Abstract
Epigenetic silencing of the DNA repair protein O6-methylguanine-DNA methyltransferase (MGMT) by promoter methylation predicts successful alkylating agent therapy, such as with temozolomide, in glioblastoma patients. Stratified therapy assignment of patients in prospective clinical trials according to tumor MGMT status requires a standardized diagnostic test, suitable for high-throughput analysis of small amounts of formalin-fixed, paraffin-embedded tumor tissue. A direct, real-time methylation-specific PCR (MSP) assay was developed to determine methylation status of the MGMT gene promoter. Assay specificity was obtained by selective amplification of methylated DNA sequences of sodium bisulfite-modified DNA. The copy number of the methylated MGMT promoter, normalized to the β-actin gene, provides a quantitative test result. We analyzed 134 clinical glioma samples, comparing the new test with the previously validated nested gel-based MSP assay, which yields a binary readout. A cut-off value for the MGMT methylation status was suggested by fitting a bimodal normal mixture model to the real-time results, supporting the hypothesis that there are two distinct populations within the test samples. Comparison of the tests showed high concordance of the results (82/91 [90%]; Cohen's kappa = 0.80; 95% confidence interval, 0.82−0.95). The direct, real-time MSP assay was highly reproducible (Pearson correlation 0.996) and showed valid test results for 93% (125/134) of samples compared with 75% (94/125) for the nested, gel-based MSP assay. This high-throughput test provides an important pharmacogenomic tool for individualized management of alkylating agent chemotherapy.
O6-Methylguanine-DNA methyltransferase (MGMT) is a cellular DNA repair protein that rapidly reverses alkylation (eg, methylation) at the O6 position of guanine, thereby neutralizing the cytotoxic effects of alkylating agent therapy such as temozolomide (TMZ) and carmustine.1,2,3 It has been shown that epigenetic silencing of the MGMT gene by promoter methylation shuts down gene transcription4,5 and reflects a common alteration in primary human tumors leading to MGMT deficiency.6 Epigenetic silencing of the MGMT gene has been shown to correlate with improved survival in several studies of glioma patients treated with alkylating agent therapy7,8 and has been substantiated in two clinical trials.9,10 A recent randomized clinical trial suggests that the MGMT methylation status has a very good predictive value for benefit from the addition of the alkylating agent TMZ to radiotherapy.10,11 This finding has important clinical implications for stratified therapy.12 Although this trial has established the new standard of care for glioblastoma patients,11 the benefit of the addition of TMZ chemotherapy to radiotherapy was heavily weighted to patients whose tumors had a methylated MGMT promoter, with 46% still alive at 2 years, compared with only 14% of the patients with non-methylated MGMT promoter.10 Hence, this epigenetic alteration in tumors can now be exploited in a diagnostic test to predict the benefit from alkylating agent therapy for individualized management of patients. Besides glioblastoma, there is published evidence that the MGMT methylation status may also predict the benefit from alkylating agent-containing therapy in patients with low grade glioma, oligodendroglioma, and diffuse large B-cell lymphoma.13,14,15
Methylation-specific PCR (MSP) with visualization of the results on a gel (gel-based MSP) is widely used to determine epigenetic silencing of genes,16 and in particular for testing MGMT promoter methylation in glioma,8,14 although tests using other technologies have been developed.17,18,19 Nested, gel-based MSP has been used to establish the predictive value of the methylation status of the MGMT gene promoter in the clinical trials detailed above.9,10 This methodology is highly sensitive and accurate, but has drawbacks for routine clinical use.26 Here we describe a standardized direct, real-time MSP assay for methylated MGMT promoter (m_MGMT) on a high-throughput platform and its comparison to the nested, gel-based MSP with known predictive value in the clinical setting. The study reveals good reproducibility of the direct, real-time MSP assay and high congruence between the results of the two assays, and provides information for the definition of thresholds for diagnostic purposes.
