Abstract
The observation of heterogeneous protein folding kinetics has been widely interpreted in terms of multiple independent unrelated pathways (IUP model), both experimentally and in theoretical calculations. However, direct structural information on folding intermediates and their properties now indicates that all of a protein population folds through essentially the same stepwise pathway, determined by cooperative native-like foldon units and the way that the foldons fit together in the native protein. It is essential to decide between these fundamentally different folding mechanisms. This article shows, contrary to previous supposition, that the heterogeneous folding kinetics observed for the staphylococcal nuclease protein (SNase) does not require alternative parallel pathways. SNase folding kinetics can be fit equally well by a single predetermined pathway that allows for optional misfolding errors, which are known to occur ubiquitously in protein folding. Structural, kinetic, and thermodynamic information for the folding intermediates and pathways of many proteins is consistent with the predetermined pathway–optional error (PPOE) model but contrary to the properties implied in IUP models.
Keywords: foldons, PPOE model, staphylococcal nuclease
That proteins might fold by way of some predetermined pathway was first suggested by C. Levinthal (1), who reasoned that folding through a random search process would take far longer than the subsecond time scale often observed. Experimental work driven by this concept found evidence for distinct pathway intermediates (2). However, theoretical work showed that the energetically downhill nature of the conformational search might by itself be sufficient to drive random folding on a realistic time scale (3). No specific pathway would then be necessary. This scenario has been formalized in the funneled energy landscape view for protein folding (4–8). Following this precedent, experimental results for a number of proteins have been similarly interpreted in terms of multiple unrelated parallel pathways (9–16). The critical observation is that protein folding can be kinetically heterogeneous. Different fractions of a folding population fold at different rates and exhibit different populated intermediates, suggesting alternative pathways. This view is often represented in terms of multiple tracks through the funneled energy landscape. We refer to this view as the independent unrelated pathways (IUP) model to emphasize that intermediate structures in the different tracks have no particular relationship.
A much more determinate pathway is supported by structural information derived experimentally for folding intermediates in many proteins (17–20). This information indicates that protein folding intermediates are definitively native-like, constructed from cooperative foldon units of the target native protein. It appears that native-like foldon units are formed and docked into place one after another in a stepwise sequence, each one guided and stabilized by prior complementary structure just as the foldon units fit together in the native protein. These two principles—the cooperativity of the structural elements that compose protein molecules and the energetically favorable association of complementary structures—are well recognized in many other molecular processes. Acting together, they can cause all of the molecules in a refolding population to fold through essentially the same predetermined pathway.
However, one often finds that different population fractions of a given protein can fold with different kinetics (9–16). To account for heterogeneous kinetic folding, it is necessary to modify the predetermined pathway model to recognize the ubiquity of optional misfolding errors. The resulting predetermined pathway–optional error (PPOE) model (21) attributes heterogeneous folding to chance misfolding errors within a given pathway rather than to multiple alternative pathways. In this view, two-state folding occurs when the misfolding probability is zero, as is often observed for small proteins. Three-state folding dominates when the misfolding probability at some pathway point is large. More generally, heterogeneous folding occurs because error formation is probabilistic rather than intrinsic to the folding process. Different fractions of the refolding population can commit different errors and therefore block at different pathway points, populate different corrupted intermediates, and reach the native state at different rates (21). This behavior gives the appearance of different pathways even though all of the population moves through the same productive on-pathway intermediates.
This article considers the heterogeneous folding behavior of the well studied staphylococcal nuclease (SNase) protein, which has been thought to require more than one independent pathway (12). We repeated the kinetic studies that led to the IUP model for SNase and compared the ability of the IUP and PPOE models to explain the folding kinetics observed. The results show that both models are able to fit kinetic folding behavior equally well. This capability is general (21). Thus, unlike previous supposition, the observation of kinetic heterogeneity allows but does not require an IUP model.
This result raises a more fundamental issue. The two different models project fundamentally different mechanisms for the protein folding process. It is essential to decide between them. Kinetic behavior alone is unable to make this important distinction. It is necessary to compare the properties of intermediates and pathways implied by the different models with structural and mechanistic information and not just kinetic behavior. We discuss available information in this light.
Results
Kinetic Folding and Unfolding by Stopped-Flow.
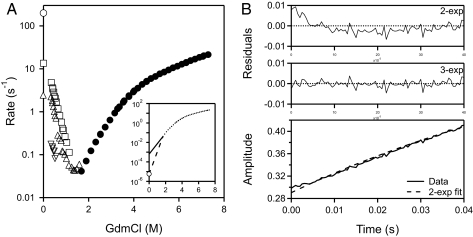
Fig. 1A shows kinetic chevron plots for SNase folding and unfolding measured by change in fluorescence of the single near-C-terminal Trp-140 residue. Refolding by dilution from denaturing GdmCl produces two major kinetic folding phases with similar rate constants and relative amplitudes at low denaturant of 55% faster phase to 40% slower phase. As denaturant concentration is increased the two major phases tend to merge. At near zero denaturant a lag phase in native-state formation is also measured.
