Abstract
The yeast Saccharomyces cerevisiae has developed specialized mechanisms that enable growth on suboptimal nitrogen sources. Exposure of yeast cells to poor nitrogen sources or treatment with the Tor kinase inhibitor rapamycin elicits activation of Gln3 and transcription of nitrogen catabolite-repressed (NCR) genes whose products function in scavenging and metabolizing nitrogen. Here, we show that mutations in class C and D Vps components, which mediate Golgi-to-endosome vesicle transport, impair nuclear translocation of Gln3, NCR gene activation, and growth in poor nitrogen sources. In nutrient-replete conditions, a significant fraction of Gln3 is peripherally associated with light membranes and partially colocalizes with Vps10-containing foci. These results reveal a role for Golgi-to-endosome vesicular trafficking in TORC1-controlled nuclear translocation of Gln3 and support a model in which Tor-mediated signaling in response to nutrient cues occurs in these compartments. These findings have important implications for nutrient sensing and growth control via mTor pathways in metazoans.
Keywords: rapamycin action, Tor signaling
Exposure of Saccharomyces cerevisiae to nitrogen limitation or poor nitrogen sources triggers the expression of nitrogen catabolite-repressed (NCR) genes, whose products function in scavenging and metabolizing nitrogen (1). Expression of the NCR genes is controlled by the GATA-like transcription factors Gln3 and Gat1. Regulation of these transactivators is exerted at the level of cellular localization, which in some cases correlates with their phosphorylation status (2). The nutrient-sensing Tor pathway, via regulation of Tap42–Sit4 phosphatase activity, influences Gln3 phosphorylation and thereby its interaction with the Ure2 cytoplasmic repressor. Inhibition of Tor by rapamycin leads to Gln3 dephosphorylation, release from Ure2, and subsequent nuclear translocation (3, 4). However, little is known about the mechanism by which poor nitrogen sources influence Tor activity.
A prominent role for endogenous membranes of the protein secretory pathway as a platform for Tor signaling has begun to emerge: (i) different components of the Tor protein complexes, termed TORC1 and TORC2, as well as the Tap42–Sit4 phosphatase complex, have been localized to endosomal and vacuolar compartments (5–12); (ii) a function for the Golgi Ca2+/Mn2+ ATPase Pmr1 in negatively regulating TORC1 signaling has been demonstrated (13); and (iii) recent studies have shown genetic interactions between TORC1 and different components of the protein-sorting machinery, including those of the class C Vps complex that functions in docking and fusion of vesicles with the Golgi, endosomes, and vacuoles (refs. 9, 14, and reviewed in ref. 15). In particular, the class C Vps complex was proposed to provide amino acid homeostasis for efficient Tor signaling (14).
We have examined in detail the effects of class C vps mutations on the expression of TORC1-regulated genes and find a marked defect in nuclear translocation of Gln3 and impaired induction of the NCR genes in response to poor nitrogen sources but not in response to rapamycin. Moreover, class D vps mutants exhibit similar defects, implicating Golgi-to-endosome trafficking as a critical event for Gln3 regulation. We show that Gln3 is peripherally associated with light membranes and partially colocalizes with Vps10 in Golgi and endosomal compartments. We conclude that Golgi-to-endosome trafficking is an obligate step for the Gln3 route to the nucleus, and we suggest a model in which Tor signaling to nitrogen-regulated transactivators occurs on endosomal membranes.
Results
Class C vps Mutants Are Defective for Activating NCR in Poor Nitrogen Conditions, but Not in Response to Rapamycin.
We recently reported that mutations in class C VPS genes exhibit synthetic lethality (SL) when combined with tor1 mutations (14). This defect is remedied by supplementation of the growth medium with glutamine, and we proposed that the SL phenotype derives from an alteration in amino acid homeostasis caused by class C vps mutations (14). In addition, class C vps mutants are hypersensitive to rapamycin, have growth defects in poor nitrogen sources, and fail to survive nitrogen starvation.
To gain further insight into the functional defects that underlie these phenotypes, we examined TOR-regulated expression of NCR genes in these mutants. Expression of NCR genes was compared in nutrient-replete and rapamycin-treated cells or cells shifted from ammonium to a poor nitrogen source, proline-containing media. Interestingly, whereas either rapamycin exposure or shift into proline medium induced expression of the NCR genes MEP2 and GAP1 in WT cells, the class C vps mutants pep5, vps16, and vps33 respond differently to these treatments (Fig. 1A). These mutants induced NCR gene expression in the presence of rapamycin, similar to the WT strain; however, when shifted to proline, they failed to induce expression of GAP1 or MEP2 (Fig. 1A). This observation supports earlier reports suggesting that rapamycin treatment and poor nitrogen sources induce NCR gene expression via different mechanisms (2). Closer examination of the kinetics of induction of MEP2 revealed that expression in the WT strain reached a maximum at 15 min and was sustained for the 2-h period examined, whereas in the pep3 mutant a markedly lower level of expression was detected at 15 min, and this level was not sustained (Fig. 1B). Reduced expression of NCR genes in class C vps mutants in comparison to WT cells also was observed in low-nitrogen medium (SLAD) (data not shown).
