Abstract
To confirm studies suggesting that HIV-1 infects neurons and to determine whether CD8+ T lymphocytes traffic to HIV-1-infected neurons, we used laser capture microdissection to remove hippocampal neurons with and without perineuronal CD8+ T cells from AIDS patients with HIV-1 encephalitis (HIVE) or without HIVE and from normal controls. We used hyperbranched multidisplacement amplification for whole gene amplification (MDA-WGA) plus two rounds of PCR to amplify housekeeping sequences (HK+) and, in HK+ samples, to amplify HIV-1 gag, nef, and pol sequences. Sample size and, in single neurons, MDA-WGA correlated with housekeeping gene amplification (P < 0.05), whereas patient group and postmortem interval did not (P > 0.05). Neuronal viral sequences correlated with HIVE (43% vs. 13% and 0 in non-HIVE and controls, respectively) and, in HIVE cases, with perineuronal CD8+ T lymphocytes (70% in CD8+ samples vs. 37% of CD8− samples). Our results suggest that MDA-WGA is a useful technique when analyzing DNA from single cells from autopsy brains, supporting prior studies that show that neurons may contain HIV-1 neuronal sequences in vivo. The association between neuronal infection and perineuronal CD8+ T cells supports our hypothesis that these cells specifically traffic to infected neurons but raises the possibility that CD8+ T cells, if infected, could transmit virus to neurons.
Activated macrophages, viral proteins, and restricted HIV-1 infection of astrocytes and neurons are among the mechanisms that may cause HIV-1-induced brain injury.1,2 Although fetally derived neuronal cells in vitro transiently can sustain productive infection when exposed to HIV-1,3,4 actual infection in vivo is more controversial since studies examining neuronal infection in autopsy brains give conflicting results. Two initial studies detected viral sequences in neurons in tissue sections of AIDS brains by combining in situ hybridization with gene amplification by PCR5,6; subsequent studies, using similar techniques, were negative for neuronal HIV-1.7,8,9 More recently, Canto- Nogules et al10 used reverse transcription-PCR and in situ hybridization on paraffin-embedded brain sections and detected viral RNA in cortical neurons in two of four pediatric patients with AIDS-related encephalopathy.
We also examined the potential for neuronal infection but used laser capture microdissection to remove small groups of neurons from autopsy brain samples of AIDS and control patients. After extracting neuronal DNA, we detected HIV-1 gene sequences in many of the AIDS neuronal samples after double-round PCR amplification.11 Neuronal as well as glial and monocytic viral infection was found with similar technology of microdissection and PCR amplification in one subsequent study12 but not in a second.13
In the present study, we reasoned that amplification of viral sequences in single neurons would provide additional evidence of neuronal infection since it would minimize the likelihood of specimen contamination by adjacent infected cells. We also theorized that separate microdissections of neurons with, and without, perineuronal CD8+ T cells would allow us to determine whether the perineuronal T cells in AIDS brains specifically trafficked to infected neurons, as we previously hypothesized.14 Since the amount of DNA in single microdissected neurons is low, and since autopsy DNA already is reduced by agonal hypoxia, postmortem autolysis, and formalin fixation,15,16,17 we increased the amount of genomic DNA (gDNA) by applying whole genome amplification (WGA) before primer-specific PCR for viral sequences.
Both PCR- and non-PCR-based methods are available for WGA. Since PCR-based WGA uses repeated thermocycling to 96°C, and since our preliminary data with one of the PCR-based WGA methods gave false-positive results from microdissected cells (personal observation), we selected isothermal WGA to avoid thermal-induced DNA breaks during WGA amplification. The isothermal hyperbranched multidisplacement amplification method (MDA) for WGA, first described by Blanco et al,18 is ideal for microdissected cell samples for several reasons. First, by using the bacteriophage Φ29-DNA polymerase, high fidelity is maintained during replication and a high strand displacement activity remains stable at 30°C for 12 hours or longer.18,19 Second, isothermal WGA avoids the cyclical changes in temperature that occur in all PCR-based methods, such as degenerate oligonucleotide-primed PCR or ligation-mediated PCR. The isothermic method for WGA may be of specific importance when amplifying DNA from microdissected cell, or because mis-amplification artifacts increase with such small sample sizes.20,21 The MDA method for WGA uses exonuclease-resistant primers that allow higher product stability and larger fragment synthesis during amplification.18,19,22 Lastly, the hyperbranched version of MDA is more suitable for clinical material that amplifies chromosomal genomic DNA. In fact, this version is a derivation of the original rolling circle amplification method, which also uses MDA but is designed specifically to amplify shorter DNA lengths from generic circular DNA molecules rather than long fragments of genomic DNA.22,23,24 Restriction and circularization-aided rolling circle amplification requires the use of considerable enzymatic manipulations, which is not advisable when working with very small samples.