Fig. 1.
SNase folding and unfolding kinetics. (A) Chevron plot for folding and unfolding rates against final GdmCl concentration, measured as the change in fluorescence between the unfolded state and the native state (intermediate fluorescence is equal to the U state). Folding rates were obtained by multiphase fitting of kinetic data for native-state formation like that shown in B. (Inset) Unfolding rate at zero denaturant measured by kinetic native-state hydrogen exchange (24). (B) Kinetic folding data for native-state formation at zero denaturant, shown at the earliest folding times to exhibit the lag phase. The dashed curve is the two-exponential fit. The plot of residuals shows the reality of the lag phase, which is confirmed by similar results at other conditions and in other laboratories (see Table 1).
No very slow proline-dependent phase is seen because a crucial proline has been removed in the mutant SNase used here (P117G/H124L). A minor very slow folding phase (≈6% amplitude), on the time scale of seconds at low denaturant, attributed to cis-trans isomers of nonprolyl residues (12, 22), was ignored in subsequent model fitting.
Unfolding is simpler. The removal of Pro-117, which exists in slowly interconverting cis and trans conformers in wild-type SNase makes unfolding kinetics monoexponential (23). However, the chevron unfolding arm (d(log k)/d[GdmCl]) is not linear. It curves continually to slower rates. Fig. 1A Inset includes the unfolding rate measured at zero denaturant by kinetic native-state hydrogen exchange (24). The curvature is due to the changing dominance of a succession of on-pathway barriers, indicating the presence of several on-pathway intermediates.
Fig. 1B shows a kinetic folding trace starting with the protein fully unfolded at pH 2 (shown by CD) and then jumping to higher pH to initiate refolding at zero denaturant. A burst phase increase in fluorescence (not shown), initially attributed to the formation of a fast burst phase intermediate, is an artifact due to fluorescence quenching at the acid unfolding pH (25, 26). The refolding kinetics at zero denaturant shows an initial lag phase to which standard multiexponential fitting assigns a negative amplitude (−5%). The lag phase signals the transient accumulation of an obligatory blocked intermediate before the rate-limiting step. The lag phase rate increases from 45 s−1 at pH 5.3 to 200 s−1 at pH 8.5. These results agree closely with published data as noted in Table 1(12, 26, 27).
Table 1.
Comparison with published kinetic folding rates
| Construct | λ1, s−1 | A1 (lag) | λ2, s−1 | A2 | λ3, s−1 | A3 | λ4, s−1 | A4 |
|---|---|---|---|---|---|---|---|---|
| H124L† | 38 ± 9 | −0.07 ± 0.01 | 6.2 ± 0.3 | 0.53 ± 0.01 | 0.93 ± 0.07 | 0.22 ± 0.01 | 0.025 ± 0.001 | 0.17 ± 0.003 |
| Pro-† | 99 ± 15 | −0.08 ± 0.01 | 7.8 ± 0.4 | 0.69 ± 0.03 | 2.3 ± 0.2 | 0.24 ± 0.03 | — | — |
| Pro-‡ | 134 ± 21 | −11.3 ± 0.1% | 17.2 ± 0.5 | 57.3 ± 0.9% | 4.7 ± 0.1 | 35.3 ± 1.1% | 0.82 ± 0.04 | 4.5 ± 0.2% |
| WT*§ | 40.6 | −0.23 | 10.4 | 1.78 | 1.35 | 0.4 | 0.1 | 0.26 |
| WT*/Trp76§ | 41.3 | −0.32 | 11.3 | 1.51 | 2.3 | 0.43 | 0.01 | 0.24 |
| P117G/H124L¶ | 47 ± 3 | −0.10 ± 0.01 | 9.7 ± 0.2 | 0.63 ± 0.01 | 2.4 ± 0.1 | 0.17 ± 0.01 | 0.30 ± 0.04 | 0.12 ± 0.01 |
| P117G/H124L‖ | 200 ± 41 | −0.016 ± 0.002 | 13.5 ± 0.2 | 0.26 ± 0.003 | 2.4 ± 0.1 | 0.153 ± 0.003 | — | — |
Values listed are at zero denaturant.
†Unfold at pH 2.2, measure folding at pH 5.3, 15°C, 0.1 M sodium acetate (27).
‡Unfold at pH 2.0, measure folding at pH 6.0, 20°C, 0.1 M sodium cacodylate, 2 mM EGTA (12). “%” denotes relative amplitudes.
§Unfold at pH 2.0, measure folding at pH 5.2, 15°C, 0.1 M sodium acetate (26). WT* is SNase P47G/P117G/H124L.
¶This work. Same conditions as †.
‖This work. Unfold at pH 2.2, measure folding at pH 8.5, 50 mM CHES, 20°C, 0.1 M NaCl.
The Independent Unrelated Pathways (IUP) Model.