Materials and Methods
Sample Preparation
Formalin-fixed, paraffin-embedded (FFPE) glioma tissue samples were obtained from patients who had given their informed consent. Patients were enrolled in clinical trials coordinated at the University Hospitals in Lausanne, Switzerland; Rotterdam, The Netherlands; and Regensburg, Germany. All studies received appropriate approval from the relevant Ethical Committee. For each tumor sample, eight 5-μm consecutive sections were prepared on glass slides. An additional slide stained with H&E was used to define the viable tumor area that was subsequently marked on the unstained serial sections. The equivalent of two sections was used for each MSP. Prepared sample sections were processed in parallel at OncoMethylome Sciences (direct, real-time MSP) and Centre Hospitalier Universitaire Vaudois (nested, gel-based MSP), according to each laboratory's respective protocols. The laboratories were each blinded to the results obtained in the other.
Nested, Gel-Based MSP
This assay was conducted as published previously.10 In brief, DNA was isolated from the tumor tissue macrodissected from histologically marked slides using the Ex-Wax DNA extraction kit (S4530; Chemicon, Temecula, CA), limiting the proteinase digestion to 6 hours. After the bisulfite treatment step, purified DNA was subjected to MSP using a two-step approach with nested primers.20 The first round of PCR amplifies both the methylated (m_MGMT) and the non-methylated (u_MGMT) version of the MGMT sequence. The resulting PCR product of 289 bp served as a template for the second MSPs, amplifying either m_MGMT or u_MGMT, yielding PCR products of 81 and 93 bp, respectively.7 The specific primers for m_MGMT recognize the fully methylated sequence. The u_MGMT assay served as a control for the PCR. The first PCR consisted of 35 cycles. Two μl of the first PCR product was diluted 1:20 and input to the second PCR of 30 cycles. The products from the second PCR were visualized on 4% agarose gels (NuSieve 3:1) to determine the MGMT methylation status. The outcome of the nested, gel-based MSP was considered valid when the following four criteria were fulfilled: i) PCR products of the expected sizes were detected on agarose gel (single bands); ii) a band for either u_MGMT, m_MGMT, or both was present; iii) routinely included positive and negative controls, including a no-template control, gave the expected result, and iv) the result was confirmed in an independent experiment starting with the bisulfite treatment of DNA. The valid results of all clinical samples were classified as methylated or non-methylated depending on the presence or absence of a band for m_MGMT. This procedure was used for the analysis of all samples in the laboratory at Centre Hospitalier Universitaire Vaudois.
Direct, Real-Time MSP
DNA Isolation
Two sections (for single assays) or four sections (for duplicate assays) were deparaffinized and DNA was extracted, using the classical phenol/chloroform extraction method, and resuspended in 50 μl of LoTE (3 mmol/L Tris, 0.2 mmol/L EDTA, pH 8.0). The DNA was quantified using the PicoGreen dsDNA quantitation kit (Molecular Probes, Invitrogen, Carlsbad, CA), following the manufacturer's directions, and up to 1.5 μg of DNA was input to the modification reaction.
DNA Modification
The bisulfite reaction was performed using the EZ DNA Modification Kit (Zymo, Orange, CA; D5002). This reaction selectively deaminates unmethylated cytosine residues resulting in a conversion to uracil, whereas 5-methyl cytosine residues are not modified. The modified DNA was eluted into 25 μl of Tris-HCl (1 mmol/L, pH 8.0) and then stored at −80°C. To perform duplicate assays for 105 samples, isolated DNA was divided into two independent aliquots for bisulfite modification. These modified DNA samples were each used for direct, real-time MSP.
Direct, Real-Time MSP
Analyte (m_MGMT and β-actin [ACTB]) quantification was performed by real-time MSP assays. These consisted of parallel amplification/quantification processes using specific primer and primer/detector pairs for each analyte using the Amplifluor assay format on an ABI Prism 7900HT instrument (Applied Biosystems, Foster City, CA). The analyte defined in the direct, real-time MSP was the MGMT promoter sequence and detects the fully methylated version. ACTB was used as a reference gene in the assay, using primers that are outside any CpG islands. The Amplifluor direct forward primers are preceded by the detection elements (underlined). The amplicon size is 136 bp for the m_MGMT analyte and 125 bp for the ACTB analyte, including the Amplifluor detection sequence. Sequence details for both forward and reverse primers are as follows: forward primer m_MGMT: 5′-AGCGATGCGTTCGAGCATCGCUTTTCGACGTTCGTAGGTTTTCGC-3′; reverse primer m_MGMT: 5′-CTCGAAACTACCACCGTCCCGA-3′; forward primer ACTB: 5′-AGCGATGCGTTCGAGCATCGCUTAGGGAGTATATAGGTTGGGGAAGTT-3′; reverse primer ACTB: 5′-AACACACAATAACAAACACAAATTCAC-3′.