Fig. 1.
Class C VPS genes are required for efficient induction of Tor-regulated NCR response. (A) Exponentially growing cultures of WT (BY4742) and isogenic class C vps mutants were treated with drug vehicle (−) or 100 nM rapamycin (R) for 30 min or shifted from YPD to proline medium for 1 h (P). RNA was prepared and analyzed by Northern blotting with 32P-labeled DNA probes for GAP1, MEP2, CIT2, SOD2, and ACT1 as loading control. (B) Cultures of WT and pep3 strains grown as in A were shifted into proline medium. Culture aliquots were removed before shifting into proline or after 15, 30, 60, and 120 min, and RNA was isolated and analyzed by Northern blotting with CIT2 and MEP2 probes. (C) Cultures of isogenic WT (BY4742), pep3 (14105), gln3 (10173), and pep3 gln3 (RPY77) strains were shifted from YPD to proline medium for 15 and 60 min. RNA was isolated and GAP1 expression analyzed. (D) Exponentially growing WT (TB123) and pep3 (RPY30) strains were treated as described in A. Gln3-Myc electrophoretic mobility was assayed by Western blot analysis with Myc antibody. (E) Isogenic WT (TB123) and pep3 (RPY30) strains were grown in YPD and treated as in A. Cells were processed for indirect immunofluorescence with anti-Myc. Nuclei were stained with DAPI.
The expression of other TOR-regulated transcripts, such as the STRE genes HOR2 and SOD2 and the ribosomal protein gene RPS26, did not show any substantial difference compared with WT (Fig. 1A) (data not shown). However, expression of the retrograde response gene CIT2 was induced in pep3 cells, even in nitrogen-rich medium, confirming earlier observations that vps class C mutants show mitochondrial defects (Fig. 1B) (16).
Regulation of NCR genes is under the control of the GATA transcription factors Gln3 and Gat1. Although Gln3 and Gat1 were thought to be functionally redundant, recent studies have suggested that they respond differentially to various nitrogen conditions (17). For some genes, abolishment of the NCR response is achieved only by mutation of both GLN3 and GAT1 (18). To assess the contribution of each transcription factor to the NCR response in class C vps mutants, GLN3 was deleted in the pep3 strain. Expression of GAP1 and MEP2 was completely blocked in the pep3 gln3 double-mutant strain, indicating that Gat1 does not contribute to the expression of these genes in the pep3 strain (Fig. 1C). Also, the low expression of GAP1 and MEP2 in the gln3 mutant, compared with WT, suggested that Gln3 largely controls most of the expression of these NCR genes induced by proline.
Inhibition of TORC1 with rapamycin induces Gln3 dephosphorylation and translocation into the nucleus (3, 4). However, Gln3 regulation is more complex in nonpreferred nitrogen sources. When WT cells are shifted into proline medium, dephosphorylation of Gln3 is undetectable, although under these conditions NCR gene expression is activated (Fig. 1D) (2). We tested the influence of the pep3 mutation on the Gln3-Myc phosphorylation state. Rapamycin treatment resulted in Gln3 dephosphorylation (as assessed by a shift in electrophoretic mobility) in both the WT and the pep3 strain; however, no visible dephosphorylation was observed upon shift into proline medium (Fig. 1D).
The class C Vps complex has a major role in protein sorting, and intracellular localization of Gln3 is the mechanism by which NCR-sensitive transcription is regulated (1). We therefore investigated whether the class C Vps complex is required for nuclear translocation of Gln3 in response to poor nitrogen conditions. In WT cells, Gln3 was restricted to the cytosol in nitrogen-rich medium (YPD) and was largely nuclear in cells treated with rapamycin or shifted to proline medium. A similar localization was observed in pep3 mutants without or with rapamycin (Fig. 1E). In contrast to WT cells, Gln3 nuclear localization was impaired in pep3 mutants shifted to proline as a nitrogen source. This observation can explain the marked NCR defect in class C vps mutants and indicates that, in contrast to rapamycin, efficient proline-induced Gln3 nuclear translocation requires class C VPS complex function.
Golgi-to-Endosome Trafficking Is Required for Efficient Gln3 Translocation to the Nucleus in Response to Poor Nitrogen Source.