Kamagata et al. (12) used direct and interrupted folding experiments to show that the fluorescence changes observed in SNase folding directly measure acquisition of the native state. This occurs because the populated kinetic intermediates of SNase have fluorescence that is identical to the unfolded state, and the C-terminal segment that harbors the lone SNase tryptophan, Trp-140, is the last to fold (24).
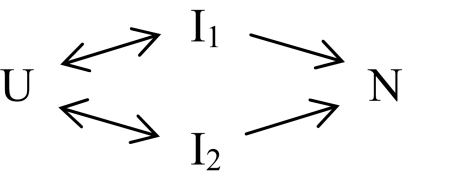
In a conventional pathway (U to I to N), the native state is reached in a single kinetic phase rate-limited by the slowest pathway step, with the possible addition of a kinetic lag phase (negative amplitude). However, SNase reaches the native state in three kinetic phases (lag phase plus two major phases; Fig. 1). Kinetic principles dictate that the three kinetic phases require the significant population of four states, namely U, N, and two intermediates. The two positive amplitude phases indicate two different rate-limiting steps for reaching N. These considerations led Kamagata et al. (12) to write the minimal reaction in Scheme 1 thought necessary to explain the SNase kinetic folding data (IUP model). A global fit of Scheme 1 to the kinetic data is shown in Fig. 2 A–C.
Scheme 1.
Fig. 2.
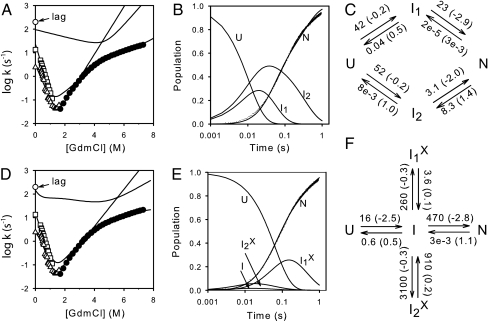
Global fit of the measured SNase kinetic folding and unfolding data by an IUP model (A–C) and a PPOE model (D–F). Shown are the chevron display of denaturant-dependent data (from Fig. 1), measured data for native-state acquisition at zero denaturant, and reaction schemes for the two different models with best global fit parameters (denaturant-dependent m values in parentheses). The curves drawn are the curves predicted by the best fit parameters for the denaturant-dependent chevron and for the time-dependent populations (zero denaturant) of the unfolded and native states and the two intermediates that significantly populate in the different models. A minor folding phase (amplitude ≈6%) on the 1-sec time scale (see Fig. 1A) was ignored.
Scheme 1 and other IUP reaction schemes in general are written to symbolize that different population fractions flow through different intermediates in different pathways. An inherent assumption is that the specified intermediates (Ii) will accumulate and will go forward at different rates.
The Predetermined Pathway–Optional Error (PPOE) model.
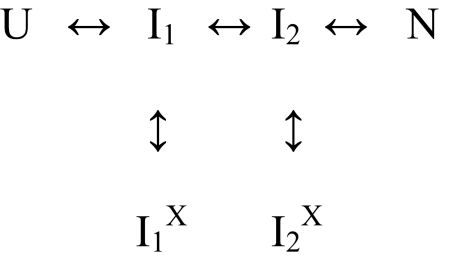
Contrary to the conventional assumption, heterogeneous folding does not require that proteins fold through independent parallel pathways. A single pathway that is traversed by all of the protein molecules in common will produce heterogeneous folding behavior if different population fractions can optionally experience different blocking errors. The general PPOE model is given by Krishna and Englander (21). A simplified “double T” version can be written as in Scheme 2.
Scheme 2.
PPOE models picture that the entire refolding population moves through the same obligatory productive on-pathway intermediates (Ii). A chance misfolding error can occur at any step to produce IiX. The probability for any given error, and therefore the fractional population that will experience it, is determined by the misfolding rate constant (Ii to IiX) divided by the sum of the three rate constants away from Ii. If the return error-repair rate (IiX to Ii) is slow, then IiX will accumulate and that population fraction will exhibit slowed folding.
Fig. 2 D–F shows a global fit to the SNase kinetic data by an even more minimal PPOE model called the Plus model. The Plus model has one productive intermediate and two misfolded variants. It is kinetically equivalent to the reaction in Scheme 2 because, as the fitting of Scheme 2 to the data in Fig. 2 shows, I1 and I2 do not kinetically accumulate. Thus, the two rate-limiting phases are independent of the I1-to-I2 forward and reverse rates.
The PPOE and IUP models both fit the data equally well, albeit imperfectly, with the same number of fitting constants. For more perfect fitting, some more complex reaction scheme would be required for either model. The important result of this exercise is that, contrary to previous supposition, heterogeneous folding does not require multiple independent pathways. A single predetermined pathway with optional errors fits the data just as well.
Discussion
This article starts by asking whether the experimental observation of kinetically heterogeneous folding in the SNase protein requires multiple parallel pathways. It does not.
