PTLD, it is costly to prospectively monitor EBV viral load in all allogeneic transplant recipients. Typically, only patients at highest risk of PTLD are screened on a regular basis. Risk factors include the degree of T-cell depletion (eg, anti-thymocyte globulin or fludarabine use), prior immunity as demonstrated by EBV serology before transplant, an HLA-mismatched or unrelated donor, and reduced intensity conditioning.99,100 High-risk patients are monitored as frequently as once per week in the first few months after transplant.100 Since low-level viremia does not correlate with progression to PTLD, some investigators have set cutoff levels beyond which they intervene.96,100,101,102,103,104 An alternative approach is to complement EBV DNA measurement with another predictive assay, and the following assays have been proposed in limited studies: absolute number of CD8 or CD4 T cells, EBV-specific T cell response as measured by a peptide tetramer assay or by ELISPOT, ATP release, EA serology, cytokine gene polymorphism, and viral gene expression pattern by real-time PCR.22,68,105,106,107,108,109,110,111 Successful preemptive intervention is accompanied by a drop in circulating EBV DNA.
Beyond its role in prevention of PTLD, EBV viral load testing is also indicated in any transplant recipient who presents with lymphadenopathy, fever, or other signs and symptoms suggestive of PTLD. A high EBV load should trigger the search for mass lesions or organ dysfunction pinpointing putative sites of disease, followed by biopsy to diagnose the lesion. Importantly, one should not diagnose infectious mononucleosis in an immunosuppressed transplant recipient since that diagnosis implies a self-limited process not requiring active intervention.
Early diagnosis is critical for successful clinical management of PTLD. Management is aimed primarily at restoring natural immune recognition and destruction of infected cells by reducing the degree of iatrogenic immunosuppression. Even monoclonal tumors may respond. Other therapies include CD20 antibody (eg, rituximab), donor lymphocyte infusion, infusion of EBV-specific cytotoxic T cells that were expanded ex vivo by exposing T cells to EBV antigens,112,113,114 radiation, and multidrug chemotherapy. Successful therapy for frank PTLD is accompanied by a rapid drop in plasma EBV levels reflecting diminished disease burden.94,98,115
CD20 antibody therapy may fail due to outgrowth of a neoplastic subclone lacking CD20.116,117 Indeed, EBV infection contributes to survival of defective B cells by rescuing them from programmed cell death despite their aberrant lack of CD20 or lack of immunoglobulin production.118,119 Crippled B lymphocytes have been identified in some cases of PTLD, Hodgkin lymphoma, AIDS lymphoma, pyothorax-associated lymphoma, and AILT.120,121
Rare PTLDs of T or NK cell lineage are less likely to be EBV-related compared to the more typical B lineage cases.122 EBV-negative PTLD tends to occur late after transplant, whereas nearly all PTLD occurring within the first year are EBV-related.122 While some EBV-negative tumors undoubtedly represent lymphoma of the conventional type, anecdotal reports of response to withdrawal of immunosuppression support using PTLD terminology for all lymphoid neoplasms occurring in immunosuppressed transplant recipients.98
There is controversy over whether plasma, whole blood, or blood mononuclear cells is the preferred specimen type, and all three appear to be informative to some extent.94,100,123,124,125,126,127 Plasma was more specific for lymphoproliferative disease in one study, and was more informative for monitoring efficacy of therapy in another study.94,128 Plasma is the specimen type evaluated in NPC patients and in the two commercial proficiency surveys (College of American Pathologists [Northfield, IL] and Quality Control for Molecular Diagnostics [Glasgow, Scotland]).
Guidelines for EBV testing in solid organ transplant recipients in clinical trials call for frequent monitoring (at least once a month) in the first year after transplantation when the risk of PTLD is highest.98,129 Afterward, continued monitoring should be considered for patients with a history of persistently high EBV loads or who are particularly immunosuppressed.129 The European Best Practice Guideline for Renal Transplantation calls for serological testing of EBV immune status before transplant, antiviral prophylaxis, pathologist diagnosis of PTLD, and use of blood levels to gauge treatment.130 Primary EBV infection is a contraindication to renal transplantation, while remote EBV infection is protective against PTLD.130 After transplant, those renal and lung recipients having asymptomatic EBV viremia are at risk for other adverse outcomes such as graft dysfunction, acute rejection, and late onset PTLD.131,132,133 Surprisingly, there was no correlation between EBV viral load and complications caused by other pathogens.131
EBV Viral Load in Hodgkin and Non-Hodgkin Lymphomas
EBV viral load in plasma serves as a marker of tumor burden in patients with sporadic EBV-related lymphoma including B cell, T cell, NK cell and Hodgkin subtypes.35,134,135,136 EBV DNA is detectable before cancer diagnosis, and the level of EBV DNA at diagnosis may predict outcome and efficacy of therapy.135,136 The findings argue in favor of routine plasma EBV viral load testing on initial diagnosis of Hodgkin lymphoma as well as any of the non-Hodgkin lymphomas that are likely to be EBV-related.
EBV Viral Load in Human Immunodeficiency Virus-Infected Patients
Immunocompromised hosts have higher baseline levels of circulating EBV than do healthy individuals.137,138 AIDS patients who subsequently develop EBV-related lymphoma have high blood and plasma EBV levels that fall on initiation of therapy.137,139 An exception is primary brain lymphoma; although EBV is nearly always present within the malignant cells, blood and plasma levels are not elevated because the blood-brain barrier limits dispersion.137,140 Instead, cerebrospinal fluid contains EBV DNA,40,141 and it has been suggested that cerebrospinal fluid EBV substitutes for brain biopsy in making a diagnosis of lymphoma in an AIDS patient who has clinical and radiographic evidence supporting the diagnosis.142 Serial EBV DNA levels in cerebrospinal fluid reflect therapeutic efficacy.143 Gancyclovir alone can lower EBV levels in cerebrospinal fluid,141 and gancyclovir may synergize with traditional antineoplastic therapy in managing a wide spectrum of EBV-related malignancies.144
Measuring EBV Viral Load in Biopsy Tissues
EBV DNA can be quantified by real-time PCR in paraffin-embedded tissue or in cytologic specimens.33 Advantages over EBER in situ hybridization are PCR's relatively low cost, its applicability to specimens with poor quality RNA, and its ability to detect (pure lytic) infection lacking EBER transcripts. It is important that the PCR assay be quantitative and that parallel Q-PCR of an endogenous human gene be used to normalize for the number of nucleated cells represented in the reaction.33
EBV load in NK/T lymphoma tissue is touted as a prognostic indicator.145 Likewise, a biopsy or fine needle aspirate that is suspicious for NPC contains high levels EBV if the cancer is indeed present and is EBV-related.33,146 On the other hand, low to undetectable EBV DNA by Q-PCR is consistent with scant to absent EBV-related neoplastic cells, or rare infected B lymphocytes that might be present in any viral carrier, emphasizing the need to interpret EBV Q-PCR results in the context of histopathological findings.
Future Perspectives
EBV is one of the best tumor markers yet discovered. EBV viral load testing has been incorporated into routine care of patients with PTLD, nasopharyngeal carcinoma, and AIDS lymphoma of the brain. An international effort is underway to establish a standard by which to calibrate EBV DNA measurement. Further evidence and consensus building is needed on clinical indications for testing, optimum specimen types and handling procedures, assay design and scope, units for reporting, thresholds for intervention, and management strategies. Serology is the best way to confirm a diagnosis of infectious mononucleosis, and EBER in situ hybridization is the single best histochemical assay for defining EBV-related neoplasia. It is likely that emerging technologies such as gene expression profiling and proteomics will identify patterns of viral and human gene expression correlating with diagnosis, prognosis, and outcome in response to therapy. A coordinated effort by basic scientists and clinical investigators will improve our arsenal of laboratory methods and better define their clinical utility.
