Abstract
Protein conformational diseases are associated with the aberrant accumulation of amyloid protein aggregates, but whether amyloid formation is cytotoxic or protective is unclear. To address this issue, we investigated a normally benign amyloid formed by the yeast prion [RNQ+]. Surprisingly, modest overexpression of Rnq1 protein was deadly, but only when preexisting Rnq1 was in the [RNQ+] prion conformation. Molecular chaperones protect against protein aggregation diseases and are generally believed to do so by solubilizing their substrates. The Hsp40 chaperone, Sis1, suppressed Rnq1 proteotoxicity, but instead of blocking Rnq1 protein aggregation, it stimulated conversion of soluble Rnq1 to [RNQ+] amyloid. Furthermore, interference with Sis1-mediated [RNQ+] amyloid formation exacerbated Rnq1 toxicity. These and other data establish that even subtle changes in the folding homeostasis of an amyloidogenic protein can create a severe proteotoxic gain-of-function phenotype and that chaperone-mediated amyloid assembly can be cytoprotective. The possible relevance of these findings to other phenomena, including prion-driven neurodegenerative diseases and heterokaryon incompatibility in fungi, is discussed.
Keywords: Hsp40, neurodegenerative disease, Sis1, Rnq1, yeast prion
Alzheimer's disease, transmissible spongiform encephalopathies, and polyglutamine diseases are representatives of a large group of neurodegenerative disorders that are associated with the misfolding and conversion of particular proteins into amyloids (1). Amyloids form in response to many perturbations in protein homeostasis, namely mutations in the amino acid sequence of a disease-related protein, expansion of simple sequence elements in disease genes, elevated protein levels, and age-associated cell stress (2). Amyloid fibrils share a cross-β structural motif, in which β-strands run perpendicular to the long fiber axis and accumulate in intra- and extracellular inclusions (3, 4). Fibril formation requires that a misfolded protein expose a pleated β-surface that is capable of serving as a template and hydrogen-bonding partner with an extra β-strand (1). Biochemical parameters for the classification of protein aggregates as amyloids include resistance to solubilization by the detergent SDS and the ability to bind indicator dyes such as thioflavin-T (2).
Amyloid deposits in the brain are a hallmark of protein conformational disease, but often there is only a poor correlation between the detection of amyloid fibrils and other markers of neurodegeneration (5). Thus, there is still intense debate about whether amyloids are the causative toxic protein species in neurodegenerative diseases. In fact, recent, still controversial work suggests that amyloids might be benign or cytoprotective and that difficult-to-characterize soluble oligomeric conformers are the toxic species of disease-causing proteins (6–8).
Cells buffer proteotoxic events related to intracellular protein misfolding via chaperone-mediated partitioning of nonnative conformers between pathways for proper folding, inclusion body formation, and degradation (9). Molecular chaperones also play a critical role in the propagation of yeast prions (10), which are examples of intracellular amyloids that, in general, are not inherently toxic (11, 12). However, the conversion of active soluble Sup35 and Ure2 into their prion states [PSI+] and [URE3], respectively, inactivates these proteins (11, 12). Yeast prion formation occurs spontaneously at a low frequency, and the prion state is then perpetuated through the templating of newly synthesized prion proteins by preexisting amyloid-like prions (13). Templated prion proteins then undergo stable changes in structure and function to enter an amyloid-like state that is propagated and passed from mother cells to their daughters in a molecular chaperone-dependent manner (10). Yeast prions thereby constitute cytoplasmically transmitted, protein-based elements of inheritance that are dominant in genetic crosses (prions are denoted by brackets, italics, and capital letters to reflect these properties).