The class C Vps protein complex is required for membrane docking and fusion of Golgi-derived vesicles in the multiple protein-sorting routes to the vacuole (19, 20). We hypothesized that if proline-induced nuclear translocation of Gln3 requires one of the trafficking routes assisted by the class C Vps complex, then mutations in this route should confer similar NCR defects as vps class C mutations. Genes whose function is required for trafficking from late Golgi to vacuole occur in six phenotypic classes (21). Mutants of these different classes exhibit blocks at discrete membrane-trafficking steps. To test this hypothesis, we examined NCR gene expression in class A–F vps mutants and in the vac8 mutant that blocks cytosol to vacuole transport. Rapamycin treatment did not result in any significant difference in the expression of GAP1 in all tested strains. Remarkably, class D (vps45) and, to a lesser extent, class E (vps27) vps mutants showed reduced expression of GAP1 on shift into proline medium [supporting information (SI) Fig. S1A]. Furthermore, all class D vps mutants tested (pep12, vps45, vps3, vps9, vps19, and vps34) showed impaired expression of GAP1 and MEP2 (Fig. 2A). A time course expression analysis with pep3 and vps45 mutants in proline medium showed that these mutants have a similar defect in GAP1 and MEP2 expression (Fig. 2 B and C) (data not shown). Furthermore, similarly to class C vps mutants, class D vps mutants are hypersensitive to rapamycin and grow poorly in proline medium (Fig. S1B).
Fig. 2.
Class D vps mutations impair Gln3 function. (A) Isogenic WT, pep3, and the class D vps mutants (pep12, vps45, vps3, vps9, vps19, and vps34) were tested for NCR gene response as in Fig. 1A. (B) Isogenic WT and the pep3 and vps45 mutants were assayed for GAP1 expression over 2-h shift from YPD into proline medium. (C) Northern blot signals for GAP1 were quantified in WT (dots), pep3 (squares), and vps45 (triangles) strains and normalized to ACT1 loading control. Results shown are the relative percentage of gene expression with maximal level of expression at 15 min as 100%. (D) Gln3 was detected in WT (TB123) and vps45 (RPY46) strains as in Fig. 1E. (E) Percentage of total cells in which Gln3 was nuclear localized in the conditions analyzed in D. Values represent mean ± SD of three independent determinations.
To avoid secondary effects due to the broader effects of class C vps mutations, further studies focused on the vps45 mutant. Consistent with previous observations with WT and pep3 strains, no detectable difference in Gln3 phosphorylation was observed when vps45 cells were shifted from YPD to proline media (data not shown). Moreover, although in the WT strain the shift into proline medium resulted in Gln3 nuclear translocation in 90% of cells, in the vps45 mutant, only 30–40% of cells had a fraction of Gln3 in the nucleus (Fig. 2 D and E). Class D VPS genes function in the trans Golgi-to-endosome protein sorting route and play roles in fusion of Golgi-derived vesicles to late endosomes (22). Our results and these observations suggest that Golgi-to-endosome trafficking is an obligate step in the Gln3 trafficking route to the nucleus. Mutations in class D VPS genes result in the accumulation of Golgi-derived vesicles in the cytosol (23, 24). Our working model is that the failure of Gln3 to translocate into the nucleus in class D vps mutants results from entrapment of Gln3 within (or on) these vesicles.
A Fraction of Gln3 Associates with Light Membranes via a Noncontiguous Amino Acid Sequence.
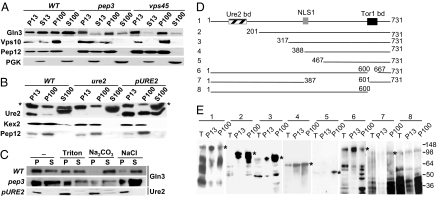
Cellular localization studies of TORC1 and TORC2, as well as the Tap42-Sit4 phosphatase, indicate that these complexes associate with internal light membranes (9, 12). These findings, and our results that vps mutants show defects in the NCR response, raise the possibility that TORC1 signaling to Gln3 occurs on membranes of the protein trafficking pathway. An earlier study localized Gln3 to undefined punctate structures in the cytoplasm (25). These observations prompted us to examine the cellular localization of Gln3 in further detail. In WT cells, Gln3 fractionated in approximately equivalent proportions with heavy (P13) membranes known to contain plasma membrane and endoplasmic reticulum and, interestingly, with lighter (P100) membranes along with the Golgi and endosomal markers Vps10 and Pep12. In addition, a significant portion of Gln3 was recovered in the cytosolic S100 fraction (Fig. 3A). In contrast, Gln3 largely fractionated with P13 and P100 fractions in pep3 and vps45 mutant strains, and only a residual fraction was present in the cytosol. This finding suggests that, in these mutants, the bulk of Gln3 is less soluble and is associated with endoplasmic reticulum and Golgi membranes or Golgi-derived vesicles (discussed in Gln3 Partially Colocalizes with Vps10-Containing Foci.).