References
- 1.Babcock GJ, Decker LL, Freeman RB, Thorley-Lawson DA. Epstein-Barr virus-infected resting memory B cells, not proliferating lymphoblasts, accumulate in the peripheral blood of immunosuppressed patients. J Exp Med. 1999;190:567–576. doi: 10.1084/jem.190.4.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maurmann S, Fricke L, Wagner HJ, Schlenke P, Hennig H, Steinhoff J, Jabs WJ. Molecular parameters for precise diagnosis of asymptomatic Epstein-Barr virus reactivation in healthy carriers. J Clin Microbiol. 2003;41:5419–5428. doi: 10.1128/JCM.41.12.5419-5428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gulley ML, Fan H, Elmore SH. Validation of Roche LightCycler Epstein-Barr virus quantification reagents in a clinical laboratory setting. J Mol Diagn. 2006;8:589–597. doi: 10.2353/jmoldx.2006.050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fafi-Kremer S, Morand P, Brion JP, Pavese P, Baccard M, Germi R, Genoulaz O, Nicod S, Jolivet M, Ruigrok RW, Stahl JP, Seigneurin JM. Long-term shedding of infectious Epstein-Barr virus after infectious mononucleosis. J Infect Dis. 2005;191:985–989. doi: 10.1086/428097. [DOI] [PubMed] [Google Scholar]
- 5.Faulkner GC, Burrows SR, Khanna R, Moss DJ, Bird AG, Crawford DH. X-Linked agammaglobulinemia patients are not infected with Epstein-Barr virus: implications for the biology of the virus. J Virol. 1999;73:1555–1564. doi: 10.1128/jvi.73.2.1555-1564.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frangou P, Buettner M, Niedobitek G. Epstein-Barr virus (EBV) infection in epithelial cells in vivo: rare detection of EBV replication in tongue mucosa but not in salivary glands. J Infect Dis. 2005;191:238–242. doi: 10.1086/426823. [DOI] [PubMed] [Google Scholar]
- 7.Halvorsen JA, Brevig T, Aas T, Skar AG, Slevolden EM, Moi H. Genital ulcers as initial manifestation of Epstein-Barr virus infection: two new cases and a review of the literature. Acta Dermatol Venereol. 2006;86:439–442. doi: 10.2340/00015555-0140. [DOI] [PubMed] [Google Scholar]
- 8.Thomas R, Macsween KF, McAulay K, Clutterbuck D, Anderson R, Reid S, Higgins CD, Swerdlow AJ, Harrison N, Williams H, Crawford DH. Evidence of shared Epstein-Barr viral isolates between sexual partners, and low level EBV in genital secretions. J Med Virol. 2006;78:1204–1209. doi: 10.1002/jmv.20682. [DOI] [PubMed] [Google Scholar]
- 9.Kusuhara K, Takabayashi A, Ueda K, Hidaka Y, Minamishima I, Take H, Fujioka K, Imai S, Osato T. Breast milk is not a significant source for early Epstein-Barr virus or human herpesvirus 6 infection in infants: a seroepidemiologic study in 2 endemic areas of human T-cell lymphotropic virus type I in Japan. Microbiol Immunol. 1997;41:309–312. doi: 10.1111/j.1348-0421.1997.tb01206.x. [DOI] [PubMed] [Google Scholar]
- 10.Raab-Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47:883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 11.Gulley ML, Pulitzer DR, Eagan PA, Schneider BG. Epstein-Barr virus infection is an early event in gastric carcinogenesis and is independent of bcl-2 expression and p53 accumulation. Hum Pathol. 1996;27:20–27. doi: 10.1016/s0046-8177(96)90133-1. [DOI] [PubMed] [Google Scholar]
- 12.Locker J, Nalesnik M. Molecular genetic analysis of lymphoid tumors arising after organ transplantation. Am J Pathol. 1989;135:977–987. [PMC free article] [PubMed] [Google Scholar]
- 13.Reisinger J, Rumpler S, Lion T, Ambros PF. Visualization of episomal and integrated Epstein-Barr virus DNA by fiber fluorescence in situ hybridization. Int J Cancer. 2006;118:1603–1608. doi: 10.1002/ijc.21498. [DOI] [PubMed] [Google Scholar]
- 14.Thorley-Lawson DA, Duca KA, Shapiro M. Epstein-Barr virus: a paradigm for persistent infection—for real and in virtual reality. Trends Immunol. 2008;29:195–201. doi: 10.1016/j.it.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc Natl Acad Sci USA. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niens M, Jarrett RF, Hepkema B, Nolte IM, Diepstra A, Platteel M, Kouprie N, Delury CP, Gallagher A, Visser L, Poppema S, te Meerman GJ, van den Berg A. HLA-A*02 is associated with a reduced risk and HLA-A*01 with an increased risk of developing EBV+ Hodgkin lymphoma. Blood. 2007;110:3310–3315. doi: 10.1182/blood-2007-05-086934. [DOI] [PubMed] [Google Scholar]
- 17.Keegan TH, Glaser SL, Clarke CA, Gulley ML, Craig FE, Digiuseppe JA, Dorfman RF, Mann RB, Ambinder RF. Epstein-Barr virus as a marker of survival after Hodgkin's lymphoma: a population-based study. J Clin Oncol. 2005;23:7604–7613. doi: 10.1200/JCO.2005.02.6310. [DOI] [PubMed] [Google Scholar]
- 18.Jarrett RF, Stark GL, White J, Angus B, Alexander FE, Krajewski AS, Freeland J, Taylor GM, Taylor PR. Impact of tumor Epstein-Barr virus status on presenting features and outcome in age-defined subgroups of patients with classic Hodgkin lymphoma: a population-based study. Blood. 2005;106:2444–2451. doi: 10.1182/blood-2004-09-3759. [DOI] [PubMed] [Google Scholar]
- 19.van Beek J, Brink AA, Vervoort MB, van Zijp MJ, Meijer CJ, van den Brule AJ, Middeldorp JM. In vivo transcription of the Epstein-Barr virus (EBV) BamHI-A region without associated in vivo BARF0 protein expression in multiple EBV-associated disorders. J Gen Virol. 2003;84:2647–2659. doi: 10.1099/vir.0.19196-0. [DOI] [PubMed] [Google Scholar]
- 20.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, Toomey NL, Brites C, Dittmer DP, Harrington WJ., Jr EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1–3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhaduri-McIntosh S, Rotenberg MJ, Gardner B, Robert M, Miller G. Repertoire and frequency of immune cells reactive to Epstein-Barr virus-derived autologous lymphoblastoid cell lines. Blood. 2008;111:1334–1343. doi: 10.1182/blood-2007-07-101907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeytinoglu A, Altuglu I, Karatas E, Yazan Sertoz R. Comparison of immunofluorescence assay and multiplexed microparticle-based immunoassay for detecting Epstein-Barr virus viral capsid antigen antibodies. J Virol Methods. 2008;148:300–302. doi: 10.1016/j.jviromet.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Gartner BC, Hess RD, Bandt D, Kruse A, Rethwilm A, Roemer K, Mueller-Lantzsch N. Evaluation of four commercially available Epstein-Barr virus enzyme immunoassays with an immunofluorescence assay as the reference method. Clin Diagn Lab Immunol. 2003;10:78–82. doi: 10.1128/CDLI.10.1.78-82.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luderer R, Kok M, Niesters HG, Schuurman R, de Weerdt O, Thijsen SF. Real-time Epstein-Barr virus PCR for the diagnosis of primary EBV infections and EBV reactivation. Mol Diagn. 2005;9:195–200. doi: 10.1007/BF03260091. [DOI] [PubMed] [Google Scholar]