The yeast prion [RNQ+] is determined by the conformational state of the Rnq1 protein, which contains a C-terminal asparagine- and glutamine-rich prion domain and an N-terminal non-prion-forming domain (14, 15). The native form of Rnq1 has no known normal biological function and is nonessential. Yet the [RNQ+] prion can have important effects on yeast cells because it influences certain other proteins to convert to amyloid-like states (16–18). For example, [RNQ+] prions are required for the initial conversion of native Sup35 to the [PSI+] state. Indeed, [RNQ+] constitutes the cytoplasmically inherited factor known as [PIN+] and is the only known yeast prion that is commonly found in wild strains (19, 20). [RNQ+] prions also cause the exon 1 fragment of huntingtin protein, containing glutamine repeats, to become toxic in yeast (16). Thus, [RNQ+] prions can interact with other amyloid-forming proteins and thereby help drive their conversion into benign or toxic amyloid-like species.
Results
Overexpression of Rnq1 Is Toxic in [RNQ+] Cells.
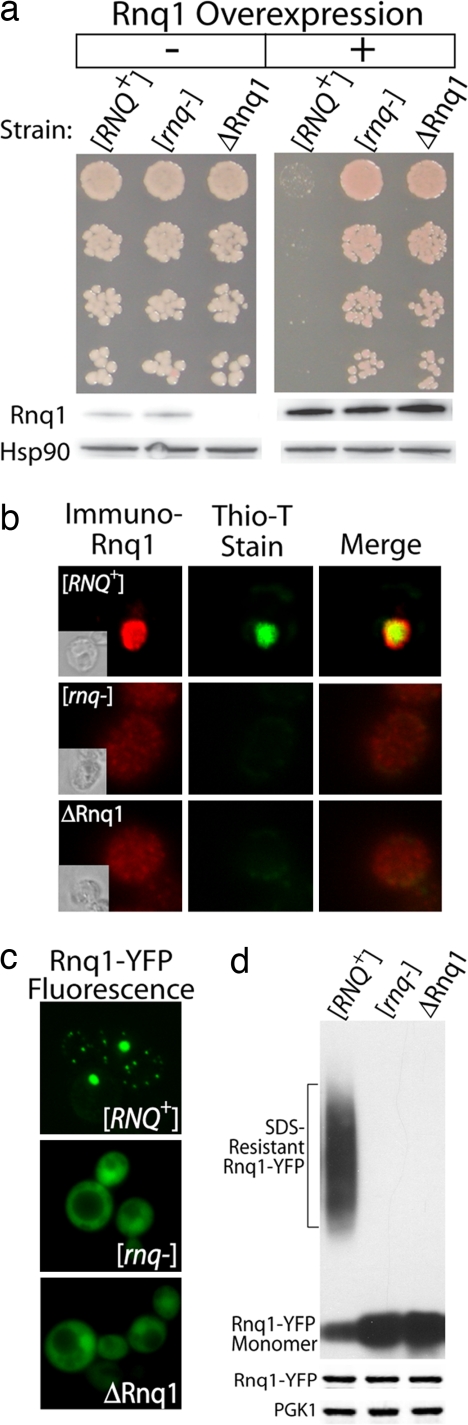
We recently discovered that moderate (i.e., ≈5- to 10-fold) overexpression of Rnq1 from the GAL1 promoter was severely toxic in cells that harbored the [RNQ+] prion (Fig. 1a (Upper) Growth of serially diluted liquid cultures on agar. (Lower) Protein levels detected by Western blotting). This finding was surprising because Rnq1 overexpression was not toxic when endogenous Rnq1 was in the [rnq−] non-prion conformation, nor was it toxic in cells carrying a deletion of the RNQ1 gene, Δrnq1 (Fig. 1a). Cell growth defects observed were more extreme than any we have observed with other misfolded proteins in yeast (21, 22). At this modest level of Rnq1 overexpression, ≈25% of [RNQ+] cells were dead within 4 h, as determined by the percentage of colony-forming units and dye exclusion (data not shown). Toxicity was accompanied by the accumulation of Rnq1 aggregates that stained with the common amyloid diagnostic dye thioflavin-T (Fig. 1b). Rnq1 overexpression was found to be toxic in [RNQ+] laboratory strains (W303, 74D-694, BY23, and BY4741), clinical strains (YJM269, YJM421, YJM436, and YJM653), a fermentation strain (Y12), and wine strains (I14, T73, and WE372) (M. Taipale and S.L., unpublished observations). Thus, Rnq1 toxicity is pervasive and not strain-specific.