Earlier work showed that a predetermined pathway model with optional errors can account for the heterogeneous folding behavior of cytochrome c (17). Recent progress shows that the multiple kinetic folding phases and intermediates of α-tryptophan synthase previously attributed to four independent folding tracks are in fact due to optional proline misisomerization errors within a single pathway (28). Extensive kinetic folding and unfolding results for hen egg lysozyme, thought to require independent parallel pathways, were found to be explained at least as well by a PPOE model (21). The present article demonstrates that the heterogeneous folding of SNase, previously thought to require alternative parallel pathways, can be explained equally well by a PPOE model. These results and additional considerations (21) show that both the multipathway IUP model and the predetermined pathway model with optional errors will be able to fit any kinetic folding behavior whether folding is heterogeneous or not.
The different models propose wholly different views of the fundamental nature of folding intermediates and folding mechanisms. Does folding proceed by an unguided search through multiple unpredictable routes driven only by the downhill nature of the energy landscape, or does it proceed through rather strictly defined intermediates and pathways predetermined by native-like structure and interactions encoded in the protein's amino acid sequence? To distinguish which model more correctly describes real protein folding behavior, it will be necessary to go past mere kinetic fitting. Fortunately, a quantity of detailed structural and mechanistic information is now available.
Determinants of IUP Models.
Theoretical investigations generally find that folding is directed by no particular guiding principles other than its energetically downhill nature. Heterogeneous folding occurs because different molecules, although they are identical, fold through different intermediates in alternative pathways that have no necessary relationship. When written to explain experimental results (e.g., Scheme 1), formal IUP models can be seen to assume that stable intermediates are absent in two-state folding, that three-state folding occurs because stable intermediates that do occur are naturally slow, and that heterogeneous folding occurs because different population fractions fold through different pathways.
The ability to measure the structure and properties of protein folding intermediates has made available a great deal of information against which folding models can be compared. Thermodynamic principle requires that proteins repeatedly unfold and refold even under native conditions, revisiting their normal folding intermediates and recapitulating their normal folding process. All or parts of this process have been observed by site-resolved hydrogen exchange (19, 29–35), by a related thiol reactivity method (36), by NMR relaxation dispersion (37–39), and by theoretical analysis (40). Stable on-pathway intermediates built from cooperative foldon elements of the native protein are seen even under two-state folding conditions. This has been shown for Cyt c (41), triose phosphate isomerase (36), RNase H (19), OspA (33), and apoCyt b562 (30, 32). Furthermore, detailed pathway information indicates that native-like foldon units are systematically put into place in well defined sequential pathways much as they fit together in the target native protein (19, 30, 36, 41–48).
These observations are contrary to IUP models. Stable intermediates (relative to U) are not generally absent in two-state folding. Three-state folding is not produced simply by the presence of stable intermediates. They are not intrinsically slow. Folding can be kinetically heterogeneous, but it appears that all of the population folds through the same sequence of structured intermediates.
Determinants of PPOE Models.
The PPOE model projects a nearly opposite view, which does match experimental information. No matter whether kinetic folding appears to be two-state, three-state, or heterogeneous, all of the population runs through essentially the same productive, on-pathway, native-like intermediates, constructed from a small number of foldon building blocks that are intrinsic to the target native protein.
In two-state folding, intermediates are kinetically invisible, but not because they are absent or unstable. In real cases as just noted and in the PPOE model, on-pathway intermediates are present and may be stable (native-like foldons). They tend to go forward efficiently because preexisting native-like structure guides and stabilizes the formation and docking of incoming complementary native-like structure (sequential stabilization). A small guided search is naturally far faster than the initial whole molecule unguided search for the nucleating native-like topology, whether intermediates are more stable than U or not. Thus, kinetic folding might be expected to be intrinsically two-state, rate-limited by the initial whole molecule search barrier (49–52).
However, proteins often fold with three-state kinetics. In the PPOE model, three-state folding is due to the chance occurrence of some slowly repaired misfolding error. This slows folding and causes the transient accumulation of the corrupted intermediate (IiX in Scheme 2). In fact, intermediate forms that accumulate in multistate folding have been widely found to incorporate some misfolding error [Cyt c (53, 54), RNase H (19), apoMb (43, 55), apoCyt b562 (30), Im7 (56), α-tryptophan synthase (28), OspA (33)], even though the misfold-containing intermediates have essentially the same native-like foldon structure as their parent on-pathway intermediates.
In the PPOE model, heterogeneous folding arises because misfolding is probabilistic. Different population fractions can experience different misfolding errors, or none at all. Any given error can slow the further folding of that particular corrupted intermediate and that particular population fraction. The ubiquity, high probability, and optional nature of misfolding errors in protein folding are thoroughly documented (prolyl and nonprolyl peptide bond misisomerization, transient aggregation, nonnative hydrophobic clustering, disulfide shuffling, heme misligation, and alternative domain docking modes). Where adequate data are available for testing, as for SNase here and hen egg lysozyme before (21), analysis shows that heterogeneous kinetic folding can be quantitatively accounted for by a predetermined pathway model together with the probabilistic occurrence of misfolding errors.