We adapted the hyperbranched multidisplacement amplification for whole gene amplification (MDA-WGA) strategy to detect DNA sequences in microdissected single neurons or small groups of 3 to 30 neurons removed from tissue sections of formalin-fixed paraffin- embedded (FFPE) autopsy brain. Before microdissection, we performed immunohistochemistry on the deparaffinized slides to identify CD8+ T cells. A preliminary report has been published in abstract form.25
Materials and Methods
We used sections from three AIDS patients with HIV-1 encephalitis (HIVE) in the hippocampus and three AIDS patients without HIVE in any brain region (HIVnE). None had brain opportunistic infections or lymphomas. All six were part of a prior study of cytotoxic CD8+ T cells in AIDS hippocampus.14 To facilitate microdissection of neurons with adherent CD8+ T cells, we chose the three HIVE cases with the most abundant perineuronal cytotoxic T lymphocytes in the hippocampus. We excluded all cases with a postmortem interval of >40 hours, and we determined that we could amplify housekeeping gene sequences from a deparaffinized 6-μm-thick tissue section of the hippocampus. Three HIV-1-negative controls with normal brains were generously donated by the National Institute of Child Health and Development's Brain and Tissue Bank for Developmental Disorders, University of Miami (NO1-HD43383 and NO1-HD43368). Between 5 and 10 serial 6-μm-thick sections of buffered formalin-fixed hippocampus were cut onto clean, plain glass slides and processed for immunohistochemistry for CD8+ T lymphocytes. Briefly, we deparaffinized the sections and sequentially incubated them with blocking solution (5% normal goat serum) for 30 minutes at room temperature, monoclonal anti-human CD8 monoclonal antibodies (Dako Corporation, Carpinteria, CA) at a 1:50 dilution overnight at 4°C, biotinylated secondary antibody for 1 hour at room temperature, and the avidin-biotin complex for 1 hour at room temperature. To avoid cross contamination between specimens, new microtome knives, cleaned with weak hydrochloric acid, and a cleaned water bath filled with new double-distilled water were used for each case.
We used the thermoplastic polymer (ethyl-vinyl-acetate)- coated CapSure TF-100 Caps and the Arcturus PixCell IIe laser capture microscope (Molecular Devices Corporation, Sunnyvale, CA) to remove nuclei from pyramidal neurons in the CA4 subregion of hippocampus from FFPE autopsy brains. We dissected two types of neuronal nuclei: those from neurons that were in direct contact with perineuronal CD8+ T cells and those from neurons that had no contact with adjacent cells. The usual postmortem shrinkage artifact usually left a clear space around neurons that facilitated dissection of their nuclei to the exclusion of other cells. Samples included single neurons and groups of 3 to 10 or 15 to 30 neurons with or without contact with perineuronal CD8+ T cells. To facilitate dissection of neuron-only samples, we adjusted the laser beam diameter to 7.5 μm and used a laser beam power of 65 mW for 500 μs. After the microdissection, we removed extraneous tissues from the TF-100 Caps with a clean Arcturus CapSure Pad and confirmed the fact that the microdissection removed only neurons by direct microscopic observation of the original slide and the TF-100 Cap's surface.
We extracted gDNA by incubating the captured cells with 20 μl of Arcturus PicoPure DNA extraction buffer for 12 hours at 65°C, followed by a 10-minute incubation at 85°C to destroy proteinase K remnants. We vacuum-dried the gDNA and reconstituted it in 3 μl of PBS. We performed MDA with the REPLI-g Mini DNA amplification kit, according to the user-developed protocol supplied by the manufacturer (QIAGEN, Valencia, CA). Briefly, we denatured gDNA in Buffer D for 10 minutes on wet ice, added stop solution, and mixed the solution by gently flicking the tube to avoid loci dropout by physical DNA breakage that may occur when the concentration and quantity of DNA is low. We added 10 μl of nuclease-free water, 29 μl of reaction buffer, and 1 μl of Φ29-DNA polymerase and incubated the reaction mix at 30°C for 16 hours. We inactivated polymerase activity by heating each sample at 65°C for 3 minutes and stored aliquots of the amplified gDNA at −20°C until the real-time PCR procedure.
We performed two rounds of real-time PCR in a Roche LightCycler (Roche Biochemicals, Indianapolis, IN) with 35 cycles for the first round and 40 cycles for the second round, using primer sets for two housekeeping (HK) genes (glyceraldehyde 3′-phosphate dehydrogenase and β2-microglobulin) (Table 1). For the first round, we used amplified gDNA template along with 10 pmol of each primer in a 10-μl SYBR Green reaction mix volume (TIB-MOLBIOL, Adelphia, NJ). For the second round, we used the same reagents, minus the amplified gDNA template, in a final volume of 20 μl. We performed double-round PCR for HIV-1 gag, nef, and pol (Table 1) on all HK-positive samples. Positive PCR controls were DNA extracted from cultured normal human monocytes and HIV-1 cDNA extracted from infected COS cells (generously donated by Dr. Charles Wood, School of Biological Sciences, University of Nebraska, Lincoln, NE). Negative controls included blank caps and tissue section-touched caps without laser capture. Results were positive when the melting curve graphs for the samples had a melting temperature (Tm) similar to those of the melting curve graphs of the control DNA.