Fig. 3.
A fraction of Gln3 is peripherally associated with light membranes along with Golgi and endosomal markers. (A) Cell-free lysates from isogenic WT (TB123), pep3 (RPY30), and vps45 (RPY46) strains were subjected to differential centrifugation to yield low-speed pellet (P13), supernatant (S13), high-speed pellet (P100), and soluble (S100) fractions. Equal cell equivalents were examined by Western blot to detect Gln3, Vps10, Pep12, and PGK. (B) The WT (TB123), ure2 (RPY64), and the TB123 strain transformed with pURE2 (pVTG20) were fractionated and analyzed by Western blot to detect Ure2 and the Golgi and endosomal markers Kex2 and Pep12 as indicated in Fig. 3A. *, Ure2-unrelated polypeptide. (C) S13 fractions from WT (TB123), pep3 (RPY30), and TB123 strain transformed with pURE2 (pVTG20) were incubated with 0.1% Triton X-100, 0.1 M Na2CO3 (pH 11), or 1 M NaCl for 30 min at 4°C. P100 and S100 fractions were isolated and analyzed for Gln3 and Ure2 by Western blot. (D) Schematic representation of the Gln3-Myc9 fusion constructs used in E. The relevant functional domains of Gln3 are shown in the full-length construct 1. (E) The gln3 (10173) mutant was transformed with the Gln3-Myc9 fusion constructs depicted in D. Gln3 in total-cell lysates and P13 and P100 were detected by Western blot. *, Gln3-Myc9 mutant proteins at the predicted molecular masses. Numbers above the fractions correspond to the Gln3-Myc9 construct number in D. Numbers at the right indicate molecular mass in KDa.
Furthermore, in the WT strain, the interaction of Gln3 with light membranes is peripheral as indicated by its sensitivity to extraction with 0.1 M Na2CO3 (pH 11) or 1 M NaCl (Fig. 3C). In the pep3 mutant, association of Gln3 with light membranes appears to be more stable because it could not be completely released by any of the treatments, including Na2CO3.
Structure–function analyses of Gln3 have identified specific sequences for nuclear localization, nuclear export, and interaction with Ure2 and Tor1 (26, 27). We find that a fraction of Ure2, expressed from its endogenous locus or from a low-copy centromeric plasmid to improve detection, also is tightly associated with light membranes and resistant to salt extraction (Fig. 3 B and C). Next, we sought to identify Gln3 sequences that mediate its association with light membranes. N-terminal, internal, and C-terminal deletion alleles of Gln3–Myc fusion proteins were expressed in a gln3-deleted strain, and cell lysates were prepared and centrifuged to yield P13 and P100 fractions (Fig. 3 D and E). Our results were in accord with the observed localization of these Gln3 mutants reported previously (27). Gln3 lacking the initial 200 amino acids (except construct 5) predominantly localized to the nucleus (27). Accordingly, these Gln3 mutants were recovered in the P13 and P100 membrane fractions (Fig. 3E). Deletions that led to cytosolic localization of Gln3 due to the loss of the nuclear localization sequence (NLS1) (constructs 5 and 7) (27) were predominantly restricted to the P100 fraction (Fig. 3E). These results suggest that Gln3 membrane association is mediated by noncontiguous amino acid sequences.
Tor1 regulates Gln3 by promoting its association with Ure2 (3, 4). In addition, an interaction of Tor1 with Gln3 has been reported (27). Thus, it was important to test whether the Ure2- and Tor1-binding domains mediate Gln3 interaction with light membranes. Deletion of the Ure2- (amino acid residues 100–150) or Tor1- (construct 6) binding domains did not disrupt Gln3 membrane association (Fig. 3 D and E).
Gln3 Partially Colocalizes with Vps10-Containing Foci.