The Principles of Protein Folding.
The variety of protein folding behavior can seem to represent an incoherent collection of bewildering complexity. As organized in the PPOE model, observed behavior can be seen to flow from three basic principles. Folding intermediates are constructed by the association of intrinsically cooperative foldon units of the target native protein. Their formation and docking are driven by a sequential stabilization process in which prior native-like structure acts as a template that guides and stabilizes the formation of incoming native-like foldons with complementary structure. During kinetic folding any intermediate can become corrupted and taken off the main pathway by some slowly repaired optional misfolding error.
Each of these three motivating principles is well recognized in many other molecular contexts. One has long understood that secondary structural elements tend to act as cooperative folding units (57, 58). When built into three-dimensional proteins, their cooperative unit behavior may well be modified, but it seems unlikely that the cooperative property will be lost. Experiment shows that measured foldon units do mimic elements of the native protein. Thus, it may be expected that the unit of protein folding is not the individual amino acid but rather intrinsically cooperative structural units.
Once some native-like intermediate structure is in place, it will naturally tend to act as a template that can guide and stabilize the formation of subsequent native-like complementary structure. It is obvious that complementary structures are mutually stabilizing (59, 60). Folding by sequential stabilization is simply an intramolecular version of analogous observations in bimolecular interactions, for example the folding of intrinsically disordered peptides upon encounter with their target proteins (61), domain swapping (62), and amyloid growth reactions.
These two principles, the cooperativity of native-like foldon units and their sequential stabilization by previously formed complementary structure, can be expected to generate a well defined, stepwise, sequential, native-like, kinetically two-state pathway. However, the often observed heterogeneity of protein folding points to a third factor. Protein folding is difficult (1), and errors in the protein folding process are ubiquitous. As shown here and before (21), the optional nature of misfolding errors will populate alternative intermediates and produce kinetic heterogeneity.
Conclusions
Results and analysis shown here and previously demonstrate that experimentally observed protein folding kinetics do not require multiple independent pathways. Folding kinetics, whether two-state, three-state, or heterogeneous, can be quite generally accounted for by the PPOE model. Furthermore, the properties of intermediates, barriers, and pathways, experimentally determined for many proteins, contradict the properties expected for IUP models but are intrinsic to the PPOE model. These considerations consistently support the view that the folding process sequentially forms and assembles foldon units of the target native protein in a predetermined pathway sequence that is governed by native-like foldon–foldon interactions but is subject to chance misfolding errors. Strong support comes also from the fact that this folding behavior can be seen to stem from three well recognized principles: the cooperativity of structural elements (foldon units), the mutual stabilization of complementary structures (sequential stabilization), and the tendency of folding proteins to commit misfolding errors.
Materials and Methods
Protein Preparation.
The plasmid pTSN2cc containing the SNase gene with two stabilizing mutations, P117G/H124L (provided by John L. Markley, University of Wisconsin, Madison), was transformed into E. coli BL21(DE3)/pLysS cells. SNase was expressed and purified according to Royer et al. (63) with some modifications.
Folding and Unfolding.
Kinetic folding and unfolding were measured by stopped-flow fluorescence (50 mM Tris at pH 8.0; BioLogic Science Instruments SFM-400). The change in fluorescence directly measures native-state acquisition because the fluorescence of intermediates is identical to that of the initial unfolded state (12). Multiexponential fitting of the time course of native-state formation yielded the several kinetic folding phases shown in the figures.
The refolding of SNase initially unfolded in 4 M GdmCl was measured at various final GdmCl concentrations, with final protein concentration of 20 μM. For folding in zero denaturant, SNase initially unfolded at pH 2 was mixed into pH 8.5, 50 mM CHES buffer (2-[N-cyclohexylamino]ethanesulfonic acid). Experiments were done in 0.1 M NaCl at 20°C.
Unfolding measured at various concentrations of GdmCl was monoexponential.
Model-Dependent Data Fitting.
Folding and unfolding data for SNase were fit to the kinetic schemes in Fig. 2, according to the procedure described in ref. 21. Each microscopic rate constant kij, connecting species i and j, was assumed to depend on denaturant concentration D according to the usual relationship,
where kij0 is the rate at zero denaturant concentration and mij is the slope of the dependence on denaturant (D) of the term RT ln kij.
Because the measured fluorescence intensity is only from the native state (see above), the native-state population was calculated from the measured kinetics by using the relationship
where Ft, F∞, and F0 are the measured fluorescence intensities at time t, at the end, and at the zero time along the kinetic reaction. Fig. 2B shows the native population with time and was used in data fitting.
Goodness of the chevron and kinetic fit was judged by using the global reduced χ2 parameter defined as described in ref. 21.
Acknowledgments.