- 26.de Sanjose S, Bosch R, Schouten T, Verkuijlen S, Nieters A, Foretova L, Maynadie M, Cocco PL, Staines A, Becker N, Brennan P, Benavente Y, Boffetta P, Meijer CJ, Middeldorp JM. Epstein-Barr virus infection and risk of lymphoma: immunoblot analysis of antibody responses against EBV-related proteins in a large series of lymphoma subjects and matched controls. Int J Cancer. 2007;121:1806–1812. doi: 10.1002/ijc.22857. [DOI] [PubMed] [Google Scholar]
- 27.Deng H, Zeng Y, Lei Y, Zhao Z, Wang P, Li B, Pi Z, Tan B, Zheng Y, Pan W. Serological survey of nasopharyngeal carcinoma in 21 cities of south China. Chinese Med Journal. 1995;108:300–303. [PubMed] [Google Scholar]
- 28.Fan H, Nicholls J, Chua D, Chan KH, Sham J, Lee S, Gulley ML. Laboratory markers of tumor burden in nasopharyngeal carcinoma: a comparison of viral load and serologic tests for Epstein-Barr virus. Int J Cancer. 2004;112:1036–1041. doi: 10.1002/ijc.20520. [DOI] [PubMed] [Google Scholar]
- 29.Paramita DK, Fachiroh J, Artama WT, van Benthem E, Haryana SM, Middeldorp JM. Native early antigen of Epstein-Barr virus, a promising antigen for diagnosis of nasopharyngeal carcinoma. J Med Virol. 2007;79:1710–1721. doi: 10.1002/jmv.20987. [DOI] [PubMed] [Google Scholar]
- 30.Chan KH, Gu YL, Ng F, Ng PS, Seto WH, Sham JS, Chua D, Wei W, Chen YL, Luk W, Zong YS, Ng MH. EBV specific antibody-based and DNA-based assays in serologic diagnosis of nasopharyngeal carcinoma. Int J Cancer. 2003;105:706–709. doi: 10.1002/ijc.11130. [DOI] [PubMed] [Google Scholar]
- 31.Tierney RJ, Edwards RH, Sitki-Green D, Croom-Carter D, Roy S, Yao QY, Raab-Traub N, Rickinson AB. Multiple Epstein-Barr virus strains in patients with infectious mononucleosis: comparison of ex vivo samples with in vitro isolates by use of heteroduplex tracking assays. J Infect Dis. 2006;193:287–297. doi: 10.1086/498913. [DOI] [PubMed] [Google Scholar]
- 32.Pai PC, Tseng CK, Chuang CC, Wei KC, Hao SP, Hsueh C, Chang KP, Tsang NM. Polymorphism of C-terminal activation region 2 of Epstein-Barr virus latent membrane protein 1 in predicting distant failure and post-metastatic survival in patients with nasopharyngeal carcinoma. Head Neck. 2007;29:109–119. doi: 10.1002/hed.20483. [DOI] [PubMed] [Google Scholar]
- 33.Ryan JL, Fan H, Glaser SL, Schichman SA, Raab-Traub N, Gulley ML. Epstein-Barr virus quantitation by real-time PCR targeting multiple gene segments: a novel approach to screen for the virus in paraffin-embedded tissue and plasma. J Mol Diagn. 2004;6:378–385. doi: 10.1016/S1525-1578(10)60535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan KC, Yeung SW, Lui WB, Rainer TH, Lo YM. Effects of preanalytical factors on the molecular size of cell-free DNA in blood. Clin Chem. 2005;51:781–784. doi: 10.1373/clinchem.2004.046219. [DOI] [PubMed] [Google Scholar]
- 35.Chan KC, Zhang J, Chan AT, Lei KI, Leung SF, Chan LY, Chow KC, Lo YM. Molecular characterization of circulating EBV DNA in the plasma of nasopharyngeal carcinoma and lymphoma patients. Cancer Res. 2003;63:2028–2032. [PubMed] [Google Scholar]
- 36.Ryan JL, Fan H, Swinnen LJ, Schichman SA, Raab-Traub N, Covington M, Elmore S, Gulley ML. Epstein-Barr Virus (EBV) DNA in plasma is not encapsidated in patients with EBV-related malignancies. Diagn Mol Pathol. 2004;13:61–68. doi: 10.1097/00019606-200406000-00001. [DOI] [PubMed] [Google Scholar]
- 37.Gulley ML, Glaser SL, Craig FE, Borowitz M, Mann RB, Shema SJ, Ambinder RF. Guidelines for interpreting EBER in situ hybridization and LMP1 immunohistochemical tests for detecting Epstein-Barr virus in Hodgkin lymphoma. Am J Clin Pathol. 2002;117:259–267. doi: 10.1309/MMAU-0QYH-7BHA-W8C2. [DOI] [PubMed] [Google Scholar]
- 38.Glaser SL, Hsu JL, Gulley ML. Epstein-Barr virus and breast cancer: state of the evidence for viral carcinogenesis. Cancer Epidemiol Biomarkers Prev. 2004;13:688–697. [PubMed] [Google Scholar]
- 39.Junying J, Herrmann K, Davies G, Lissauer D, Bell A, Timms J, Reynolds GM, Hubscher SG, Young LS, Niedobitek G, Murray PG. Absence of Epstein-Barr virus DNA in the tumor cells of European hepatocellular carcinoma. Virology. 2003;306:236–243. doi: 10.1016/s0042-6822(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 40.Ivers LC, Kim AY, Sax PE. Predictive value of polymerase chain reaction of cerebrospinal fluid for detection of Epstein-Barr virus to establish the diagnosis of HIV-related primary central nervous system lymphoma. Clin Infect Dis. 2004;38:1629–1632. doi: 10.1086/420934. [DOI] [PubMed] [Google Scholar]
- 41.Dupuis J, Emile JF, Mounier N, Gisselbrecht C, Martin-Garcia N, Petrella T, Bouabdallah R, Berger F, Delmer A, Coiffier B, Reyes F, Gaulard P. Prognostic significance of Epstein-Barr virus in nodal peripheral T-cell lymphoma, unspecified: a Groupe d'Etude des Lymphomes de l'Adulte (GELA) study. Blood. 2006;108:4163–4169. doi: 10.1182/blood-2006-04-017632. [DOI] [PubMed] [Google Scholar]
- 42.Attygalle AD, Chuang SS, Diss TC, Du MQ, Isaacson PG, Dogan A. Distinguishing angioimmunoblastic T-cell lymphoma from peripheral T-cell lymphoma, unspecified, using morphology, immunophenotype and molecular genetics. Histopathology. 2007;50:498–508. doi: 10.1111/j.1365-2559.2007.02632.x. [DOI] [PubMed] [Google Scholar]
- 43.Tan BT, Warnke RA, Arber DA. The frequency of B- and T-cell gene rearrangements and Epstein-Barr virus in T-cell lymphomas: a comparison between angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified with and without associated B-cell proliferations. J Mol Diagn. 2006;8:466–475. doi: 10.2353/jmoldx.2006.060016. quiz 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Y, Attygalle AD, Chuang SS, Diss T, Ye H, Liu H, Hamoudi RA, Munson P, Bacon CM, Dogan A, Du MQ. Angioimmunoblastic T-cell lymphoma: histological progression associates with EBV and HHV6B viral load. Br J Haematol. 2007;138:44–53. doi: 10.1111/j.1365-2141.2007.06620.x. [DOI] [PubMed] [Google Scholar]
- 45.Niedobitek G, Herbst H. In situ detection of Epstein-Barr virus and phenotype determination of EBV-infected cells. Methods Mol Biol. 2006;326:115–137. doi: 10.1385/1-59745-007-3:115. [DOI] [PubMed] [Google Scholar]