Table 1.
Primer Sequences for Housekeeping and HIV-1 DNA Sequences
| Gene sequence | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| β2-Microglobulin | 5′-GATGAGTATGCCTGCCGTGTG-3′ | 5′-CAATCCAAATGCGGCATCT-3′ |
| GAPDH | 5′-GAAGGTGAAGGTCGGAGTC-3′ | 5-GAAGATGGTGATGGGATTTC-3′ |
| HIV-1-gag | 5′-TTTGGCTGAGGCAATGAGT-3′ | 5′-TTTCTCTGGCCCCTAAAATT-3′ |
| HIV-1-pol | 5′-TGGGAAGTTCAATTAGGAATACCAC-3′ | 5-CCTACATACAAATCATCCATGTATTG-3′ |
| HIV-1-nef | 5′TGGATCTACCACACACAAGG-3′ | 5′-CAACTGGTACTAGCTTGAAGCA-3′ |
GAPDH, glyceraldehyde 3′-phosphate dehydrogenase.
We correlated HK gene amplification with postmortem intervals using analysis of variance and with sample size and patient group using the χ2 test.
Results
The average age of the AIDS patients was slightly higher than that of controls (43 ± 8 years versus 34 ± 3 years); their postmortem intervals were similar (20 ± 10 hours and 17 ± 10 hours); all were men. The inflammatory lesions of HIVE were characterized by perivascular and parenchymal lymphocytes, activated microglia and perivascular macrophages, and the characteristic multinucleated giant cell of HIVE. Reactive astrocytosis was present in the more severe lesions as previously described.26 CD8+ T cells were numerous in perivascular spaces, brain parenchyma, and perineuronal sites (Figure 1, A and B) and were readily visualized with the laser capture microscope (Figure 2A). We previously showed that these CD8+ T cells were cytotoxic T cells since they co-labeled with granzyme B and perforin.14 We did not perform immunohistochemistry for CD4+ T cells since they usually are confined to perivascular spaces27 and since most of the CD3+ T cells were of the CD8+ subtype.14 This corresponds to the relative distribution of T-cell subtypes in a non-human primate model of HIVE.28 One of the three HIVnE brains contained sparse perivascular infiltrates of lymphocytes. The three control cases had normal brains on gross and microscopic examination.
Figure 1.
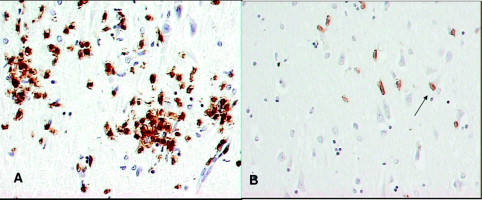
CD8+ T lymphocyte immunohistochemistry of an AIDS patient with HIV-1 encephalitis in the CA4 subregion of the hippocampus. The CD8+ T lymphocytes localize to the microglial nodules of HIV-1 encephalitis (A) and diffusely infiltrate adjacent brain (B). Arrow indicates the cell-to-cell contacts between pyramidal neurons and perineuronal CD8+ T cells. Hematoxylin counterstain; original magnification ×400.
Figure 2.
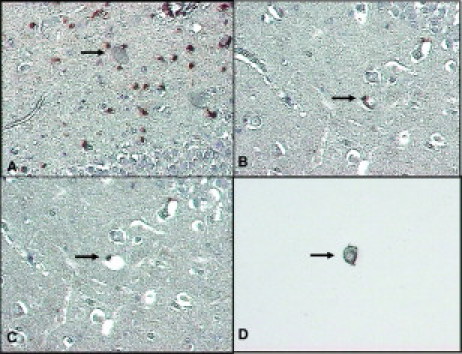
A: HIV encephalitis with infiltrating CD8+ T lymphocytes (brown) in the CA4 region of hippocampus, as seen with the laser capture microscope. B–D: Successful dissection and capture of a CD8+ pyramidal neuron (arrows). B shows the neuron and its perineuronal CD8+ T cell before microdissection. C shows the empty space on the tissue section that remains after successful microdissection; note that the perineuronal T cell has not been captured. D shows the Transfer Cap to which only the neuron adheres. Hematoxylin counterstain; original magnifications ×200 (A); ×400 (B–D).
The large size of the pyramidal neurons, combined with the customary postmortem shrinkage artifacts, facilitated the removal of neurons without contamination from adjacent cells or tissues (Figure 2, B–D). We confirmed the absence of non-neuronal tissue by microscopic examination of the FFPE section and the surface of the Transfer Cap after the microdissection (Figure 2, C and D). Only 1 of 24 microdissections of single neurons from the HIVE cases was removed with an adherent CD8+ T cell. We used this sample as part of the evaluation of WGA in single-neuron dissection but did not include it in subsequent PCRs for viral sequences. After isothermal WGA and double-round PCR, HK or HIV-1 gene sequences were identified by a fluorescence activity that was similar to their positive control (Figure 3).