This work was supported by National Institutes of Health Grants GM031847 and GM075105 (to S.W.E.), the Mathers Charitable foundation (S.W.E.), and scholarships from the Natural Sciences and Engineering Research Council of Canada and the Fonds Québécois de la Recherche sur la Nature et les Technologies (to S.B.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Levinthal C. Are there pathways for protein folding? J Chim Phys. 1968;65:44–45. [Google Scholar]
- 2.Kim PS, Baldwin RL. Intermediates in the folding reactions of small proteins. Annu Rev Biochem. 1990;59:631–660. doi: 10.1146/annurev.bi.59.070190.003215. [DOI] [PubMed] [Google Scholar]
- 3.Zwanzig R, Szabo A, Bagchi B. Levinthal's paradox. Proc Natl Acad Sci USA. 1992;89:20–22. doi: 10.1073/pnas.89.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leopold PE, Montal M, Onuchic JN. Protein folding funnels: A kinetic approach to the sequence-structure relationship. Proc Natl Acad Sci USA. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolynes PG, Onuchic JN, Thirumalai D. Navigating the folding routes. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin RL. The nature of protein folding pathways: The classical versus the new view. J Biomol NMR. 1995;5:103–109. doi: 10.1007/BF00208801. [DOI] [PubMed] [Google Scholar]
- 7.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 8.Plotkin SS, Onuchic JN. Understanding protein folding with energy landscape theory. Part I: Basic concepts. Q Rev Biophys. 2002;35:111–167. doi: 10.1017/s0033583502003761. [DOI] [PubMed] [Google Scholar]
- 9.Radford SE, Dobson CM, Evans PA. The folding of hen lysozyme involves partially structured intermediates and multiple pathways. Nature. 1992;358:302–307. doi: 10.1038/358302a0. [DOI] [PubMed] [Google Scholar]
- 10.Wildegger G, Kiefhaber T. Three-state model for lysozyme folding: Triangular folding mechanism with an energetically trapped intermediate. J Mol Biol. 1997;270:294–304. doi: 10.1006/jmbi.1997.1030. [DOI] [PubMed] [Google Scholar]
- 11.Bieri O, Wildegger G, Bachmann A, Wagner C, Kiefhaber T. A salt-induced kinetic intermediate is on a new parallel pathway of lysozyme folding. Biochemistry. 1999;38:12460–12470. doi: 10.1021/bi9909703. [DOI] [PubMed] [Google Scholar]
- 12.Kamagata K, Sawano Y, Tanokura M, Kuwajima K. Multiple parallel-pathway folding of proline-free staphylococcal nuclease. J Mol Biol. 2003;332:1143–1153. doi: 10.1016/j.jmb.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Bilsel O, Zitzewitz JA, Bowers KE, Matthews CR. Folding mechanism of the α-subunit of tryptophan synthase, an α/β barrel protein: Global analysis highlights the interconversion of multiple native, intermediate, and unfolded forms through parallel channels. Biochemistry. 1999;38:1018–1029. doi: 10.1021/bi982365q. [DOI] [PubMed] [Google Scholar]
- 14.Wallace LA, Matthews CR. Sequential versus parallel protein-folding mechanisms: Experimental tests for complex folding reactions. Biophys Chem. 2002;101/102:113–131. doi: 10.1016/s0301-4622(02)00155-2. [DOI] [PubMed] [Google Scholar]
- 15.Wu Y, Matthews CR. Parallel channels and rate-limiting steps in complex protein folding reactions: Prolyl isomerization and the alpha subunit of Trp synthase, a TIM barrel protein. J Mol Biol. 2002;323:309–325. doi: 10.1016/s0022-2836(02)00922-1. [DOI] [PubMed] [Google Scholar]
- 16.Gianni S, et al. Parallel pathways in cytochrome c551 folding. J Mol Biol. 2003;330:1145–1152. doi: 10.1016/s0022-2836(03)00689-2. [DOI] [PubMed] [Google Scholar]
- 17.Englander SW, et al. Fast and slow folding in cytochrome c. Acc Chem Res. 1998;31:737–744. [Google Scholar]
- 18.Englander SW. Protein folding intermediates and pathways studied by hydrogen exchange. Annu Rev Biophys Biomol Struct. 2000;29:213–238. doi: 10.1146/annurev.biophys.29.1.213. [DOI] [PubMed] [Google Scholar]
- 19.Chamberlain AK, Marqusee S. Comparison of equilibrium and kinetic approaches for determining protein folding mechanisms. Adv Protein Chem. 2000;53:283–328. doi: 10.1016/s0065-3233(00)53006-x. [DOI] [PubMed] [Google Scholar]
- 20.Englander SW, Mayne L, Krishna MMG. Protein folding and misfolding: Mechanism and principles. Q Rev Biophys. 2008 doi: 10.1017/S0033583508004654. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishna MMG, Englander SW. A unified mechanism for protein folding: Predetermined pathways with optional errors. Protein Sci. 2007;16:449–464. doi: 10.1110/ps.062655907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maki K, Ikura T, Mohs A, Kuwajima K. Equilibrium and kinetics of folding of staphylococcal nuclease and its proline mutants. Int Congr Ser. 1999;1194:271–278. [Google Scholar]