The MGMT target sequence is located on chromosome 10 between positions 131155505 and 131155619 (RefSeq, NM_002412), whereas the ACTB target sequence is located on chromosome 7 between positions 5538428 and 5538325 (RefSeq, NM_001101), based on version 36.1 of the NCBI human genome.
The following thermal profile was used: stage 1, 50°C for 2 minutes; stage 2, 95°C for 10 minutes; stage 3, 95°C for 15 seconds, 62°C for 1 minute (=plateau data collection) for 45 cycles.
Quantification
The results were generated using the SDS 2.2 software (Applied Biosystems, Foster City, CA), exported as Ct values (cycle number at which the amplification curves cross the threshold value, set automatically by the software), and then used to calculate copy numbers based on a linear regression of the values plotted on a standard curve of 20 − 2 × 106 gene copy equivalents, using plasmid DNA containing the bisulfite-modified sequence of interest. Cell lines SW48 and HT29 (ATCC, Teddington, UK) were included in each experiment as positive and negative controls, respectively, and entered the procedure at the DNA extraction step.
A run was considered valid when the following five criteria were met: i) slopes of both standard curves were above −4 (PCR efficiency >77.8%); ii) r2 of at least 4 relevant data points above 0.990; iii) routinely included no-template control was not amplified; iv) 10% of a 1-μg conversion reaction of the positive cell line assay control SW48 was detectable; and v) 10% of a 1-μg conversion reaction of the negative cell line assay control HT29 was not detected within the standard curve.
In addition, results were considered invalid if criteria of DNA quantity and quality were not met. These were based on minimal copy numbers of ACTB and m_MGMT detected, as illustrated in Figure 1.
Figure 1.
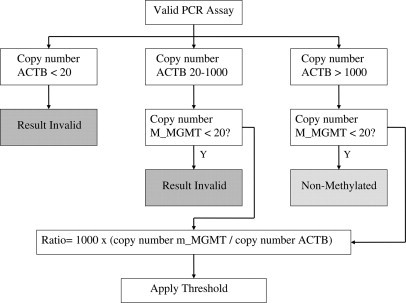
Decision tree for real-time MSP results.
Normalization of the Results
To compensate for variations in copy number due to differences in sample volume, sample handling, DNA isolation, and tumor heterogeneity, the m_MGMT copy numbers derived were divided by the ACTB copy numbers for that sample. This value was multiplied by 1000 for convenient handling, and the result was referred to as the ratio value. Cutoffs can then be applied to this ratio value to determine whether a sample is methylated or not.
Statistics
Statistical analyses were performed with R, a free software environment available at http://www.r-project.org/.
Dichotomization of the Direct, Real-Time MSP Results
The ratio values were log2 transformed for statistical analysis. The evaluation of the distribution of the MGMT measurements in the density plot was performed using 94 of 99 valid, duplicate samples with Ct values <40. Gaussian mixture models were fitted to the average data log2(1000 * m_MGMT/ACTB) of the direct, real-time MSP duplicates.21,22,23
Results
FFPE glioma tissue samples were analyzed in parallel to determine the methylation status of the MGMT promoter in independent laboratories using two distinct technical approaches. Both assays interrogate the same region, although using distinct reverse primers. The gel-based assay includes nine CpGs, whereas the real-time assay includes eight. This blinded study compared the results obtained by a direct, real-time MSP on a high-throughput platform to the nested, gel-based MSP assay previously shown to predict the benefit from the addition of the alkylating agent TMZ to the treatment of newly diagnosed glioblastoma in two clinical trials.9,10
Assay Validity
One hundred twenty-five samples were measured in duplicate and classified using the gel-based method, and 94 yielded valid results (75%). Results from 125 of 134 samples (29 single and 105 duplicate) analyzed by direct, real-time MSP assays were valid (93%).