Figure 3.
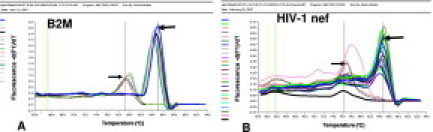
Real-time PCR SYBR Green Tm graph for β2-microglobulin (A) and HIV-1 nef (B). Vertical lines are used to point out PCR product Tm. The blue line with arrow shows the Tm of one positive control and several positive neuronal samples. The red line with arrow shows the Tm from negative control and several negative samples. Default green and yellow lines were not used.
HK gene sequence amplification in single neurons required MDA-WGA before PCR. As shown in Figure 4A, 37% of 14 single-neuron samples contained housekeeping gene sequences when MDA-WGA preceded double-round PCR. The crossing threshold for these single neuronal samples was 28.3 ± 0.6. In contrast, none of 17 single neuronal samples contained HK sequences when double-round PCR was performed in the absence of MDA-WGA.
Figure 4.

Housekeeping gene sequence amplification was related to whole-gene amplification before double-round PCR in single neurons (A) and to sample size in the HIVE and control groups (B) but was unaffected by postmortem intervals (C).
Twenty-four of the 34 groups of 15 to 30 neuronal samples (44%) contained HK gene sequences; their crossing threshold was 13.2 ± 0.9. Three of the four groups of 3 to 10 neuronal samples (75%) also contained HK sequences. Successful amplification correlated with the amount of DNA in the original sample (P < 0.01; Figure 4B) but not with patient group (P > 0.05; data not shown) or with postmortem intervals up to 40 hours (P > 0.05; Figure 4C).
HIV-1 viral sequences in HK+ neuronal samples correlated with patient group (Table 2 and Figure 5A). Nine of 22 (40%) HK+ neuronal samples from the three HIVE cases contained HIV-1 DNA sequences. Gag and nef, alone or in combination, were most common. Pol was found twice only, once in combination with nef and once in combination with nef and gag. Two of 15 (13%) HK+ neuronal samples from the three HIVnE cases contained HIV-1 DNA for gag. Both were single-neuron samples from HIVnE cases without hippocampal lymphocytes (patients 5 and 6 in Table 2). Viral sequences were absent in all HK+ neuronal samples from controls.
Table 2.
Gene Amplification in AIDS and Control Hippocampal Neuronal Samples
| No. of samples with housekeeping sequences |
No. of HK+ samples with viral sequences |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | Group | Age (y) | PMI (h) | HIVE | 1 Neuron | 3–10 Neurons | 15–30 Neurons | 1 Neuron | 3–10 Neurons | 15–30 Neurons |
| 1 | AIDS | 36 | 12 | X | 3 of 5 | n.d. | 3 of 3 | n.d. | n.d. | 1 of 3 |
| 2 | AIDS | 37 | 16 | X | 3 of 12 | 3 of 3 | 1 of 3 | 1 of 3 | 1 of 3 | 0 of 3 |
| 3 | AIDS | 42 | 23 | X | 3 of 7 | 0 of 1 | 6 of 7 | 2 of 3 | n.d. | 4 of 6 |
| 4 | AIDS | 41 | 40 | O | 2 of 5 | n.d. | 3 of 3 | n.d. | n.d. | 0 of 4 |
| 5 | AIDS | 47 | 17 | O | 3 of 5 | n.d. | 2 of 5 | 0 of 3 | n.d. | 0 of 2 |
| 6 | AIDS | 57 | 14 | O | 4 of 5 | n.d. | 2 of 4 | 0 of 4 | n.d. | 0 of 2 |
| 7 | Control | 35 | 6 | O | 2 of 5 | n.d. | 3 of 3 | 0 of 2 | n.d. | 0 of 3 |
| 8 | Control | 36 | 22 | O | 1 of 5 | n.d. | 2 of 3 | 0 of 1 | n.d. | 0 of 2 |
| 9 | Control | 30 | 23 | O | 2 of 4 | n.d. | 2 of 3 | 0 of 2 | n.d. | 0 of 2 |
PMI, postmortem interval; HK, housekeeping gene sequences (β 2-microglobulin and glyceraldehyde 3′-phosphate dehydrogenase); n.d., not done; X, present; O, absent.
Figure 5.

In housekeeping gene-positive neurons, HIV-1 viral sequences were more common in the HIVE cases than in the HIVnE cases; they were absent in uninfected controls (A). HIV-1 viral sequences were more common in housekeeping gene-positive neuronal samples that were in direct contact with perineuronal CD8+ T lymphocytes than in neuronal samples that had no contacts with adjacent cells (B).