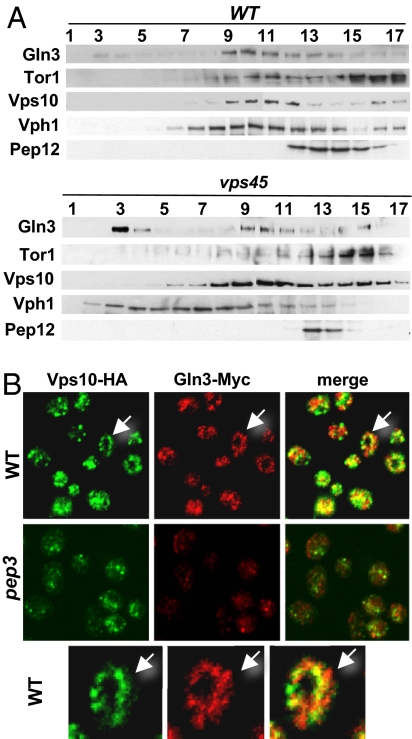
To characterize the light membranes with which Gln3 is associated, P100 fractions from WT and vps45 strains were subjected to sucrose density gradient fractionation. In both WT and vps45 strains, Gln3 migrated in the middle and lower fractions of the gradient, together with the Golgi and endosomal marker Vps10 (Fig. 4A). A residual fraction of Gln3 also was observed in the upper fractions of the gradient (fractions 3 and 4) in WT cells; interestingly, this fraction was markedly increased in the vps45 strain and represented ≈25% of the total Gln3 protein recovered. A shift in Vph1 toward the top of the gradient also was observed in the vps45 mutant compared with WT (Fig. 4A). This difference is attributable to Vph1 entrapment in small vesicles generated in class D mutants (28). Thus, it is possible that the fraction of Gln3 in the top gradient fractions is associated with a subpopulation of these vesicles. Interestingly, Tor1 also is present in the same Gln3- and Vps10-containing fractions in the middle portion of the gradient but was more concentrated toward the lowest region of the gradient (Fig. 4A). In contrast to Gln3, mutation of vps45 did not markedly alter Tor1 migration.
Fig. 4.
Gln3 cofractionates with Vps10. (A) P100 membranes from WT and vps45 strains were loaded on 18–54% sucrose density step gradients. Gradients were centrifuged and fractionated from top to bottom, and Gln3, Tor1, Vps10, Vph1, and Pep12 were detected by Western blot analysis in equal aliquots from each fraction. (B) Gln3 partially colocalizes with Vps10-containing foci. Cells of WT (RPY73) and pep3 (RPY75), expressing the Gln3-Myc and Vps10-HA fusion proteins from their chromosomal loci, were double stained with monoclonal anti-HA and polyclonal anti-Myc antibodies. Primary antibodies were detected and images were obtained as indicated in Materials and Methods. Arrows in Top indicate a cell magnified in Bottom.
To further substantiate these subcellular fractionation results, we examined whether Gln3 colocalizes with Vps10. To stabilize Vps10, the pep4–3 mutation was introduced in the WT and pep3 strains. Gln3–Myc was distributed throughout the cytoplasm in small punctate foci frequently concentrated near the vacuolar membrane in WT cells (Fig. 4B). Moreover, Gln3–Myc foci partially colocalized with Vps10-specific foci. In the pep3 mutant, Gln3 was more diffuse and less concentrated at any specific intracellular location, although partial colocalization of Gln3 with Vps10 was observed, but it was less evident than in WT cells (Fig. 4B). We conclude that a fraction of Gln3 partially colocalizes with Vps10-containing compartments.
Discussion
We have shown that mutations in class C and D Vps components lead to marked defects in: (i) Gln3 nuclear translocation, (ii) expression of NCR genes, and (iii) growth in poor nitrogen sources. Class C and D Vps proteins mediate docking and fusion of Golgi-derived vesicles with endosomes and globally affect protein sorting (19, 20, 22). Intriguingly, treatment of vps class C and D mutants with rapamycin bypassed the need for Golgi-to-endosome transport, resulting in Gln3 nuclear translocation. This result is in agreement with previous observations that the mechanism of nuclear translocation induced by rapamycin differs from that induced by proline (2). Moreover, this result argues that the Gln3 nuclear translocation machinery is intact in class C and D vps mutants. Taken together, these observations indicate that Golgi-to-endosome transport is an obligate step for Gln3 routing to the nucleus in response to nutrient cues.
We demonstrated that a significant fraction of Gln3 is peripherally associated with membranes enriched for the Golgi and endosomal markers Vps10 and Pep12. In addition, Gln3 fractionates with Vps10 in sucrose gradients and partially colocalizes with Vps10-containing foci. Mutations in class C and D Vps components conferred subtle but detectable Gln3 phenotypes: an increase in the insoluble Gln3 fraction rendering its membrane association more resistant to salt extraction and a shift in Gln3 and Vph1 migration to lighter fractions of the sucrose gradient predicted to contain the small vesicles that accumulate in these mutants (28). This effect resulted in more diffuse Gln3 staining and less colocalization with Vps10 foci. These results strongly support that Gln3 localizes to Golgi and endosomal compartments.