- 23.Hinck AP, Eberhardt ES, Markley JL. NMR strategy for determining Xaa-Pro peptide bond configurations in proteins: Mutants of staphylococcal nuclease with altered configuration at proline-117. Biochemistry. 1993;32:11810–11818. doi: 10.1021/bi00095a009. [DOI] [PubMed] [Google Scholar]
- 24.Bédard S, Mayne L, Peterson RW, Wand AJ, Englander SW. The foldon substructure of staphylococcal nuclease. J Mol Biol. 2008;376:1142–1154. doi: 10.1016/j.jmb.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robbins RJ, et al. Photophysics of aqueous tryptophan: pH and temperature effects. J Am Chem Soc. 1980;102:6271–6279. [Google Scholar]
- 26.Maki K, Cheng H, Dolgikh DA, Shastry MCR, Roder H. Early events during folding of wild-type staphylococcal nuclease and a single-tryptophan variant studied by ultrarapid mixing. J Mol Biol. 2004;338:383–400. doi: 10.1016/j.jmb.2004.02.044. [DOI] [PubMed] [Google Scholar]
- 27.Walkenhorst WF, Green SM, Roder H. Kinetic evidence for folding and unfolding intermediates in staphylococcal nuclease. Biochemistry. 1997;36:5795–5805. doi: 10.1021/bi9700476. [DOI] [PubMed] [Google Scholar]
- 28.Wu Y, Matthews CR. Proline replacements and the simplification of the complex, parallel channel folding mechanism for the alpha subunit of Trp synthase, a TIM barrel protein. J Mol Biol. 2003;330:1131–1144. doi: 10.1016/s0022-2836(03)00723-x. [DOI] [PubMed] [Google Scholar]
- 29.Chu R, Pei W, Takei J, Bai Y. Relationship between native-state hydrogen exchange and folding pathways of a four-helix bundle protein. Biochemistry. 2002;41:7998–8003. doi: 10.1021/bi025872n. [DOI] [PubMed] [Google Scholar]
- 30.Feng H, Zhou Z, Bai Y. A protein folding pathway with multiple folding intermediates at atomic resolution. Proc Natl Acad Sci USA. 2005;102:5026–5031. doi: 10.1073/pnas.0501372102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vu NT, Feng H, Bai Y. The folding pathway of barnase: The rate-limiting transition state and a hidden intermediate under native conditions. Biochemistry. 2004;43:3346–3356. doi: 10.1021/bi0362267. [DOI] [PubMed] [Google Scholar]
- 32.Fuentes EJ, Wand AJ. Local stability and dynamics of apocytochrome b562 examined by the dependence of hydrogen exchange on hydrostatic pressure. Biochemistry. 1998;37:9877–9883. doi: 10.1021/bi980894o. [DOI] [PubMed] [Google Scholar]
- 33.Yan S, Kennedy SD, Koide S. Thermodynamic and kinetic exploration of the energy landscape of Borrelia burgdorferi OspA by native-state hydrogen exchange. J Mol Biol. 2002;323:363–375. doi: 10.1016/s0022-2836(02)00882-3. [DOI] [PubMed] [Google Scholar]
- 34.Bollen YJM, Kamphuis MB, van Mierlo CPM. The folding energy landscape of apoflavodoxin is rugged: Hydrogen exchange reveals nonproductive misfolded intermediates. Proc Natl Acad Sci USA. 2006;103:4095–4100. doi: 10.1073/pnas.0509133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krishna MMG, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods. 2004;34:51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Silverman JA, Harbury PB. The equilibrium unfolding pathway of a (β/α)8 barrel. J Mol Biol. 2002;324:1031–1040. doi: 10.1016/s0022-2836(02)01100-2. [DOI] [PubMed] [Google Scholar]
- 37.Korzhnev DM, et al. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature. 2004;430:586–590. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 38.Korzhnev DM, Neudecker P, Zarrine-Afsar A, Davidson AR, Kay LE. Abp1p and Fyn SH3 domains fold through similar low-populated intermediate states. Biochemistry. 2006;45:10175–10183. doi: 10.1021/bi0611560. [DOI] [PubMed] [Google Scholar]
- 39.Korzhnev DM, Religa TL, Lundstrom P, Fersht AR, Kay LE. The folding pathway of an FF domain: Characterization of an on-pathway intermediate state under folding conditions by 15N, 13Cα and 13C-methyl relaxation dispersion and 1H/2H-exchange NMR spectroscopy. J Mol Biol. 2007;372:497–512. doi: 10.1016/j.jmb.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 40.Pletneva EV, Gray HB, Winkler JR. Snapshots of cytochrome c folding. Proc Natl Acad Sci USA. 2005;102:18397–18402. doi: 10.1073/pnas.0509076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maity H, Maity M, Krishna MMG, Mayne L, Englander SW. Protein folding: The stepwise assembly of foldon units. Proc Natl Acad Sci USA. 2005;102:4741–4746. doi: 10.1073/pnas.0501043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rumbley J, Hoang L, Mayne L, Englander SW. An amino acid code for protein folding. Proc Natl Acad Sci USA. 2001;98:105–112. doi: 10.1073/pnas.98.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura C, Dyson HJ, Wright PE. Identification of native and non-native structure in kinetic folding intermediates of apomyoglobin. J Mol Biol. 2006;355:139–156. doi: 10.1016/j.jmb.2005.10.047. [DOI] [PubMed] [Google Scholar]