Viral sequences in the HK+ neuronal samples correlated with the presence of perineuronal CD8+ T cells in the three HIVE cases (Figure 5B). Ten of 19 neuronal samples in direct contact with CD8+ T cells contained HK sequences. Seven of these 10 HK+ samples contained viral sequences (70%). Eleven of 21 neuronal samples without perineuronal CD8+ T cells contained HK sequences. Four of these 11 HK+ neuronal samples contained viral sequences (37%).
Discussion
Microdissected single-cell samples from formalin-fixed paraffin-embedded tissue blocks of autopsy brains are important in studying certain human diseases. They allow the analysis of cell-specific alterations in DNA and RNA and facilitate sampling of brain lesions that are not uniformly distributed in the brain, as occurs with the inflammatory lesions of HIVE. Furthermore, premortem brain biopsies usually are not available for disorders other than tumors and experimental studies may not directly model the clinical condition.
Our current study clearly shows that application of MDA-WGA enhances the chances to detect amplified gene sequences from single neurons removed from FFPE brain sections. Housekeeping gene sequences never were amplified by double-round PCR alone but only were detected when PCR was preceded by whole genomic amplification by MDA. As reviewed in the introduction, this non-PCR-based method for WGA protocol offers distinct advantages over PCR-based WGA or PCR-alone methods since it permits amplification of up to 70,000 nucleotides19 and avoids possible thermal-induced physical breaks of naked DNA that may cause a dropout of loci representation.
The present study confirms earlier ones that detected viral sequences in brain neurons of AIDS patients. Approximately one-third of all HK+ samples of single as well as small groups of neurons contained at least one of three HIV-1 gene sequences. By finding viral genes in single neurons, we reduced the potential for false-positive PCR due to tissue contamination at the time of laser capture of multiple cells on a Transfer Cap. In addition, contamination from adjacent, potentially infected cells was reduced by using a laser beam size of smaller diameter than a neuronal nucleus and by directly observing the surface of the Transfer Cap to ensure the absence of non-neuronal material. Furthermore, the low variance in the crossing thresholds for the PCRs from the single neuronal samples suggests that equal amounts of DNA were extracted and supports the fact that only neuronal nuclei were captured. We also excluded false-positive amplification for HK and viral DNA sequences by including two negative controls (blank Transfer Cap and Transfer Cap applied to tissue section without laser beam exposure) in the studies. The DNA extraction procedure that we used does not yield RNA, so it is unlikely that the positive results were due to amplification of viral RNA from neurons or from any adjacent, infected cells.
Although our microdissection protocol principally captured neuronal nuclei, it is likely that small amounts of neuronal cytoplasm were included as well. Thus, the bulk of neuronal viral sequences represent integrated proviral DNA, but the inclusion of cytoplasm indicates the potential for some or all of the DNA to include unintegrated linear DNA or even circular DNA with its one or two long terminal repeats.29,30
Several factors could underlie the disparate results in those studies that used microdissected autopsy brain neurons to amplify neuronal DNA for viral sequences. First, technical differences in amplification techniques included MDA-WGA plus double-round PCR (current study), double-round PCR,11 and triple-nested PCR.12,13 Neuronal viral sequences may be sufficiently low, when compared to those in monocytic or astrocytic cells, that their detection in microdissected autopsy neurons requires more amplification than is needed to detect viral sequences in infected monocytes or astrocytes. Biological variations among the studies included intrinsic differences in neurons (hippocampus versus cortex) and the presence or absence of local HIVE, with its attendant productive viral infection and intense pro-inflammatory milieu. The present study and that of Trillo-Pazos et al12 removed neurons from hippocampus or cortex with local HIVE, whereas two other studies removed neurons from seemingly normal hippocampus or cortex.11,13 Lastly, if neuronal infection is defective, only certain viral genes may be incorporated into host cell DNA. This might explain why neuronal viral sequences are detected when primer pairs include those for HIV-1 gag,12 gag plus nef11, or gag, nef and pol (current study), whereas neuronal sequences were not detected when primer pairs were confined to HIV-1 env.13
The neuronal infection is either latent or restricted since there is no evidence that neurons produce viral proteins in vivo. Latent infection might impair neuronal functioning, as is seen with other viruses that can latently infect neurons.31,32 Latent neuronal infection also could be a potential viral reservoir and develop into productive infection with inflammation, as occurs with astrocytes and neural progenitor cells in culture.33 Latent neuronal infection also could injury cells if viral antigen is produced and presented to cytotoxic T lymphocytes. Indirect evidence for this mode of neuronal killing comes from our studies that show accumulations of perineuronal CD8+ and granzyme B+ cytotoxic T lymphocytes that form direct cell-to-cell contact with neurons14 and from clinical studies that correlate worsening of HIV-1-associated dementia with accumulations of CD8+ T cells in the central nervous system.34
The high correlation between neuronal viral sequences and perineuronal CD8+ T cells supports our prior hypothesis that CD8+ T cells traffic specifically to infected neurons.14 The initial adherence likely occurs via neuronal intercellular adhesion molecule-5, which has high expression levels in brain neurons, especially hippocampal neurons.35 We anticipate that studies in progress will identify neuronal major histocompatibility antigen I, since this CD8+ T cell antigen-presenting molecule is induced in neurons in other inflammatory brain disorders or after exposure to cytokines.36,37
Alternatively, the correlation between neuronal infection and perineuronal CD8+ T cells raises the possibility that neuronal infection could be secondary to infection of the perineuronal CD8+ T cells. We previously considered the possibility that perineuronal CD4+ T cells might infect neurons.27 However, recent studies have shown that CD8+ T cells also are infected by HIV-1.38,39 The large numbers of CD8+ T cells in brains of highly active antiretroviral therapy-exposed patients, combined with their perineuronal locations, offers the potential for neuronal infection by the CD8+ T cell. This is supported by the current study, which positively correlated infected neurons with the presence of perineuronal CD8+ T lymphocytes. Infection in CD4 receptor-negative neurons may occur by trans-receptor mechanisms, as occurs when CD4− astrocytes are co-cultured with infected CD4+ T cells40 or with infected monocytes.41 Additional support for the potential for neurons to be infected are studies such as the one by Li et al,3 which demonstrate infection of CD4− fetal neurons when exposed to free virus in vitro.