Under nitrogen-replete conditions, Ure2 sequesters Gln3 in the cytoplasm (1), which raised the possibility that Ure2 mediates association of Gln3 with light membranes. We find that Ure2 tightly associates with P100 membranes. However, deletion of the Ure2-binding domain of Gln3 did not affect Gln3 interaction with these membranes. Similarly, we excluded the possibility that the Tor1–Gln3 interaction previously characterized mediates Gln3 light membrane association (27).
Several components of the TORC1 and the Tap42–Sit4 complex localize to membranes of the protein secretory pathway (5–12). Moreover, the EGO–GSE complex, which, in response to amino acids, controls sorting of the general amino acid permease Gap1, and, in combination with TORC1, regulates microautophagy, resides in prevacuolar compartments (29, 30). These and other studies underscore a prominent role for membranes of the protein secretory pathway as a portal for amino acid sensing and Tor-signaling events. Our results and these observations support a model in which Gln3 regulation by TORC1 in response to nitrogen sources occurs on endosomal membranes and depends on Golgi-to-endosome vesicular trafficking assisted by class C and D Vps protein function.
These findings and recent reports demonstrating that the Rim101-mediated pH response and MAPK activation via the Gα subunit Gpa1 and the PI3K Vps34 signal on endosomes highlight a role for these organelles as prominent platforms from which diverse cues are translated into appropriate physiological effects (31, 32). It is possible that the involvement of vesicular trafficking in nuclear translocation in response to diverse stimuli is more general than anticipated.
Our studies provide insights into nutrient sensing via Tor and suggest that analogous mechanisms may operate in mammals, including humans, in which class C and D Vps orthologs are conserved and mTor has been found in the Golgi compartment (33). Targeting these orthologs may allow more precise pharmacological intervention in selected mTor-signaling events.
Materials and Methods
Yeast Strains, Plasmids, and Media.
Strains in this study are listed in Table S1. With the exception of LCY294, all strains are isogenic derivatives of either TB123 (3) or BY4742 and, unless otherwise indicated, were constructed by the Saccharomyces Genome Deletion Project (Invitrogen). RPY30 and RPY46 strains were constructed by the deletion of PEP3 and VPS45 in TB123 by using the NAT resistance gene. GLN3 and URE2 were deleted by using the HYG resistance gene in strains #14105 and TB123 to produce RPY77 and RPY64, respectively. RPY73 was obtained by crossing strains LCY294 (34) and TB123, and resulting diploids were sporulated and dissected. Meiotic segregants were selected for G418 resistance, and the pep4–3 mutation was confirmed by examining Cpy deficiency by using the APE (N-acetyl-dl-phenylalanine β-naphthyl ester) overlay method (35). The presence of GLN3-Myc and VPS10-HA alleles was confirmed by Western blot with antibodies against Myc and HA tags, respectively. Strain RPY75 was obtained by deleting PEP3 in RPY73 with NAT. Mutant yeast strains were constructed by PCR-mediated gene disruption as described previously (36, 37), and gene deletions were PCR confirmed. The pRS315-GLN3-Myc9 deletion alleles and the centromeric plasmid pVTG20 for Ure2 expression were obtained from Steven Zheng (27) and Reed Wickner (38).
Unless otherwise indicated, strains were grown to exponential phase in yeast extract-peptone dextrose (YEPD) or synthetic complete (SC) media. Yeast synthetic media (YNB) with either ammonium sulfate or, where indicated, 0.1% proline was supplemented with 2% glucose. SC media was supplemented with the required amino acids to satisfy auxotrophic requirements. Rapamycin was added to the media from a concentrated stock solution in 90% ethanol and 10% Tween-20.
Northern Blot.
Yeast strains were grown overnight in YPD medium to exponential phase (OD600 of 0.6–0.8). For gene expression studies in poor nitrogen, source cells were washed twice with YNB 0.1% proline and incubated in this medium for the indicated periods of times at 30°C. RNA isolation and Northern blot analysis were as described previously (39), and specific signals were quantified with a typhoon 9200 variable mode imager using image Quantifier 5.2 software (Molecular Dynamics).
Western Blot.
Whole-cell extracts were prepared from exponentially growing cultures and treated as indicated. Cells were disrupted by using glass beads in lysis buffer containing 50 mM KHPO4 (pH 7.4), 50 mM KCl, 2 mM EDTA, 25 mM β-glycerophosphate, 25 mM NaF, 2 mM benzamidine, 0.5% Triton X-100, and 1 mM DTT supplemented with the protease inhibitors leupeptin, aprotinin, and pepstatin added to 1 μg/ml and 0.5 mM phenylmethylsulfunyl fluoride. Whole-cell extracts and subcellular and sucrose gradient fractions were subjected to SDS electrophoresis, followed by Western blot with monoclonal antibodies for c-Myc (9E10) (Santa Cruz Biotechnology), Vps10, Pep12, Kex2, PGK1 (Molecular Probes), and Tor1, Ure2, and Vph1 polyclonal antibodies (40–42).