- 46.Kurokawa M, Ghosh SK, Ramos JC, Mian AM, Toomey NL, Cabral L, Whitby D, Barber GN, Dittmer DP, Harrington WJ., Jr Azidothymidine inhibits NF-kappaB and induces Epstein-Barr virus gene expression in Burkitt lymphoma. Blood. 2005;106:235–240. doi: 10.1182/blood-2004-09-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ballout M, Germi R, Fafi-Kremer S, Guimet J, Bargues G, Seigneurin JM, Morand P. Real-time quantitative PCR for assessment of antiviral drug effects against Epstein-Barr virus replication and EBV late mRNA expression. J Virol Methods. 2007;143:38–44. doi: 10.1016/j.jviromet.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Kubota N, Wada K, Ito Y, Shimoyama Y, Nakamura S, Nishiyama Y, Kimura H. One-step multiplex real-time PCR assay to analyse the latency patterns of Epstein-Barr virus infection. J Virol Methods. 2008;147:26–36. doi: 10.1016/j.jviromet.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 49.Stevens SJ, Verkuijlen SA, Hariwiyanto B, Harijadi, Paramita DK, Fachiroh J, Adham M, Tan IB, Haryana SM, Middeldorp JM. Noninvasive diagnosis of nasopharyngeal carcinoma: nasopharyngeal brushings reveal high Epstein-Barr virus DNA load and carcinoma-specific viral BARF1 mRNA. Int J Cancer. 2006;119:608–614. doi: 10.1002/ijc.21914. [DOI] [PubMed] [Google Scholar]
- 50.Hayes DP, Brink AA, Vervoort MB, Middeldorp JM, Meijer CJ, van den Brule AJ. Expression of Epstein-Barr virus (EBV) transcripts encoding homologues to important human proteins in diverse EBV associated diseases. Mol Pathol. 1999;52:97–103. doi: 10.1136/mp.52.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.zur Hausen A, Brink AA, Craanen ME, Middeldorp JM, Meijer CJ, van den Brule AJ. Unique transcription pattern of Epstein-Barr virus (EBV) in EBV-carrying gastric adenocarcinomas: expression of the transforming BARF1 gene. Cancer Res. 2000;60:2745–2748. [PubMed] [Google Scholar]
- 52.Houali K, Wang X, Shimizu Y, Djennaoui D, Nicholls J, Fiorini S, Bouguermouh A, Ooka T. A new diagnostic marker for secreted Epstein-Barr virus encoded LMP1 and BARF1 oncoproteins in the serum and saliva of patients with nasopharyngeal carcinoma. Clin Cancer Res. 2007;13:4993–5000. doi: 10.1158/1078-0432.CCR-06-2945. [DOI] [PubMed] [Google Scholar]
- 53.Tong JH, Tsang RK, Lo KW, Woo JK, Kwong J, Chan MW, Chang AR, van Hasselt CA, Huang DP, To KF. Quantitative Epstein-Barr virus DNA analysis and detection of gene promoter hypermethylation in nasopharyngeal (NP) brushing samples from patients with NP carcinoma. Clin Cancer Res. 2002;8:2612–2619. [PubMed] [Google Scholar]
- 54.Nicholls JM, Chua D, Chiu PM, Kwong DL. The detection of clinically occult nasopharyngeal carcinoma in patients following radiotherapy–an analysis of 69 patients. J Laryngol Otol. 1996;110:496–499. doi: 10.1017/s0022215100134097. [DOI] [PubMed] [Google Scholar]
- 55.Delecluse HJ, Feederle R, O'Sullivan B, Taniere P. Epstein Barr virus-associated tumours: an update for the attention of the working pathologist. J Clin Pathol. 2007;60:1358–1364. doi: 10.1136/jcp.2006.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hennard C, Pfuhl T, Buettner M, Becker KF, Knofel T, Middeldorp J, Kremmer E, Niedobitek G, Grasser F. The antibody 2B4 directed against the Epstein-Barr virus (EBV)-encoded nuclear antigen 1 (EBNA1) detects MAGE-4: implications for studies on the EBV association of human cancers. J Pathol. 2006;209:430–435. doi: 10.1002/path.1996. [DOI] [PubMed] [Google Scholar]
- 57.Jebbink J, Bai X, Rogers BB, Dawson DB, Scheuermann RH, Domiati-Saad R. Development of real-time PCR assays for the quantitative detection of Epstein-Barr virus and cytomegalovirus, comparison of TaqMan probes, and molecular beacons. J Mol Diagn. 2003;5:15–20. doi: 10.1016/S1525-1578(10)60446-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stevens SJ, Verkuijlen SA, Brule AJ, Middeldorp JM. Comparison of quantitative competitive PCR with LightCycler-based PCR for measuring Epstein-Barr virus DNA load in clinical specimens. J Clin Microbiol. 2002;40:3986–3992. doi: 10.1128/JCM.40.11.3986-3992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin JC, Wang WY, Liang WM, Chou HY, Jan JS, Jiang RS, Wang JY, Twu CW, Liang KL, Chao J, Shen WC. Long-term prognostic effects of plasma Epstein-Barr virus DNA by minor groove binder-probe real-time quantitative PCR on nasopharyngeal carcinoma patients receiving concurrent chemoradiotherapy. Int J Radiat Oncol Biol Phys. 2007;68:1342–1348. doi: 10.1016/j.ijrobp.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 60.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JD, Wengenack NL, Rosenblatt JE, Cockerill FR, 3rd, Smith TF. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fafi-Kremer S, Morand P, Barranger C, Bargues G, Magro S, Bes J, Bourgeois P, Joannes M, Seigneurin JM. Evaluation of the Epstein-Barr virus R-gene quantification kit in whole blood with different extraction methods and PCR platforms. J Mol Diagn. 2008;10:78–84. doi: 10.2353/jmoldx.2008.070054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hill CE, Harris SB, Culler EE, Zimring JC, Nolte FS, Caliendo AM. Performance characteristics of two real-time PCR assays for the quantification of Epstein-Barr virus DNA. Am J Clin Pathol. 2006;125:665–671. doi: 10.1309/ABEY-V2VK-E6DH-XAAA. [DOI] [PubMed] [Google Scholar]
- 63.Ruiz G, Pena P, de Ory F, Echevarria JE. Comparison of commercial real-time PCR assays for quantification of Epstein-Barr virus DNA. J Clin Microbiol. 2005;43:2053–2057. doi: 10.1128/JCM.43.5.2053-2057.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perandin F, Cariani E, Pollara CP, Manca N. Comparison of commercial and in-house Real-time PCR assays for quantification of Epstein-Barr virus (EBV) DNA in plasma. BMC Microbiol. 2007;7:22. doi: 10.1186/1471-2180-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayden RT, Hokanson KM, Pounds SB, Bankowski MJ, Belzer SW, Carr J, Diorio D, Forman MS, Joshi Y, Hillyard D, Hodinka RL, Nikiforova MN, Romain CA, Stevenson J, Valsamakis A, Balfour HH., Jr Multicenter comparison of different real-time PCR assays for quantitative detection of epstein-barr virus. J Clin Microbiol. 2008;46:157–163. doi: 10.1128/JCM.01252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng CC, Chang LY, Shao PL, Lee PI, Chen JM, Lu CY, Lee CY, Huang LM. Clinical manifestations and quantitative analysis of virus load in Taiwanese children with Epstein-Barr virus-associated infectious mononucleosis. J Microbiol Immunol Infect. 2007;40:216–221. [PubMed] [Google Scholar]