In conclusion, our results demonstrate that gDNA from microdissected single cells from FFPE tissues can be efficiently amplified using isothermal WGA-MDA followed by two rounds of PCR. They show that brain neurons contain viral sequences and suggest that viral infection of neurons may be related to the presence of perineuronal CD8+ T lymphocytes.
Acknowledgements
We gratefully acknowledge the helpful advice of Dr. Maximimo Redondo in the statistical analyses and the generosity of the NICHD Brain and Tissue Bank in providing the HIV-1-negative control samples (NO1-HD-4-3383 and NO1-HD-4- 3368).
Footnotes
Supported in part by the National Institutes of Health (RO1-NS-39117).
References
- 1.Jones G, Power C. Regulation of neural cell survival by HIV-1 infection. Neurobiol Dis. 2006;21:1–17. doi: 10.1016/j.nbd.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 2.Kramer-Hammerle S, Rothenaigner I, Wolff H, Bell JE, Brack-Werner R. Cells of the central nervous system: targets and reservoirs of the human immunodeficiency virus. Virus Res. 2005;111:194–213. doi: 10.1016/j.virusres.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Li XL, Moudgil T, Vinters HV, Ho DD. CD4-independent productive infection of a neuronal cell line by human immunodeficiency virus-type 1. J Virol. 1990;64:1383–1387. doi: 10.1128/jvi.64.3.1383-1387.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trillo-Pazos G, Kandanearatchi A, Eyeson J, King D, Vyakarnam A, Everall IP. Infection of stationary human brain aggregates with HIV-1 SF162 and IIIB results in transient neuronal damage and neurotoxicity. Neuropathol Appl Neurobiol. 2004;30:136–147. doi: 10.1046/j.0305-1846.2003.00519.x. [DOI] [PubMed] [Google Scholar]
- 5.Bagasra O, Lavi E, Bobroski L, Khalili K, Pestaner JP, Tawadros R, Pomerantz RJ. Cellular reservoirs of HIV-1 in the central nervous system of infected individuals: identification by the combination of in situ polymerase chain reaction and immunohistochemistry. AIDS. 1996;10:573–585. doi: 10.1097/00002030-199606000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Nuovo GJ, Gallery F, MacConnell P, Braun A. In situ detection of polymerase chain reaction-amplified HIV-1 nucleic acids and tumor necrosis factor mRNA in the central nervous system. Am J Pathol. 1994;144:659–666. [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Wesselingh SL, Griffin DE, McArthur JC, Johnson RT, Glass JD. Localization of HIV-1 in human brain using polymerase chain reaction/in situ hybridization and immunocytochemistry. Ann Neurol. 1996;39:705–711. doi: 10.1002/ana.410390606. [DOI] [PubMed] [Google Scholar]
- 8.Sharer LR, Saito Y, Cunha AD, Ung PC, Gelbard HA, Epstein LG, Blumberg BM. In situ amplification and detection of HIV-1 DNA in fixed pediatric AIDS brain tissue. Hum Pathol. 1996;27:614–617. doi: 10.1016/s0046-8177(96)90172-0. [DOI] [PubMed] [Google Scholar]
- 9.An SF, Groves M, Giometto B, Beckett AAJ, Scaravilli F. Detection and localization of HIV-1 DNA and RNA in fixed adult AIDS brain by polymerase chain reaction-in situ technique. Acta Neuropathol. 1999;98:481–487. doi: 10.1007/s004010051113. [DOI] [PubMed] [Google Scholar]
- 10.Canto-Nogules C, Sanchez-Ramon S, Alvarez S, Lacruz C, Munoz-Fernande MA. HIV-1 infection of neurons might account for progressive HIV-1-associated encephalopathy in children. J Mol Neurosci. 2005;27:79–90. doi: 10.1385/JMN:27:1:079. [DOI] [PubMed] [Google Scholar]
- 11.Torres-Muñoz J, Stockton P, Tacoronte N, Roberts B, Maronpot RR, Petito CK. Detection of HIV-1 gene sequences in hippocampal neurons isolated from postmortem AIDS brains by laser capture microdissection. J Neuropathol Exp Neurol. 2001;60:885–892. doi: 10.1093/jnen/60.9.885. [DOI] [PubMed] [Google Scholar]