Indirect Immunofluoroscence Microscopy.
Gln3-Myc was visualized by indirect immunofluorescence on fixed cells as described previously employing monoclonal antibody c-Myc (9E10) (6). For colocalization studies, Vps10-HA was detected with a monoclonal anti-HA antibody (Santa Cruz Biotechnology) and Gln3-Myc9 with a polyclonal anti-c-Myc antibody from rabbit (Sigma). Secondary antibodies used for detection were Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 594 goat anti-rabbit (Molecular Probes). Cells were imaged by using a Zeiss LSM510 inverted confocal microscope.
Subcellular Fractionation and Sucrose Gradients.
Cells were lysed in buffer containing 50 mM Tris (pH 7.5), 0.2 M sorbitol, 1 mM EDTA, 1 mM DTT, protease, and phosphatase inhibitor cocktails as indicated for whole-cell extract preparation. Unbroken cells were removed by centrifugation at 500 × g for 10 min. The cell-free extract was centrifuged at 13,000 × g for 15 min to yield P13 (pellet) fraction. The supernatant was then centrifuged at 100,000 × g for 30 min to obtain P100 (pellet) and S100 (supernatant). In all experiments, equal cell equivalents of total-cell lysate and the different subcellular fractions were analyzed by Western blot. For some experiments, the P100 fraction was resuspended in 600 μl of lysis buffer and layered on an 18–54% sucrose step gradient. Gradients were centrifuged at 130,000 × g for 18 h in an SW41-Ti rotor and fractionated from top to bottom, and aliquots of each fraction were analyzed by Western blotting.
Supplementary Material
Acknowledgments.
We thank Joseph Heitman and Raphael Valdivia for helpful suggestions and comments and Steven Zheng (University of Medicine and Dentistry of New Jersey, Piscataway, NJ), Reed Wickner (National Institutes of Health, Bethesda, MD), Raphael Valdivia (Duke University Medical Center, Durham, NC), Michael Hall (Biozentrum, University of Basel, Basel, Switzerland), and Elizabeth Conibear (University of British Columbia, Vancouver, BC, Canada) for generous gifts of plasmids, antisera, and strains. This work was supported by National Cancer Institute Grant CA114107 (to M.E.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 7111.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801087105/DCSupplemental.
References
- 1.Magasanik B, Kaiser CA. Nitrogen regulation in Saccharomyces cerevisiae. Gene. 2002;290:1–18. doi: 10.1016/s0378-1119(02)00558-9. [DOI] [PubMed] [Google Scholar]
- 2.Cox KH, Kulkarni A, Tate JJ, Cooper TG. Gln3 phosphorylation and intracellular localization in nutrient limitation and starvation differ from those generated by rapamycin inhibition of Tor1/2 in Saccharomyces cerevisiae. J Biol Chem. 2004;279:10270–10278. doi: 10.1074/jbc.M312023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 4.Bertram PG, et al. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J Biol Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- 5.Wedaman KP, et al. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:1204–1220. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardenas ME, Heitman J. FKBP12-rapamycin target TOR2 is a vacuolar protein with an associated phosphatidylinositol-4 kinase activity. EMBO J. 1995;14:5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunz J, Schneider U, Howald I, Schmidt A, Hall MN. HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J Biol Chem. 2000;275:37011–37020. doi: 10.1074/jbc.M007296200. [DOI] [PubMed] [Google Scholar]
- 8.Chen EJ, Kaiser CA. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J Cell Biol. 2003;161:333–347. doi: 10.1083/jcb.200210141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aronova S, Wedaman K, Anderson S, Yates J, III, Powers T. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2779–2794. doi: 10.1091/mbc.E07-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinke A, et al. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- 11.Araki T, Uesono Y, Oguchi T, Toh EA. LAS24/KOG1, a component of the TOR complex 1 (TORC1), is needed for resistance to local anesthetic tetracaine and normal distribution of actin cytoskeleton in yeast. Genes Genet Syst. 2005;80:325–343. doi: 10.1266/ggs.80.325. [DOI] [PubMed] [Google Scholar]