- 67.Bauer CC, Aberle SW, Popow-Kraupp T, Kapitan M, Hofmann H, Puchhammer-Stockl E. Serum Epstein-Barr virus DNA load in primary Epstein-Barr virus infection. J Med Virol. 2005;75:54–58. doi: 10.1002/jmv.20237. [DOI] [PubMed] [Google Scholar]
- 68.Yamashita N, Kimura H, Morishima T. Virological aspects of Epstein-Barr virus infections. Acta Med Okayama. 2005;59:239–246. doi: 10.18926/AMO/31961. [DOI] [PubMed] [Google Scholar]
- 69.Niesters HG. Molecular and diagnostic clinical virology in real time. Clin Microbiol Infect. 2004;10:5–11. doi: 10.1111/j.1469-0691.2004.00699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hadinoto V, Shapiro M, Greenough TC, Sullivan JL, Luzuriaga K, Thorley-Lawson DA. On the dynamics of acute EBV infection and the pathogenesis of infectious mononucleosis. Blood. 2008;111:1420–1427. doi: 10.1182/blood-2007-06-093278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Piriou E, van Dort K, Otto S, van Oers MH, van Baarle D. Tight regulation of the Epstein-Barr virus setpoint: interindividual differences in Epstein-Barr virus DNA load are conserved after HIV infection. Clin Infect Dis. 2008;46:313–316. doi: 10.1086/524079. [DOI] [PubMed] [Google Scholar]
- 72.Okano M, Kawa K, Kimura H, Yachie A, Wakiguchi H, Maeda A, Imai S, Ohga S, Kanegane H, Tsuchiya S, Morio T, Mori M, Yokota S, Imashuku S. Proposed guidelines for diagnosing chronic active Epstein-Barr virus infection. Am J Hematol. 2005;80:64–69. doi: 10.1002/ajh.20398. [DOI] [PubMed] [Google Scholar]
- 73.Kimura H, Hoshino Y, Hara S, Sugaya N, Kawada J, Shibata Y, Kojima S, Nagasaka T, Kuzushima K, Morishima T. Differences between T cell-type and natural killer cell-type chronic active Epstein-Barr virus infection. J Infect Dis. 2005;191:531–539. doi: 10.1086/427239. [DOI] [PubMed] [Google Scholar]
- 74.Endo R, Yoshioka M, Ebihara T, Ishiguro N, Kikuta H, Kobayashi K. Clonal expansion of multiphenotypic Epstein-Barr virus-infected lymphocytes in chronic active Epstein-Barr virus infection. Med Hypotheses. 2004;63:582–587. doi: 10.1016/j.mehy.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 75.Kimura H. Pathogenesis of chronic active Epstein-Barr virus infection: is this an infectious disease, lymphoproliferative disorder, or immunodeficiency? Rev Med Virol. 2006;16:251–261. doi: 10.1002/rmv.505. [DOI] [PubMed] [Google Scholar]
- 76.Mischler M, Fleming GM, Shanley TP, Madden L, Levine J, Castle V, Filipovich AH, Cornell TT. Epstein-Barr virus-induced hemophagocytic lymphohistiocytosis and X-linked lymphoproliferative disease: a mimicker of sepsis in the pediatric intensive care unit. Pediatrics. 2007;119:e1212–e1218. doi: 10.1542/peds.2006-1534. [DOI] [PubMed] [Google Scholar]
- 77.Gulley ML, Amin MB, Nicholls JM, Banks PM, Ayala AG, Srigley JR, Eagan PA, Ro JY. Epstein-Barr virus is detected in undifferentiated nasopharyngeal carcinoma but not in lymphoepithelioma-like carcinoma of the urinary bladder. Hum Pathol. 1995;26:1207–1214. doi: 10.1016/0046-8177(95)90195-7. [DOI] [PubMed] [Google Scholar]
- 78.Yip KW, Shi W, Pintilie M, Martin JD, Mocanu JD, Wong D, MacMillan C, Gullane P, O'Sullivan B, Bastianutto C, Liu FF. Prognostic significance of the Epstein-Barr virus, p53. Bcl-2, and survivin in nasopharyngeal cancer. Clin Cancer Res. 2006;12:5726–5732. doi: 10.1158/1078-0432.CCR-06-0571. [DOI] [PubMed] [Google Scholar]
- 79.Bortolin MT, Pratesi C, Dolcetti R, Bidoli E, Vaccher E, Zanussi S, Tedeschi R, De Paoli P. Clinical value of Epstein-Barr virus DNA levels in peripheral blood samples of Italian patients with undifferentiated carcinoma of nasopharyngeal type. Cancer Lett. 2006;233:247–254. doi: 10.1016/j.canlet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 80.Leung SF, Chan AT, Zee B, Ma B, Chan LY, Johnson PJ, Lo YM. Pretherapy quantitative measurement of circulating Epstein-Barr virus DNA is predictive of posttherapy distant failure in patients with early-stage nasopharyngeal carcinoma of undifferentiated type. Cancer. 2003;98:288–291. doi: 10.1002/cncr.11496. [DOI] [PubMed] [Google Scholar]
- 81.Lo YMD, Chan LYS, A.T.C. C, S.F. L, Lo KW, Zhang J, Lee JCK, Hjelm NM, Johnson PJ, Huang DP. Quantitative and temporal correlation between circulating cell-free Epstein-Barr virus DNA and tumor recurrence in nasopharyngeal carcinoma. Cancer Res. 1999;59:1188–1191. [PubMed] [Google Scholar]
- 82.Lin JC, Wang WY, Chen KY, Wei YH, Liang WM, Jan JS, Jiang RS. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med. 2004;350:2461–2470. doi: 10.1056/NEJMoa032260. [DOI] [PubMed] [Google Scholar]
- 83.Ma BB, King A, Lo YM, Yau YY, Zee B, Hui EP, Leung SF, Mo F, Kam MK, Ahuja A, Kwan WH, Chan AT. Relationship between pretreatment level of plasma Epstein-Barr virus DNA, tumor burden, and metabolic activity in advanced nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys. 2006;66:714–720. doi: 10.1016/j.ijrobp.2006.05.064. [DOI] [PubMed] [Google Scholar]
- 84.Chan KC, Lo YM. Clinical applications of plasma Epstein-Barr virus DNA analysis and protocols for the quantitative analysis of the size of circulating Epstein-Barr virus DNA. Methods Mol Biol. 2006;336:111–121. doi: 10.1385/1-59745-074-X:111. [DOI] [PubMed] [Google Scholar]
- 85.Leung SF, Zee B, Ma BB, Hui EP, Mo F, Lai M, Chan KC, Chan LY, Kwan WH, Lo YM, Chan AT. Plasma Epstein-Barr viral deoxyribonucleic acid quantitation complements tumor-node-metastasis staging prognostication in nasopharyngeal carcinoma. J Clin Oncol. 2006;24:5414–5418. doi: 10.1200/JCO.2006.07.7982. [DOI] [PubMed] [Google Scholar]
- 86.Leung SF, Tam JS, Chan AT, Zee B, Chan LY, Huang DP, Van Hasselt A, Johnson PJ, Lo YM. Improved accuracy of detection of nasopharyngeal carcinoma by combined application of circulating Epstein-Barr virus DNA and anti-Epstein-Barr viral capsid antigen IgA antibody. Clin Chem. 2004;50:339–345. doi: 10.1373/clinchem.2003.022426. [DOI] [PubMed] [Google Scholar]
- 87.Yang X, Goldstein AM, Chen CJ, Rabkin CS, Chen JY, Cheng YJ, Hsu WL, Sun B, Diehl SR, Liu MY, Walters M, Shao W, Ortiz-Conde BA, Whitby D, Elmore SH, Gulley ML, Hildesheim A. Distribution of Epstein-Barr viral load in serum of individuals from nasopharyngeal carcinoma high-risk families in Taiwan. Int J Cancer. 2006;118:780–784. doi: 10.1002/ijc.21396. [DOI] [PubMed] [Google Scholar]