- 12.Trillo-Pazos G, Diamanturos A, Rislove L, Menza T, Chao W, Belem P, Sadiq S, Morgello S, Sharer L, Volsky DJ. Detection of HIV-1 DNA in microglia/macrophages, astrocytes and neurons isolated from brain tissue with HIV-1 encephalitis by laser capture microdissection. Brain Pathol. 2003;13:144–154. doi: 10.1111/j.1750-3639.2003.tb00014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson KA, Churchill MJ, Gorry PR, Sterjovski J, Oelrichs RB, Wesselingh SL, McLean CA. Astrocyte specific viral strains in HIV dementia. Ann Neurol. 2004;56:873–877. doi: 10.1002/ana.20304. [DOI] [PubMed] [Google Scholar]
- 14.Petito CK, Torres-Muñoz JE, Zielger F, McCarthy M. Brain CD8+ and cytotoxic T lymphocytes are associated with, and may be specific for, human immunodeficiency virus type 1 encephalitis in patients with acquired immunodeficiency syndrome. J NeuroVirol. 2006;12:272–283. doi: 10.1080/13550280600879204. [DOI] [PubMed] [Google Scholar]
- 15.Merkelbach S, Gehlen J, Handt S, Fuzesi L. Novel enzyme immunoassay and optimized DNA extraction for the detection of polymerase-chain-reaction-amplified viral DNA from paraffin-embedded tissue. Am J Pathol. 1997;150:1537–1546. [PMC free article] [PubMed] [Google Scholar]
- 16.Specht K, Richter T, Muller U, Walch A, Werner M, Hofler H. Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol. 2001;158:419–429. doi: 10.1016/S0002-9440(10)63985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrer I, Armstrong J, Capellari S, Parchi P, Arzberger T, Bell J, Budka H, Strobel T, Giaccone G, Rossi G, Bogdanovic N, Fakai P, Schmitt A, Riederers P, Al-Sarraj S, Ravid R, Krezschmar H. Effects of formalin fixation, paraffin embedding and time of storage on DNA preservation in brain tissue: a BrainNet Europe study. Brain Pathol. 2007;17:297–303. doi: 10.1111/j.1750-3639.2007.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanco L, Bernad A, Lazaro JM, Martin G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 19.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lasken RS, Egholm M. Whole genome amplification: abundant supplies of DNA from precious samples or clinical specimens. Trends Biotechnol. 2003;21:531–535. doi: 10.1016/j.tibtech.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 21.Lage JM, Leamon JH, Pejovic T, Hamann, Lacey M, Dillon D, Segraves R, Vossbrinck B, Gonzalez A, Pinkel D, Albertson DG, Costa J, Lizardi PM. whole genome analysis of genetic alterations in small DNA samples using hyperbranched strand displacement amplification and array–CGH. Genome Res. 2003;13:294–307. doi: 10.1101/gr.377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat Genet. 1998;19:226–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 23.Demidov VV. Rolling circle amplification (RCA) In: Fuchs J, Podda M, editors. Encyclopedia of Diagnostic Genomics and Proteomics. Boston University; Boston: 2005. pp. 1175–1179. [Google Scholar]
- 24.Janne PA, Borras AM, Kuang Y, Rogers AM, Joshi VA, Liyanage H, Lindeman N, Lee JC, Halmos B, Maher EA, Distel RJ, Meyerson M, Johnson BE. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res. 2006;12:751–758. doi: 10.1158/1078-0432.CCR-05-2047. [DOI] [PubMed] [Google Scholar]
- 25.Torres-Muñoz JT, Nunez M, Petito CK. HIV-1 viral cDNA detection in human microdissected single neurons from AIDS with encephalitis autopsy cases. J Neuropathol Exp Neurol. 2007;66:455. [Google Scholar]
- 26.Petito CK, Roberts B, Cantando JD, Rabinstein A, Duncan R. Hippocampal injury and alterations in neuronal chemokine co-receptor expression in patients with AIDS. J Neuropathol Exp Neurol. 2001;60:377–385. doi: 10.1093/jnen/60.4.377. [DOI] [PubMed] [Google Scholar]
- 27.Petito CK, Adkins B, McCarthy M, Roberts B, Khamis I. CD4+ and CD8+ cells accumulate in the brains of acquired immunodeficiency syndrome patients with human immunodeficiency virus encephalitis. J Neurovirol. 2003;9:36–44. doi: 10.1080/13550280390173391. [DOI] [PubMed] [Google Scholar]