Fig. 1.
Overexpression of Rnq1 is toxic to [RNQ+] cells. (a) The effect of Rnq1 overexpression on yeast cell viability in the presence and absence of the [RNQ+] prion. (b) Thioflavin-T staining of Rnq1 in [RNQ+], [rnq−], and ΔRnq1 cells. Fixed yeast were decorated with α-Rnq1 sera that was detected with a fluorescent secondary antibody. The same cells were simultaneously stained with the amyloid indicator dye, thioflavin-T. (c) Visualization of the aggregation state of Rnq1-YFP by fluorescence microscopy. (d) (Upper) Assembly status of Rnq1-YFP as determined by SDD-AGE. (Lower) Western blots of cell extracts.
Although not quite as deadly, a C-terminal Rnq1-YFP fusion protein behaved similarly to untagged Rnq1 and exhibited the same pattern of toxicity: toxic in [RNQ+] cells but not in [rnq−] or Δrnq1 cells. This result allowed us to correlate changes in toxicity with changes in protein distribution [Fig. 1c and supporting information (SI) Fig. S1]. While aggregated in [RNQ+] cells, Rnq1-YFP was distributed throughout the cytosol in [rnq−] or Δrnq1 cells. Using low- and high-copy plasmids that express Rnq1-YFP at different levels, we found that toxicity positively correlated with the degree of overexpression (Fig. S1a). Western blots of cell lysates separated by semidenaturing-detergent-agarose gel electrophoresis (SDD-AGE) demonstrated that, in [RNQ+] but not [rnq−] cells, Rnq1-YFP assembled into a SDS-resistant high-molecular-weight species typical of amyloid assemblies of yeast prions (Fig. 1d and Fig. S1b) (23). Yet a pool of soluble Rnq1-YFP, which ran at the position of a monomer on SDD-AGE gels, also was present in [RNQ+] cells.
Growth of [RNQ+] cells overexpressing just the non-prion-forming domain or just the prion-forming domain of Rnq1, amino acids 1–153 and 154–405, respectively, was not hindered (Fig. S1c). When the prion-forming domain is expressed on its own, it assembles into an SDS-resistant species that runs as an amyloid on SDD-AGE gels (Fig. S1d). Therefore, the mechanism for Rnq1 toxicity does not appear related to the accumulation of large quantities of [RNQ+]-like amyloid per se.
The toxicity of Rnq1 overexpression in the presence of the [RNQ+] prion might seem similar to the toxicity of overexpressed Sup35 in the presence of its prion [PSI+]. Overexpression of Sup35 is toxic in [PSI+] cells because it drives too much of the essential Sup35 protein into an inactive amyloid conformation (24). In contrast, Rnq1 toxicity cannot be due to an inhibition of Rnq1 function because deletion of the gene-encoding Rnq1 has no detectable effect on yeast growth under hundreds of conditions tested (T.F. Outeiro and S.L., unpublished data). Furthermore, in contrast to Sup35, expression of Rnq1's non-prion-forming domain does not rescue the toxicity caused by Rnq1 overexpression (data not shown).
The Hsp40 Sis1 Can Suppress Rnq1 Toxicity.