- 88.To EW, Chan KC, Leung SF, Chan LY, To KF, Chan AT, Johnson PJ, Lo YM. Rapid clearance of plasma Epstein-Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin Cancer Res. 2003;9:3254–3259. [PubMed] [Google Scholar]
- 89.Lo YM, Leung SF, Chan LY, Chan AT, Lo KW, Johnson PJ, Huang DP. Kinetics of plasma Epstein-Barr virus DNA during radiation therapy for nasopharyngeal carcinoma. Cancer Res. 2000;60:2351–2355. [PubMed] [Google Scholar]
- 90.Chan AT, Lo YM, Zee B, Chan LY, Ma BB, Leung SF, Mo F, Lai M, Ho S, Huang DP, Johnson PJ. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma. J Natl Cancer Inst. 2002;94:1614–1619. doi: 10.1093/jnci/94.21.1614. [DOI] [PubMed] [Google Scholar]
- 91.Leung SF, Lo YM, Chan AT, To KF, To E, Chan LY, Zee B, Huang DP, Johnson PJ. Disparity of sensitivities in detection of radiation-naive and postirradiation recurrent nasopharyngeal carcinoma of the undifferentiated type by quantitative analysis of circulating Epstein-Barr virus DNA1,2. Clin Cancer Res. 2003;9:3431–3434. [PubMed] [Google Scholar]
- 92.Zur Hausen A, van Rees BP, van Beek J, Craanen ME, Bloemena E, Offerhaus GJ, Meijer CJ, van den Brule AJ. Epstein-Barr virus in gastric carcinomas and gastric stump carcinomas: a late event in gastric carcinogenesis. J Clin Pathol. 2004;57:487–491. doi: 10.1136/jcp.2003.014068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lo YM, Chan WY, Ng EK, Chan LY, Lai PB, Tam JS, Chung SC. Circulating Epstein-Barr virus DNA in the serum of patients with gastric carcinoma. Clin Cancer Res. 2001;7:1856–1859. [PubMed] [Google Scholar]
- 94.Wagner HJ, Wessel M, Jabs W, Smets F, Fischer L, Offner G, Bucsky P. Patients at risk for development of posttransplant lymphoproliferative disorder: plasma versus peripheral blood mononuclear cells as material for quantification of Epstein-Barr viral load by using real-time quantitative polymerase chain reaction. Transplantation. 2001;72:1012–1019. doi: 10.1097/00007890-200109270-00006. [DOI] [PubMed] [Google Scholar]
- 95.Niesters HG, van Esser J, Fries E, Wolthers KC, Cornelissen J, Osterhaus AD. Development of a real-time quantitative assay for detection of Epstein-Barr virus. J Clin Microbiol. 2000;38:712–715. doi: 10.1128/jcm.38.2.712-715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Orentas RJ, Schauer DW, Jr, Ellis FW, Walczak J, Casper JT, Margolis DA. Monitoring and modulation of Epstein-Barr virus loads in pediatric transplant patients. Pediatr Transplant. 2003;7:305–314. doi: 10.1034/j.1399-3046.2003.00090.x. [DOI] [PubMed] [Google Scholar]
- 97.D'Antiga L, Del Rizzo M, Mengoli C, Cillo U, Guariso G, Zancan L. Sustained Epstein-Barr virus detection in paediatric liver transplantation. Insights into the occurrence of late PTLD. Liver Transplant. 2007;13:343–348. doi: 10.1002/lt.20958. [DOI] [PubMed] [Google Scholar]
- 98.Green M. Management of Epstein-Barr virus-induced post-transplant lymphoproliferative disease in recipients of solid organ transplantation. Am J Transplant. 2001;1:103–108. [PubMed] [Google Scholar]
- 99.Schubert S, Renner C, Hammer M, Abdul-Khaliq H, Lehmkuhl HB, Berger F, Hetzer R, Reinke P. Relationship of immunosuppression to Epstein-Barr viral load and lymphoproliferative disease in pediatric heart transplant patients. J Heart Lung Transplant. 2008;27:100–105. doi: 10.1016/j.healun.2007.09.027. [DOI] [PubMed] [Google Scholar]
- 100.Kinch A, Oberg G, Arvidson J, Falk KI, Linde A, Pauksens K. Post-transplant lymphoproliferative disease and other Epstein-Barr virus diseases in allogeneic haematopoietic stem cell transplantation after introduction of monitoring of viral load by polymerase chain reaction. Scand J Infect Dis. 2007;39:235–244. doi: 10.1080/00365540600978906. [DOI] [PubMed] [Google Scholar]
- 101.Aalto SM, Juvonen E, Tarkkanen J, Volin L, Haario H, Ruutu T, Hedman K. Epstein-Barr viral load and disease prediction in a large cohort of allogeneic stem cell transplant recipients. Clin Infect Dis. 2007;45:1305–1309. doi: 10.1086/522531. [DOI] [PubMed] [Google Scholar]
- 102.Lee TC, Savoldo B, Rooney CM, Heslop HE, Gee AP, Caldwell Y, Barshes NR, Scott JD, Bristow LJ, O'Mahony CA, Goss JA. Quantitative EBV viral loads and immunosuppression alterations can decrease PTLD incidence in pediatric liver transplant recipients. Am J Transplant. 2005;5:2222–2228. doi: 10.1111/j.1600-6143.2005.01002.x. [DOI] [PubMed] [Google Scholar]
- 103.van Esser JW, Niesters HG, van der Holt B, Meijer E, Osterhaus AD, Gratama JW, Verdonck LF, Lowenberg B, Cornelissen JJ. Prevention of Epstein-Barr virus-lymphoproliferative disease by molecular monitoring and preemptive rituximab in high-risk patients after allogeneic stem cell transplantation. Blood. 2002;99:4364–4369. doi: 10.1182/blood.v99.12.4364. [DOI] [PubMed] [Google Scholar]
- 104.Bakker NA, Verschuuren EA, Erasmus ME, Hepkema BG, Veeger NJ, Kallenberg CG, van der Bij W. Epstein-Barr virus-DNA load monitoring late after lung transplantation: a surrogate marker of the degree of immunosuppression and a safe guide to reduce immunosuppression. Transplantation. 2007;83:433–438. doi: 10.1097/01.tp.0000252784.60159.96. [DOI] [PubMed] [Google Scholar]
- 105.Sebelin-Wulf K, Nguyen TD, Oertel S, Papp-Vary M, Trappe RU, Schulzki A, Pezzutto A, Riess H, Subklewe M. Quantitative analysis of EBV-specific CD4/CD8 T cell numbers, absolute CD4/CD8 T cell numbers and EBV load in solid organ transplant recipients with PLTD. Transplant Immunol. 2007;17:203–210. doi: 10.1016/j.trim.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 106.Qu L, Green M, Webber S, Reyes J, Ellis D, Rowe D. Epstein-Barr virus gene expression in the peripheral blood of transplant recipients with persistent circulating virus loads. J Infect Dis. 2000;182:1013–1021. doi: 10.1086/315828. [DOI] [PubMed] [Google Scholar]
- 107.Lee TC, Goss JA, Rooney CM, Heslop HE, Barshes NR, Caldwell YM, Gee AP, Scott JD, Savoldo B. Quantification of a low cellular immune response to aid in identification of pediatric liver transplant recipients at high-risk for EBV infection. Clin Transplant. 2006;20:689–694. doi: 10.1111/j.1399-0012.2006.00537.x. [DOI] [PubMed] [Google Scholar]