- 28.Kim W-K, Corey S, Chesney G, Knight H, Klumpp S, Wuthrich C, Letvin N, Koralnik I, Lackner A, Veasey R, Williams K. Identification of T lymphocytes in simian immunodeficiency virus encephalitis: distribution of CD8+ T cells in association with central nervous system vessels and virus. J Neurovirol. 2004;10:315–325. doi: 10.1080/13550280490505382. [DOI] [PubMed] [Google Scholar]
- 29.Sharkey ME, Teo I, Greenough T, Sharova N, Luzuriaga K, Sullivan JL, Bucy RP, Kostrikis LG, Haase A, Veryard C, Davaro RE, Cheeseman SH, Daly JS, Bova C, Ellison RT, Mady B, Lai KK, Moyle G, Nelson M, Gazzard B, Shaunak S, Stevenson M. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat Med. 2000;6:76–81. doi: 10.1038/71569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharkey ME, Stevenson M. Two long terminal repeat circles and persistent HIV-1 replication. Curr Opin Infect Dis. 2001;14:5–11. doi: 10.1097/00001432-200102000-00002. [DOI] [PubMed] [Google Scholar]
- 31.De la Torre JC, Mallory M, Brot M, Gold L, Koob G, Oldstone MB, Masliah E. Viral persistence in neurons alters synaptic plasticity and cognitive functions without destruction of brain cells. Virology. 1996;220:508–515. doi: 10.1006/viro.1996.0340. [DOI] [PubMed] [Google Scholar]
- 32.Cao W, Oldstone MBA, de la Torre JC. Viral persistent infection affects both transcriptional and posttranscriptional regulation of neuron-specific molecule GAP43. Virology. 1997;230:147–154. doi: 10.1006/viro.1997.8458. [DOI] [PubMed] [Google Scholar]
- 33.Lawrence DM, Durham LC, Schwartz L, Seth P, Maric D, Major EO. Human immunodeficiency virus type 1 infection of human brain-derived progenitor cells. J Virol. 2004;78:7319–7328. doi: 10.1128/JVI.78.14.7319-7328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller RF, Isaacson PG, Hall-Craggs M, Lucas S, Gray F, Scaravilli F, An SF. Cerebral CD8+ lymphocytosis in HIV-1 infected patients with immune restoration induced by HAART. Acta Neuropathol. 2004;108:17–23. doi: 10.1007/s00401-004-0852-0. [DOI] [PubMed] [Google Scholar]
- 35.Hino H, Mori K, Yoshihara Y, Iseki E, Aklyama H, Nishimura T, Ikeda K, Kosaka K. Reduction of telencephalin immunoreactivity in the brains of patients with Alzheimer's disease. Brain Res. 1997;753:353–357. doi: 10.1016/s0006-8993(97)00158-3. [DOI] [PubMed] [Google Scholar]
- 36.Bien CG, Bauer J, Deckwerth TI, Wiendl H, Deckert M, Wiestler OD, Schramm J, Elger CE, Lassman H. Destruction of neurons by cytotoxic T-lymphocytes: a new pathogenic mechanisms in Rasmussen's encephalitis. Ann Neurol. 2002;51:311–318. doi: 10.1002/ana.10100. [DOI] [PubMed] [Google Scholar]
- 37.Nitsch R, Pohl EE, Smorodchenko A, Infante-Duarte C, Aktas O, Zipp F. Direct impact of T cells on neurons revealed by two-photon microscopy in living brain tissue. J Neurosci. 2004;24:2458–2464. doi: 10.1523/JNEUROSCI.4703-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saha K, Zhang J, Zerhouni B. Evidence of productively infected CD8+ T cells in patients with AIDS: implications for HIV-1 pathogenesis. J AIDS. 2001;26:199–207. doi: 10.1097/00042560-200103010-00001. [DOI] [PubMed] [Google Scholar]
- 39.Hughes GJ, Willey SJ, Cochrane A, Leen C, Bell JE, Simmonds P. Virus immunocapture provides evidence of CD8 lymphocyte-derived HIV-1 in vivo. AIDS. 2007;21:1507–1513. doi: 10.1097/QAD.0b013e3281e209e6. [DOI] [PubMed] [Google Scholar]
- 40.Speck FR, Esser U, Penn ML, Eckstein DA, Pulliam L, Chan SY, Goldsmith MA. Trans-receptor mechanism for infection of CD4-negative cells by human immunodeficiency virus type 1. Curr Biol. 1999;9:547–550. doi: 10.1016/s0960-9822(99)80241-3. [DOI] [PubMed] [Google Scholar]
- 41.Muratori C, Sistigu A, Ruggiero E, Falchi M, Bacigalupo I, Palladino C, Toschi E, Rederico M. Macrophages transmit human immunodeficiency virus type 1 products to CD4-negative cells: involvement of matrix metalloproteinase 9. J Virol. 2007;81:9078–9087. doi: 10.1128/JVI.00675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