- 12.Yan G, Shen X, Jiang Y. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J. 2006;25:3546–3555. doi: 10.1038/sj.emboj.7601239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devasahayam G, Ritz D, Helliwell SB, Burke DJ, Sturgill TW. Pmr1, a Golgi Ca2+/Mn2+-ATPase, is a regulator of the target of rapamycin (TOR) signaling pathway in yeast. Proc Natl Acad Sci USA. 2006;103:17840–17845. doi: 10.1073/pnas.0604303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zurita-Martinez SA, Puria R, Pan X, Boeke JD, Cardenas ME. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics. 2007;176:2139–2150. doi: 10.1534/genetics.107.072835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohde JR, Bastidas R, Puria R, Cardenas ME. Nutritional control via Tor signaling in Saccharomyces cerevisiae. Curr Opin Microbiol. 2008;11:153–160. doi: 10.1016/j.mib.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang G, Deschenes RJ. Plasma membrane localization of Ras requires class C Vps proteins and functional mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:3243–3255. doi: 10.1128/MCB.26.8.3243-3255.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulkarni A, Buford TD, Rai R, Cooper TG. Differing responses of Gat1 and Gln3 phosphorylation and localization to rapamycin and methionine sulfoximine treatment in Saccharomyces cerevisiae. FEMS Yeast Res. 2006;6:218–229. doi: 10.1111/j.1567-1364.2006.00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scherens B, Feller A, Vierendeels F, Messenguy F, Dubois E. Identification of direct and indirect targets of the Gln3 and Gat1 activators by transcriptional profiling in response to nitrogen availability in the short and long term. FEMS Yeast Res. 2006;6:777–791. doi: 10.1111/j.1567-1364.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- 19.Peterson MR, Emr SD. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic. 2001;2:476–486. doi: 10.1034/j.1600-0854.2001.20705.x. [DOI] [PubMed] [Google Scholar]
- 20.Srivastava A, Woolford CA, Jones EW. Pep3p/Pep5p complex: A putative docking factor at multiple steps of vesicular transport to the vacuole of Saccharomyces cerevisiae. Genetics. 2000;156:105–122. doi: 10.1093/genetics/156.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: Evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowers K, Stevens TH. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Piper RC, Whitters EA, Stevens TH. Yeast Vps45p is a Sec1p-like protein required for the consumption of vacuole-targeted, post-Golgi transport vesicles. Eur J Cell Biol. 1994;65:305–318. [PubMed] [Google Scholar]
- 24.Webb GC, et al. Pep7p provides a novel protein that functions in vesicle-mediated transport between the yeast Golgi and endosome. Mol Biol Cell. 1997;8:871–895. doi: 10.1091/mbc.8.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox KH, Tate JJ, Cooper TG. Cytoplasmic compartmentation of Gln3 during nitrogen catabolite repression and the mechanism of its nuclear localization during carbon starvation in Saccharomyces cerevisiae. J Biol Chem. 2002;277:37559–37566. doi: 10.1074/jbc.M204879200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulkarni AA, Abul-Hamd AT, Rai R, El Berry H, Cooper TG. Gln3p nuclear localization and interaction with Ure2p in Saccharomyces cerevisiae. J Biol Chem. 2001;276:32136–32144. doi: 10.1074/jbc.M104580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalho J, Zheng XF. Domains of Gln3p interacting with karyopherins, Ure2p, and the target of rapamycin protein. J Biol Chem. 2003;278:16878–16886. doi: 10.1074/jbc.M300429200. [DOI] [PubMed] [Google Scholar]
- 28.Gerrard SR, Bryant NJ, Stevens TH. VPS21 controls entry of endocytosed and biosynthetic proteins into the yeast prevacuolar compartment. Mol Biol Cell. 2000;11:613–626. doi: 10.1091/mbc.11.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- 30.Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 31.Boysen JH, Mitchell AP. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol Biol Cell. 2006;17:1344–1353. doi: 10.1091/mbc.E05-10-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slessareva JE, Routt SM, Temple B, Bankaitis VA, Dohlman HG. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell. 2006;126:191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Zheng XF. Endoplasmic reticulum and Golgi localization sequences for mammalian target of rapamycin. Mol Biol Cell. 2007;18:1073–1082. doi: 10.1091/mbc.E06-05-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones EW. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- 36.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 37.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 38.Edskes HK, Gray VT, Wickner RB. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci USA. 1999;96:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alarcon CM, Cardenas ME, Heitman J. Mammalian RAFT1 kinase domain provides rapamycin-sensitive TOR function in yeast. Genes Dev. 1996;10:279–288. doi: 10.1101/gad.10.3.279. [DOI] [PubMed] [Google Scholar]
- 41.Wickner RB. [URE3] as an altered Ure2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 42.Malkus P, Graham LA, Stevens TH, Schekman R. Role of Vma21p in assembly and transport of the yeast vacuolar ATPase. Mol Biol Cell. 2004;15:5075–5091. doi: 10.1091/mbc.E04-06-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.