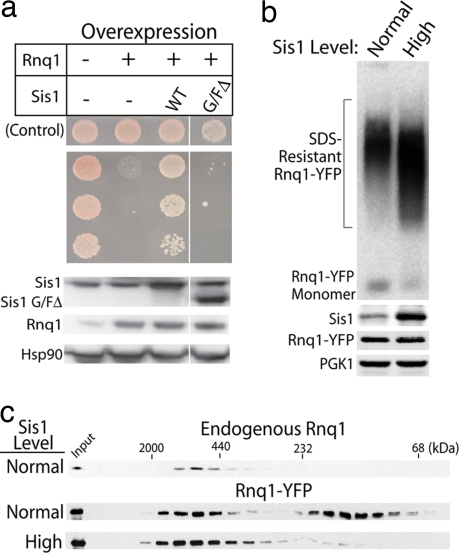
Sis1, an essential Hsp40 chaperone, is required for the propagation of the [RNQ+] prion state (25). Sis1 specifies Hsp70 function and is required for protein synthesis, protein folding, and cell stress protection (26, 27). Overexpressing Sis1 by as little as 3-fold strongly suppressed Rnq1 toxicity (Fig. 2a). To examine whether other chaperones were capable of suppressing Rnq1 toxicity, an expression library of 4,954 yeast genes was screened (21). Sis1 was the only chaperone in this library able to protect from Rnq1 toxicity (data not shown). This library includes, among many other chaperones, Ydj1, which is a member of the large Hsp40 family that is closely related to Sis1. It also includes Hsp70 Ssa1 (Hsp70) and Hsp104, which assist in shearing [RNQ+] prions to form seeds required for the propagation of the [RNQ+] state (28). Therefore, the effect of Sis1 on the toxicity of Rnq1 overexpression is unique.
Fig. 2.
Sis1 overexpression protects against Rnq1 toxicity. (a) (Upper) The effect of Sis1 or Sis1ΔG/F overexpression on Rnq1 toxicity. Control indicates strains grown under noninducing conditions. (Lower) Western blots of the indicated proteins. (b) (Upper) Effect of Sis1 overexpression on the formation of SDS-resistant [RNQ+] conformers as determined by SDD-AGE. (Lower) Western blots of the indicated proteins. (c) Gel-filtration analysis of intracellular pools of endogenous Rnq1 versus overexpressed Rnq1-YFP.
Sis1 not only promotes [RNQ+] prion formation, but it remains stably bound to the prion in a 1:1 complex (29). This finding could provide an explanation for the toxicity of overexpressed Rnq1: Elevation of [RNQ+] prion levels could kill cells by sequestering Sis1 away from its essential substrates. But this explanation is unlikely because cells in which Sis1 is depleted by nearly 100-fold grow for extended time periods and exhibit delayed lethality (30). In contrast, cells start to die within 4 h of the induction of Rnq1 overexpression.
To directly eliminate the possibility that cell death is due to the sequestration of Sis1, we deleted the domain of Sis1 that is required for interaction with the prion, the glycine- and phenylalanine-rich (G/F) region (29). A Sis1 ΔG/F variant fails to promote the propagation of [RNQ+], but can carry out Sis1's essential functions. We overexpressed Sis1 ΔG/F and found that, unlike Sis1, it could not suppress Rnq1 toxicity.
Sis1-Mediated Amyloid Formation Protects from Rnq1 Toxicity.
The toxicity produced by the overexpression of Rnq1 represents a dominant gain of function that requires endogenous Rnq1 protein to be in a [RNQ+] prion conformation. It may be Sis1's ability to facilitate [RNQ+] prion propagation that ameliorates Rnq1 toxicity. Indeed, the suppression of Rnq1 toxicity by Sis1 overexpression was accompanied by a substantial increase in the formation of SDS-resistant [RNQ+] amyloids (Fig. 2b). This seemed to be accompanied by a decrease in the pool of unassembled SDS-sensitive Rnq1. We have found, however, that although SDD-AGE is a reliable method for quantitatively detecting SDS-resistant species, it is not reliable for SDS-soluble species. To examine SDS-soluble species, we used gel-filtration chromatography. As shown in Fig. 2c, a large pool of unassembled Rnq1-YFP accumulated on Rnq1-YFP overexpression (Fig. 2c, compare Top and Middle). Suppression of Rnq1 toxicity by Sis1 correlated with a dramatic decrease in unassembled Rnq1-YFP pools and a corresponding increase in the pools of assembled forms (Fig. 2c, compare Middle and Bottom). These results suggest that cytotoxic Rnq1 conformers accumulate when levels of Rnq1 protein exceed the cell's capacity to efficiently promote the template-driven formation of the SDS-resistant [RNQ+] prion species. To test this hypothesis, we asked whether Rnq1 toxicity would be exacerbated when the efficiency of [RNQ+] amyloid assembly was reduced.