Reproducibility of the Direct, Real-Time MSP Assay
Figure 2 illustrates the reproducibility between independent replicates, especially at higher ratio values, in agreement with the characteristics of the reliability of real-time PCR (Pearson correlation 0.996). Many of the samples with very low ratio values show m_MGMT copy numbers below the lower limit of the standard curve. In practice, no ratio is calculated for these samples, and they are considered non-methylated. This reproducibility shows that the bisulfite treatment of the DNA introduced no major variability into the assay. This is in accord with a recent report quantifying precision and performance of bisulfite-converted DNA with real-time PCR.24
Figure 2.
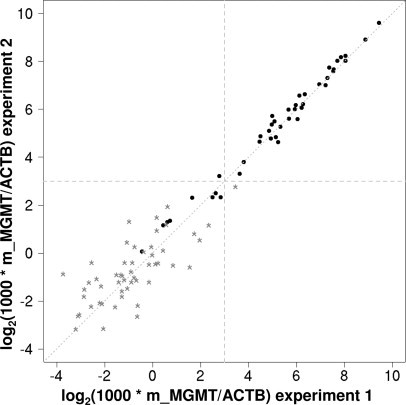
Reproducibility of duplicate measurements. Dotted line represents identity line (x = y), and dashed lines represent the cutoff between classification as methylated or non-methylated according to the mixture Gaussian model (see Figures 3 and 4). Pearson correlation 0.996, Spearman correlation 0.93, N = 94. Black dots represent samples with m_MGMT copies >20; gray stars represent samples with m_MGMT copies <20 and Ct <40.
Definition of Cutoff for the MGMT Ratio Value
Samples evaluated in duplicate by the newly established direct, real-time MSP assay revealed a clear bimodal distribution of the measurements as shown in Figure 3. This allows definition of a discriminatory cutoff for the determination of the MGMT methylation status by fitting a normal mixture model.22,23 The best model, based on 94 duplicate samples with Ct <40, yields 2 Gaussians of equal variance (σ2 = 2.3), and means of −0.52 and 6.1 in log2 units of the ratio value (Figure 4A). This corresponds to mean ratio values of 0.7 and 68.1, respectively. According to this model, which defines two classes, non-methylated and methylated MGMT, the optimal cutoff corresponds to a ratio value of 8 (log28 = 3) (Figure 4B). Close to this cutoff there are some samples for which the uncertainty of classification is high, hence the probability of belonging to one class or the other is close to 50% (Figure 4C). This suggests the use of a gray zone for diagnostic purposes and permits the definition of different thresholds depending on the clinical questions asked. The threshold for 95% probability of methylation is a ratio value of 16 (log2 ratio = 4) and that for non-methylated MGMT is 4 (log2 ratio = 2) (Figure 4D).
Figure 3.
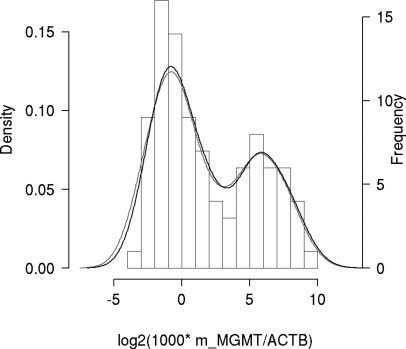
Density plot of normalized methylated MGMT copy number in glioma. Histogram of average results from 94 samples with duplicate measurements; the lines represent results from each replicate. Only samples with Ct values <40 for m_MGMT are included. The minimum of the density of the Gaussian mixture between the two local maxima is at the ratio of log2(1000 * m_MGMT/ACTB) = 3 (ratio value of 8).
Figure 4.