- 108.Smets F, Latinne D, Bazin H, Reding R, Otte JB, Buts JP, Sokal EM. Ratio between Epstein-Barr viral load and anti-Epstein-Barr virus specific T-cell response as a predictive marker of posttransplant lymphoproliferative disease. Transplantation. 2002;73:1603–1610. doi: 10.1097/00007890-200205270-00014. [DOI] [PubMed] [Google Scholar]
- 109.Weinberger B, Plentz A, Weinberger KM, Hahn J, Holler E, Jilg W. Quantitation of Epstein-Barr virus mRNA using reverse transcription and real-time PCR. J Med Virol. 2004;74:612–618. doi: 10.1002/jmv.20220. [DOI] [PubMed] [Google Scholar]
- 110.Meij P, van Esser JW, Niesters HG, van Baarle D, Miedema F, Blake N, Rickinson AB, Leiner I, Pamer E, Lowenberg B, Cornelissen JJ, Gratama JW. Impaired recovery of Epstein-Barr virus (EBV)–specific CD8+ T lymphocytes after partially T-depleted allogeneic stem cell transplantation may identify patients at very high risk for progressive EBV reactivation and lymphoproliferative disease. Blood. 2003;101:4290–4297. doi: 10.1182/blood-2002-10-3001. [DOI] [PubMed] [Google Scholar]
- 111.Carpentier L, Tapiero B, Alvarez F, Viau C, Alfieri C. Epstein-Barr virus (EBV) early-antigen serologic testing in conjunction with peripheral blood EBV DNA load as a marker for risk of posttransplantation lymphoproliferative disease. J Infect Dis. 2003;188:1853–1864. doi: 10.1086/379834. [DOI] [PubMed] [Google Scholar]
- 112.Gandhi MK, Wilkie GM, Dua U, Mollee PN, Grimmett K, Williams T, Whitaker N, Gill D, Crawford DH. Immunity, homing and efficacy of allogeneic adoptive immunotherapy for posttransplant lymphoproliferative disorders. Am J Transplant. 2007;7:1293–1299. doi: 10.1111/j.1600-6143.2007.01796.x. [DOI] [PubMed] [Google Scholar]
- 113.Haque T, Wilkie GM, Jones MM, Higgins CD, Urquhart G, Wingate P, Burns D, McAulay K, Turner M, Bellamy C, Amlot PL, Kelly D, MacGilchrist A, Gandhi MK, Swerdlow AJ, Crawford DH. Allogeneic cytotoxic T-cell therapy for EBV-positive posttransplantation lymphoproliferative disease: results of a phase 2 multicenter clinical trial. Blood. 2007;110:1123–1131. doi: 10.1182/blood-2006-12-063008. [DOI] [PubMed] [Google Scholar]
- 114.Savoldo B, Goss JA, Hammer MM, Zhang L, Lopez T, Gee AP, Lin YF, Quiros-Tejeira RE, Reinke P, Schubert S, Gottschalk S, Finegold MJ, Brenner MK, Rooney CM, Heslop HE. Treatment of solid organ transplant recipients with autologous Epstein Barr virus-specific cytotoxic T lymphocytes (CTLs) Blood. 2006;108:2942–2949. doi: 10.1182/blood-2006-05-021782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.van Esser JW, Niesters HG, Thijsen SF, Meijer E, Osterhaus AD, Wolthers KC, Boucher CA, Gratama JW, Budel LM, van der Holt B, van Loon AM, Lowenberg B, Verdonck LF, Cornelissen JJ. Molecular quantification of viral load in plasma allows for fast and accurate prediction of response to therapy of Epstein-Barr virus-associated lymphoproliferative disease after allogeneic stem cell transplantation. Br Journal of Haematol. 2001;113:814–821. doi: 10.1046/j.1365-2141.2001.02789.x. [DOI] [PubMed] [Google Scholar]
- 116.Gulley ML, Swinnen LJ, Plaisance KT, Jr, Schnell C, Grogan TM, Schneider BG. Tumor origin and CD20 expression in posttransplant lymphoproliferative disorder occurring in solid organ transplant recipients: implications for immune-based therapy. Transplantation. 2003;76:959–964. doi: 10.1097/01.TP.0000079832.00991.EE. [DOI] [PubMed] [Google Scholar]
- 117.Verschuuren EA, Stevens SJ, van Imhoff GW, Middeldorp JM, de Boer C, Koeter G, The TH, van Der Bij W. Treatment of posttransplant lymphoproliferative disease with rituximab: the remission, the relapse, and the complication. Transplantation. 2002;73:100–104. doi: 10.1097/00007890-200201150-00019. [DOI] [PubMed] [Google Scholar]
- 118.Buettner M, Greiner A, Avramidou A, Jack HM, Niedobitek G. Evidence of abortive plasma cell differentiation in Hodgkin and Reed-Sternberg cells of classical Hodgkin lymphoma. Hematol Oncol. 2005;23:127–132. doi: 10.1002/hon.764. [DOI] [PubMed] [Google Scholar]
- 119.Schauer E, Webber S, Green M, Rowe D. Surface immunoglobulin-deficient Epstein-Barr virus-infected B cells in the peripheral blood of pediatric solid-organ transplant recipients. J Clin Microbiol. 2004;42:5802–5810. doi: 10.1128/JCM.42.12.5802-5810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brauninger A, Schmitz R, Bechtel D, Renne C, Hansmann ML, Kuppers R. Molecular biology of Hodgkin's and Reed/Sternberg cells in Hodgkin's lymphoma. Int J Cancer. 2006;118:1853–1861. doi: 10.1002/ijc.21716. [DOI] [PubMed] [Google Scholar]
- 121.Takakuwa T, Tresnasari K, Rahadiani N, Miwa H, Daibata M, Aozasa K. Cell origin of pyothorax-associated lymphoma: a lymphoma strongly associated with Epstein-Barr virus infection. Leukemia. 2008;22:620–627. doi: 10.1038/sj.leu.2405059. [DOI] [PubMed] [Google Scholar]
- 122.Swerdlow SH. T-cell and NK-cell posttransplantation lymphoproliferative disorders. Am J Clin Pathol. 2007;127:887–895. doi: 10.1309/LYXN3RGF7D7KPYG0. [DOI] [PubMed] [Google Scholar]
- 123.Bakker NA, Verschuuren EA, Veeger NJ, van der Bij W, van Imhoff GW, Kallenberg CG, Hepkema BG. Quantification of Epstein-Barr virus-DNA load in lung transplant recipients: a comparison of plasma versus whole blood. J Heart Lung Transplant. 2008;27:7–10. doi: 10.1016/j.healun.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 124.Stevens SJ, Pronk I, Middeldorp JM. Toward standardization of Epstein-Barr virus DNA load monitoring: unfractionated whole blood as preferred clinical specimen. J Clin Microbiol. 2001;39:1211–1216. doi: 10.1128/JCM.39.4.1211-1216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wada K, Kubota N, Ito Y, Yagasaki H, Kato K, Yoshikawa T, Ono Y, Ando H, Fujimoto Y, Kiuchi T, Kojima S, Nishiyama Y, Kimura H. Simultaneous quantification of Epstein-Barr virus, cytomegalovirus, and human herpesvirus 6 DNA in samples from transplant recipients by multiplex real-time PCR assay. J Clin Microbiol. 2007;45:1426–1432. doi: 10.1128/JCM.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matsukura T, Yokoi A, Egawa H, Kudo T, Kawashima M, Hirata Y, Tanaka H, Kagajo K, Wada H, Tanaka K. Significance of serial real-time PCR monitoring of EBV genome load in living donor liver transplantation. Clin Transplant. 2002;16:107–112. doi: 10.1034/j.1399-0012.2002.1o112.x. [DOI] [PubMed] [Google Scholar]