Identification of the Sis1-Binding Site in Rnq1.
First, we identified and mutated the chaperone-binding motif that Sis1 uses to interact with Rnq1. A peptide array was created that contained 25 residue N-acetylated peptides spanning the entire Rnq1 amino acid sequence. This array was incubated with purified Sis1 and washed, and Sis1-interacting peptides were identified by Western blot after transfer of bound chaperone to nitrocellulose (Fig. S2a). Tight binding of Sis1 was observed only with a few neighboring Rnq1 peptides, and these peptides were located in the non-prion-forming domain.
The amino acid sequence of this region is conserved in all known Rnq1 homologues and contains a classic, hydrophobic, chaperone-binding motif, LGKLALL (Fig. 3a and Fig. S2b) (31). Hsp40 proteins stimulate the binding of their Hsp70 cochaperones to specific substrates. Indeed, Sis1 stimulated binding of its Hsp70 cochaperone, Ssa1, to the peptides containing this motif in an ATP-dependent manner (Fig. 3b). Thus, Sis1 forms a functional chaperone:substrate complex with peptides containing this chaperone-binding motif.
Fig. 3.
Sis1 binding to a conserved chaperone-binding motif in the non-prion-forming domain of Rnq1. (a) A schematic showing the domain structure of Rnq1. The underlined region in the nonprion domain of Rnq1 represents a chaperone-binding motif identified via screening a cellulose peptide array (see Fig. S2). (b) Sis1-dependent binding of Hsp70 Ssa1 to the peptide in the Rnq1 peptide array that is bound most strongly by Sis1. (c) Mutation L94A in the chaperone-binding motif reduces the ability of Sis1 to form coimmunoprecipitable complexes with Rnq1-GFP in [RNQ+] cells. Rnq1-GFP was expressed by using the CUP1 promoter. Levels of the indicated proteins in c were visualized by Western blot (WB) analysis.
Next, to reduce the efficiency of Sis1's interaction with [RNQ+], we replaced hydrophobic leucine residues in the Rnq1 chaperone-binding motif with alanines (L91A, L94A, and L97A). As demonstrated by coimmunoprecipitation, the capacity of Rnq1-GFP to interact with Sis1 was strongly, but not completely, reduced by these mutations (Fig. 3c and data not shown).
Mutations in the Sis1-Binding Site of Rnq1 Interfere with [RNQ+] Amyloid Assembly.
To determine whether the Rnq1 chaperone-binding motif mutants were defective in the assembly of [RNQ+] amyloid, we expressed them as Rnq1-GFP fusions from an extrachromosomal plasmid in cells expressing WT Rnq1 in its prion state. The L91A, L94A, and L97A mutations were expressed at levels similar to those of WT Rnq1 by using the CUP1 promoter, but they had a reduced capacity to form fluorescent foci (Fig. S2c). An L45A mutation, which also is located in the non-prion-forming domain, but lies outside of the chaperone-binding motif, had no detectable effect on the assembly of [RNQ+] prions (Fig. S2c). Further, a time-course analysis by SDD-AGE (Fig. 4a) and pulse–chase (Fig. S2d) revealed that the rate at which newly synthesized Rnq1-GFP protein was converted into SDS-resistant conformers in vivo was reduced several fold by the L94A mutation in comparison with the WT protein. In addition, in vitro-purified Rnq1 L94A could be templated by prion “seeds” present in [RNQ+] cell extracts to form SDS-resistant species (Fig. 4b). However, it was templated and converted to an SDS-resistant form with lower efficiency than the WT Rnq1 protein.
Fig. 4.