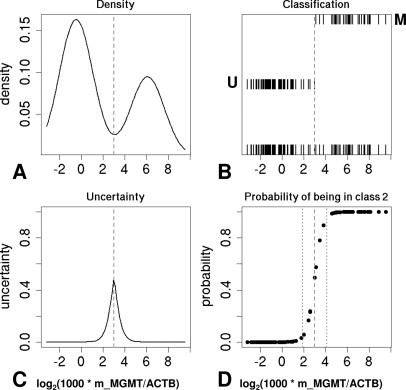
Definition of natural cut-off for methylated MGMT. Density (A), Classification (B), Uncertainty of classification (C), and posterior Probability (D) of class 2 (promoter methylation) obtained by fitting a mixture model to the average log2(1000 * m_MGMT/ACTB). In the classification plot (B), all of the data are displayed at the bottom; assignment to the class with highest posterior probability at two different levels is above (M, class 2, methylated; U, class 1, non-methylated). Uncertainty (C) was defined as the smaller of the two posterior probabilities. A gray dashed line represents the optimal cutoff according to the selected model (log2 ratio = 3; ratio value = 8). The region between the gray dotted lines (D) defines a classification “gray zone”; the thresholds for 95% posterior probability of class 1 and class 2, respectively, are 2 (ratio value = 4) and 4 (ratio value = 16).
Comparison of Direct, Real-Time MSP with Nested, Gel-Based MSP
The comparison of the results of the two tests was based on 91 samples for which both tests gave valid results. This comparison is visualized in Figure 5 and is summarized in Table 1. There is a good concordance between the two tests using a cutoff at the ratio value of 8 (82/91, 90%). Cohen's kappa coefficient of 0.80, 95% confidence interval 0.67 to 0.92, was similarly reflected in the one-sample proportions test (with continuity correction), with agreement of 0.90, with 95% confidence interval 0.82 to 0.95. The lack of a “gold standard” or other source of “true values” makes discussion of sensitivity and specificity of the real-time assay problematic. Comparison with the clinically validated nested, gel-based assay shows, at the cutoff ratio value of 8, positive and negative agreement of 0.80 (32/40) and 0.98 (50/51), respectively, and the positive and negative predictive values were 0.97 (32/33) and 0.86 (50/58) for the given prevalence of 0.56 (51/91).
Figure 5.
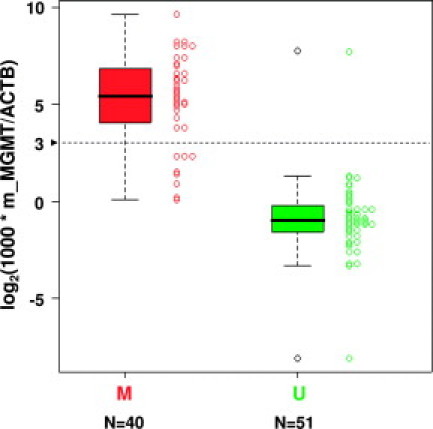
Comparison between direct, real-time MSP and nested, gel-based MSP results. Box-plots and strip-charts compare the log2(1000 * m_MGMT/ACTB) (y axis) values determined for 91 samples by direct, real-time MSP assay to the classification obtained with the nested, gel-based assay into non-methylated (green) and methylated (red) samples (x axis). A dashed line represents the cutoff defined with the Gaussian mixture model (Figures 3 and 4).
Table 1.
Concordance of Classification of the MGMT Status by the Two Techniques
| Direct, real-time MSP (ONCO) ratio value cutoff = 8 | |||
|---|---|---|---|
| Nested, gel-based MSP (CHUV) | Methylated | Non-methylated | Total |
| Methylated | 32 | 8 | 40 |
| Non-methylated | 1 | 50 | 51 |
| Total | 33 | 58 | 91 |
CHUV, Centre Hospitalier Universitaire Vaudois; ONCO, OncoMethylome Sciences.
The best concordance between the two tests would be at a lower cutoff using a ratio value of 3 with a Cohen's kappa coefficient of 0.86, 95% confidence interval 0.73 to 0.97. The agreement would also be higher if the MGMT copy number was used without normalization to the ACTB copy number, which is more similar to the principle used by the nested, gel-based assay that is not normalized. However, only few samples are impacted by such analysis.
Application for Diagnostic Purposes
Figure 1 illustrates the decision tree for interpretation of results from clinical samples for diagnostic purposes. It integrates both assay performance and the strategy for classifying the test results according to the thresholds defined.