- 44.Weinkam P, Zong C, Wolynes PG. A funneled energy landscape for cytochrome c directly predicts the sequential folding route inferred from hydrogen exchange experiments. Proc Natl Acad Sci USA. 2005;102:12401–12406. doi: 10.1073/pnas.0505274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, Mayne L, Englander SW. Evidence for an unfolding and refolding pathway in cytochrome c. Nat Struct Biol. 1998;5:774–778. doi: 10.1038/1810. [DOI] [PubMed] [Google Scholar]
- 46.Maity H, Maity M, Englander SW. How cytochrome c folds, and why: Submolecular foldon units and their stepwise sequential stabilization. J Mol Biol. 2004;343:223–233. doi: 10.1016/j.jmb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 47.Krishna MMG, Maity H, Rumbley JN, Englander SW. Branching in the folding pathway of cytochrome c. Protein Sci. 2007;16:1946–1956. doi: 10.1110/ps.072922307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krishna MMG, Maity H, Rumbley JN, Lin Y, Englander SW. Order of steps in the cytochrome c folding pathway: Evidence for a sequential stabilization mechanism. J Mol Biol. 2006;359:1411–1420. doi: 10.1016/j.jmb.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 49.Sosnick TR, Mayne L, Hiller R, Englander SW. The barriers in protein folding. Nat Struct Biol. 1994;1:149–156. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- 50.Fersht AR. Optimization of rates of protein folding: The nucleation-condensation mechanism and its implications. Proc Natl Acad Sci USA. 1995;92:10869–10873. doi: 10.1073/pnas.92.24.10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sosnick TR, Mayne L, Englander SW. Molecular collapse: The rate-limiting step in two-state cytochrome c folding. Proteins Struct Funct Genet. 1996;24:413–426. doi: 10.1002/(SICI)1097-0134(199604)24:4<413::AID-PROT1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 52.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 53.Krishna MMG, Lin Y, Englander SW. Protein misfolding: Optional barriers, misfolded intermediates, and pathway heterogeneity. J Mol Biol. 2004;343:1095–1109. doi: 10.1016/j.jmb.2004.08.098. [DOI] [PubMed] [Google Scholar]
- 54.Krishna MMG, Lin Y, Mayne L, Englander SW. Intimate view of a kinetic protein folding intermediate: Residue-resolved structure, interactions, stability, folding and unfolding rates, homogeneity. J Mol Biol. 2003;334:501–513. doi: 10.1016/j.jmb.2003.09.070. [DOI] [PubMed] [Google Scholar]
- 55.Hughson FM, Wright PE, Baldwin RL. Structural characterization of a partly folded apomyoglobin intermediate. Science. 1990;249:1544–1548. doi: 10.1126/science.2218495. [DOI] [PubMed] [Google Scholar]
- 56.Capaldi AP, Kleanthous C, Radford SE. Im7 folding mechanism: Misfolding on a path to the native state. Nat Struct Biol. 2002;9:209–216. doi: 10.1038/nsb757. [DOI] [PubMed] [Google Scholar]
- 57.Zimm GH, Bragg JK. Theory of the phase transition between helix and random coil in polypeptide chains. J Chem Phys. 1959;31:526–535. [Google Scholar]
- 58.Lifson S, Roig A. On the theory of the helix-coil transition in polypeptides. J Chem Phys. 1961;34:1963–1974. [Google Scholar]
- 59.Watson JD, Crick FHC. Molecular structure of nucleic acids. A structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 60.Pauling L. Molecular complementariness and serological specificity. Immunochemistry. 1975;12:445–447. doi: 10.1016/0019-2791(75)90062-2. [DOI] [PubMed] [Google Scholar]
- 61.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 62.Schlunegger MP, Bennett MJ, Eisenberg D. Oligomer formation by 3D domain swapping: A model for protein assembly and misassembly. Adv Protein Chem. 1997;50:61–122. doi: 10.1016/s0065-3233(08)60319-8. [DOI] [PubMed] [Google Scholar]
- 63.Royer CA, et al. Effect of amino acid substitutions on the pressure denaturation of staphylococcal nuclease as monitored by fluorescence and nuclear magnetic resonance spectroscopy. Biochemistry. 1993;32:5222–5232. doi: 10.1021/bi00070a034. [DOI] [PubMed] [Google Scholar]