- 127.Wadowsky RM, Laus S, Green M, Webber SA, Rowe D. Measurement of Epstein-Barr virus DNA loads in whole blood and plasma by TaqMan PCR and in peripheral blood lymphocytes by competitive PCR. J Clin Microbiol. 2003;41:5245–5249. doi: 10.1128/JCM.41.11.5245-5249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oertel S, Trappe RU, Zeidler K, Babel N, Reinke P, Hummel M, Jonas S, Papp-Vary M, Subklewe M, Dorken B, Riess H, Gartner B. Epstein-Barr viral load in whole blood of adults with posttransplant lymphoproliferative disorder after solid organ transplantation does not correlate with clinical course. Ann Hematol. 2006;85:478–484. doi: 10.1007/s00277-006-0109-1. [DOI] [PubMed] [Google Scholar]
- 129.Humar A, Michaels M. American Society of Transplantation recommendations for screening, monitoring and reporting of infectious complications in immunosuppression trials in recipients of organ transplantation. Am J Transplant. 2006;6:262–274. doi: 10.1111/j.1600-6143.2005.01207.x. [DOI] [PubMed] [Google Scholar]
- 130.European Best Practice Guidelines for Renal Transplantation: Section IV: Long-term management of the transplant recipient. IV. 6.1. Cancer risk after renal transplantation. Post-transplant lymphoproliferative disease (PTLD): prevention and treatment. Nephrol Dial Transplant. 2002;17(Suppl 4):31–33. doi: 10.1093/ndt/17.suppl_4.31. 35–36. [DOI] [PubMed] [Google Scholar]
- 131.Ahya VN, Douglas LP, Andreadis C, Arnoldi S, Svoboda J, Kotloff RM, Hadjiliadis D, Sager JS, Woo YJ, Pochettino A, Schuster SJ, Stadtmauer EA, Tsai DE. Association between elevated whole blood Epstein-Barr virus (EBV)-encoded RNA EBV polymerase chain reaction and reduced incidence of acute lung allograft rejection. J Heart Lung Transplant. 2007;26:839–844. doi: 10.1016/j.healun.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 132.Bingler MA, Feingold B, Miller SA, Quivers E, Michaels MG, Green M, Wadowsky RM, Rowe DT, Webber SA. Chronic high Epstein-Barr viral load state and risk for late-onset posttransplant lymphoproliferative disease/lymphoma in children. Am J Transplant. 2008;8:442–445. doi: 10.1111/j.1600-6143.2007.02080.x. [DOI] [PubMed] [Google Scholar]
- 133.Li L, Chaudhuri A, Weintraub LA, Hsieh F, Shah S, Alexander S, Salvatierra O, Jr, Sarwal MM. Subclinical cytomegalovirus and Epstein-Barr virus viremia are associated with adverse outcomes in pediatric renal transplantation. Pediatr Transplant. 2007;11:187–195. doi: 10.1111/j.1399-3046.2006.00641.x. [DOI] [PubMed] [Google Scholar]
- 134.Wagner HJ, Schlager F, Claviez A, Bucsky P. Detection of Epstein-Barr virus DNA in peripheral blood of paediatric patients with Hodgkin's disease by real-time polymerase chain reaction. Eur J Cancer. 2001;37:1853–1857. doi: 10.1016/s0959-8049(01)00152-6. [DOI] [PubMed] [Google Scholar]
- 135.Au WY, Pang A, Choy C, Chim CS, Kwong YL. Quantification of circulating Epstein-Barr virus (EBV) DNA in the diagnosis and monitoring of natural killer cell and EBV-positive lymphomas in immunocompetent patients. Blood. 2004;104:243–249. doi: 10.1182/blood-2003-12-4197. [DOI] [PubMed] [Google Scholar]
- 136.Gandhi MK, Lambley E, Burrows J, Dua U, Elliott S, Shaw PJ, Prince HM, Wolf M, Clarke K, Underhill C, Mills T, Mollee P, Gill D, Marlton P, Seymour JF, Khanna R. Plasma Epstein-Barr virus (EBV) DNA is a biomarker for EBV-positive Hodgkin's lymphoma. Clin Cancer Res. 2006;12:460–464. doi: 10.1158/1078-0432.CCR-05-2008. [DOI] [PubMed] [Google Scholar]
- 137.Fan H, Kim SC, Chima CO, Israel BF, Lawless KM, Eagan PA, Elmore S, Moore DT, Schichman SA, Swinnen LJ, Gulley ML. Epstein-Barr viral load as a marker of lymphoma in AIDS patients. J Med Virol. 2005;75:59–69. doi: 10.1002/jmv.20238. [DOI] [PubMed] [Google Scholar]
- 138.Stevens SJ, Smits PH, Verkuijlen SA, Rockx DA, van Gorp EC, Mulder JW, Middeldorp JM. Aberrant Epstein-Barr virus persistence in HIV carriers is characterized by anti-Epstein-Barr virus IgA and high cellular viral loads with restricted transcription. AIDS. 2007;21:2141–2149. doi: 10.1097/QAD.0b013e3282eeeba0. [DOI] [PubMed] [Google Scholar]
- 139.Bonnet F, Jouvencel AC, Parrens M, Leon MJ, Cotto E, Garrigue I, Morlat P, Beylot J, Fleury H, Lafon ME. A longitudinal and prospective study of Epstein-Barr virus load in AIDS-related non-Hodgkin lymphoma. J Clin Virol. 2006;36:258–263. doi: 10.1016/j.jcv.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 140.Fellner MD, Durand K, Correa RM, Redini L, Yampolsky C, Colobraro A, Sevlever G, Teyssie AR, Benetucci J, Picconi MA. Circulating Epstein-Barr virus (EBV) in HIV-infected patients and its relation with primary brain lymphoma. Int J Infect Dis. 2007;11:172–178. doi: 10.1016/j.ijid.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 141.Bossolasco S, Falk KI, Ponzoni M, Ceserani N, Crippa F, Lazzarin A, Linde A, Cinque P. Ganciclovir is associated with low or undetectable Epstein-Barr virus DNA load in cerebrospinal fluid of patients with HIV-related primary central nervous system lymphoma. Clin Infect Dis. 2006;42:e21–e25. doi: 10.1086/499956. [DOI] [PubMed] [Google Scholar]
- 142.Antinori A, De Rossi G, Ammassari A, Cingolani A, Murri R, Di Giuda D, De Luca A, Pierconti F, Tartaglione T, Scerrati M, Larocca LM, Ortona L. Value of combined approach with thallium-201 single-photon emission computed tomography and Epstein-Barr virus DNA polymerase chain reaction in CSF for the diagnosis of AIDS-related primary CNS lymphoma. J Clin Oncol. 1999;17:554–560. doi: 10.1200/JCO.1999.17.2.554. [DOI] [PubMed] [Google Scholar]
- 143.Antinori A, Cingolani A, De Luca A, Gaidano G, Ammassari A, Larocca LM, Ortona L. Epstein-Barr virus in monitoring the response to therapy of acquired immunodeficiency syndrome-related primary central nervous system lymphoma. Ann Neurol. 1999;45:259–261. [PubMed] [Google Scholar]
- 144.Israel BF, Kenney SC. Virally targeted therapies for EBV-associated malignancies. Oncogene. 2003;22:5122–5130. doi: 10.1038/sj.onc.1206548. [DOI] [PubMed] [Google Scholar]
- 145.Hsieh PP, Tung CL, Chan AB, Liao JB, Wang JS, Tseng HH, Su HH, Chang KC, Chang CC. EBV viral load in tumor tissue is an important prognostic indicator for nasal NK/T-cell lymphoma. Am J Clin Pathol. 2007;128:579–584. doi: 10.1309/MN4Y8HLQWKD9NB5E. [DOI] [PubMed] [Google Scholar]
- 146.Yap YY, Hassan S, Chan M, Choo PK, Ravichandran M. Epstein-Barr virus DNA detection in the diagnosis of nasopharyngeal carcinoma. Otolaryngol Head Neck Surg. 2007;136:986–991. doi: 10.1016/j.otohns.2006.11.027. [DOI] [PubMed] [Google Scholar]