Discussion
A sensitive and specific direct, real-time MSP assay has been developed to reliably detect the methylation status of the MGMT gene promoter in clinical samples of FFPE glioma tissue for diagnostic purposes. The results of the methylation status of this test are in good concordance with results obtained with the nested, gel-based MSP assay that established the predictive value of the MGMT methylation status in glioblastoma for the relative benefit from TMZ therapy.10
The discordance between the tests is largely confined to a few samples with values slightly lower than the cutoff defined by the mixture model. This is not surprising, since the cutoff of the gel-based assay is defined by the visual presence or absence of the PCR product without normalization. Normalization to amplifiable DNA content, measured here as a ratio to ACTB, is not an absolute measurement, since “normal tissue” contaminations are present in all clinical tumor samples, and genomic copy number aberrations are common in glioblastoma, including on chromosomes 10 and 7, where the MGMT and ACTB gene reside, respectively. However, neither homozygous deletions nor high level amplifications have been reported for MGMT or ACTB that potentially could be problematic for interpretation of results. The ACTB is also used as a control gene by other groups using direct, real-time PCR for DNA methylation analysis.18
The direct, real-time MSP assay more often yielded a valid result with FFPE tissue specimens than did the nested, gel-based MSP assay. This improved performance is likely due to the smaller amplicon size of the real-time MSP assay.
The quantitative measure obtained and the bimodal distribution of the values will allow definition of clinically relevant thresholds for stratified therapy. This study demonstrates that there is an important separation of values around that cutoff, suggesting that there are two distinct populations underlying the data, and that few samples would be expected with values near the cutoff. To arrive at the best, most clinically useful cutoff for patient management, data relating the real-time quantitative assay to patient response must be collected.
The test described here is being prospectively used for randomizing 1153 patients in an ongoing clinical phase III trial (RTOG 0525/EORTC 26052-22053) testing standard versus dose-intense adjuvant TMZ in patients with newly diagnosed glioblastoma (trial is reviewed in Stupp et al25). The working parameters used for randomization are ratio values of 5 and 12 as the lower and upper cutoffs for classification of non-methylated and methylated MGMT, respectively, with definition of a gray zone for the region with high uncertainty in between (5 to 12). These cutoffs had been defined previously in a smaller data set and are consistent with the present study suggesting the cutoff at ratio value 8. Among the goals of this clinical trial is to prospectively validate the use of MGMT methylation status for predicting benefit from alkylating agent therapy. The measurement of MGMT promoter methylation in this trial will provide further information on a clinically relevant cutoff for the MGMT ratio value.
This sensitive and robust high-throughput test for evaluating the methylation status of the MGMT gene may provide an important pharmacogenomic tool for individualized management of patients considered for treatment with TMZ or other alkylating agent chemotherapy.
Acknowledgements
We thank Drs. M.J. van den Bent, M.C.M. Kouwenhoven, R. Stupp, R.C. Janzer, I. Vajtai, and P. Hau for providing paraffin-embedded glioma samples.
Footnotes
Supported by OncoMethylome Sciences, the Nélia and Amadeo Barletta Foundation (to M.E.H.), and the National Center of Competence in Research (NCCR) Molecular Oncology (to M.D., E.M., M.E.H.). M.E.H. is also a compensated advisor for OncoMethylome Sciences.
I.V. and S.C. contributed equally to this work.
References
- 1.Pegg AE. Repair of O(6)-alkylguanine by alkyltransferases. Mutat Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 2.Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 3.Stojic L, Cejka P, Jiricny J. High doses of SN1 type methylating agents activate DNA damage signaling cascades that are largely independent of mismatch repair. Cell Cycle. 2005;4:473–4777. doi: 10.4161/cc.4.3.1528. [DOI] [PubMed] [Google Scholar]
- 4.Qian XC, Brent TP. Methylation hot spots in the 5′ flanking region denote silencing of the O6-methylguanine-DNA methyltransferase gene. Cancer Res. 1997;57:3672–3677. [PubMed] [Google Scholar]
- 5.Watts GS, Pieper RO, Costello JF, Peng YM, Dalton WS, Futscher BW. Methylation of discrete regions of the O6-methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol Cell Biol. 1997;17:5612–5619. doi: 10.1128/mcb.17.9.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 7.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 8.Criniere E, Kaloshi G, Laigle-Donadey F, Lejeune J, Auger N, Benouaich-Amiel A, Everhard S, Mokhtari K, Polivka M, Delattre JY, Hoang-Xuan K, Thillet J, Sanson M. MGMT prognostic impact on glioblastoma is dependent on therapeutic modalities. J Neurooncol. 2007;83:173–179. doi: 10.1007/s11060-006-9320-0. [DOI] [PubMed] [Google Scholar]
- 9.Hegi ME, Diserens AC, Godard S, Dietrich PY, Regli L, Ostermann S, Otten P, Van Melle G, deTribolet N, Stupp R. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. [DOI] [PubMed] [Google Scholar]
- 10.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 11.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 12.Stupp R, Hegi ME, van den Bent MJ, Mason WP, Weller M, Mirimanoff RO, Cairncross JG. Changing paradigms—an update on the multidisciplinary management of malignant glioma. Oncologist. 2006;11:165–180. doi: 10.1634/theoncologist.11-2-165. [DOI] [PubMed] [Google Scholar]
- 13.Everhard S, Kaloshi G, Criniere E, Benouaich-Amiel A, Lejeune J, Marie Y, Sanson M, Kujas M, Mokhtari K, Hoang-Xuan K, Delattre JY, Thillet MGMT methylation: a marker of response to temozolomide in low-grade gliomas. Ann Neurol. 2006;60:740–743. doi: 10.1002/ana.21044. [DOI] [PubMed] [Google Scholar]
- 14.Brandes AA, Tosoni A, Cavallo G, Reni M, Franceschi E, Bonaldi L, Bertorelle R, Gardiman M, Ghimenton C, Iuzzolino P, Pession A, Blatt V, Ermani M. Correlations between O6-methylguanine DNA methyltransferase promoter methylation status, 1p and 19q deletions, and response to temozolomide in anaplastic and recurrent oligodendroglioma: a prospective GICNO study. J Clin Oncol. 2006;24:4746–4753. doi: 10.1200/JCO.2006.06.3891. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M, Gaidano G, Goodman SN, Zagonel V, Capello D, Botto B, Rossi D, Gloghini A, Vitolo U, Carbone A, Baylin SB, Herman JG. Hypermethylation of the DNA repair gene O(6)-methylguanine DNA methyltransferase and survival of patients with diffuse large B-cell lymphoma. J Natl Cancer Inst. 2002;94:26–32. doi: 10.1093/jnci/94.1.26. [DOI] [PubMed] [Google Scholar]
- 16.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 17.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 18.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikeska T, Bock C, El-Maarri O, Hubner A, Ehrentraut D, Schramm J, Felsberg J, Kahl P, Buttner R, Pietsch T, Waha A. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn. 2007;9:1525–1578. doi: 10.2353/jmoldx.2007.060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, Belinsky SA. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 21.Fraley C, Raftery AE. Model-based clustering. Discriminant analysis, and density estimation. J Am Stat Assoc. 2002;97:611–631. [Google Scholar]
- 22.Fraley C, Raftery AE. MCLUST Version 3 for R: Normal Mixture Modeling and Model-based Clustering. Technical Report no. 504. Department of Statistics, University of Washington; 2006. [Google Scholar]
- 23.Altman DG. Practical Statistics for Medical Research. Chapman & Hall; London: 1991. [Google Scholar]
- 24.Ogino S, Kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ, Laird PW, Loda M, Fuchs CS. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stupp R, Hegi ME, Gilbert MR, Chakravarti A. Chemoradiotherapy in malignant glioma: standard of care and future directions. J Clin Oncol. 2007;25:4127–4136. doi: 10.1200/JCO.2007.11.8554. [DOI] [PubMed] [Google Scholar]
- 26.Kagan J, Srivastava S, Barker PE, Belinsky SA, Cairns P. Towards clinical application of methylated DNA sequences as cancer biomarkers: a Joint NCI's EDRN and NIST Workshop on Standards. Methods, Assays, Reagents and Tools. Cancer Res. 2007;67:4545–4549. doi: 10.1158/0008-5472.CAN-06-2888. [DOI] [PubMed] [Google Scholar]