Mutations in the chaperone-binding motif of Rnq1 reduce the efficiency of [RNQ+] amyloid assembly. (a) Kinetics of Rnq1-GFP L94A assembly into SDS-resistant aggregates in [RNQ+] yeast determined by SDD-AGE. Rnq1-GFP fusions were expressed by using the CUP1 promoter. (b) [RNQ+] seed-dependent assembly of purified Rnq1-His and Rnq1-His L94A into SDS-resistant amyloid. (c) (Upper) Growth of 5-fold serial dilutions of [RNQ+] strains in which WT or L94A Rnq1 was overexpressed from the GAL1 promoter. Where indicated, Sis1 was overexpressed from the GPD promoter. (Lower) Relative expression level of the specified proteins as determined by Western blot.
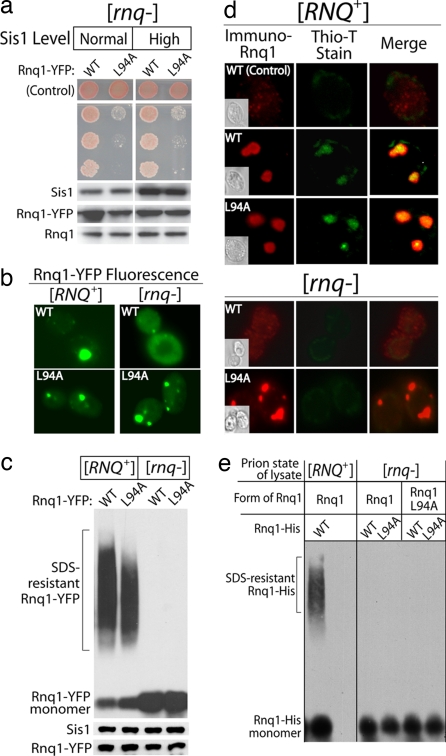
Finally, we asked whether the impairment of Rnq1 amyloid assembly increased the toxicity of Rnq1 overexpression. As we hypothesized, Rnq1 L94A was more toxic than WT Rnq1 when overexpressed in [RNQ+] cells (Fig. 4c). A triple mutant, Rnq1 L94A–L96A–L97A, was even more toxic than Rnq1 L94A (data not shown). As expected from the fact that the Rnq1 L94A mutation impaired, but did not eliminate, interaction with Sis1, the overexpression of Sis1 3-fold was still able to suppress the toxicity of the mutant protein (Fig. 4c).
Thus far, we have shown that interfering with the assembly of Rnq1 into the [RNQ+] amyloid state is extremely toxic. Toxicity occurs when Rnq1 expression is higher than normal (Figs. 1 and 2) or when mutations in Rnq1 interfere with the efficiency of Sis1 interaction (Figs. 3 and 4). In addition, depletion of Sis1 from the cytosol reduces the efficiency of [RNQ+] prion assembly and exacerbates Rnq1 toxicity (Fig. S3). These collective data indicate that the efficient conversion of native Rnq1 into its SDS-resistant amyloid form prevents the accumulation of a toxic Rnq1 conformer.
Suppression of Rnq1 Toxicity by Sis1 Requires [RNQ+] Prion Assembly.
Rnq1 L94A exhibits a higher propensity than WT Rnq1 to form SDS-soluble aggregates when [RNQ+] assembly is impeded via depletion of Sis1 (Fig. S3). The inability of cells to maintain Rnq1 L94A in a soluble state correlates with the enhanced toxicity of the L94A mutant. In this sense, Rnq1 L94A is similar to alleles of amyloidogenic proteins whose subtle defects in folding kinetics cause devastating protein conformational diseases (1). Thus, we wondered whether Rnq1 L94A would assume a toxic conformation in the absence of templating by [RNQ+] prion seeds. Indeed, the overexpression of Rnq1 L94A, but not WT Rnq1, was toxic in [rnq−] strains (Fig. 5a). Hence, a small amino acid substitution can cause Rnq1 to be toxic even in the absence of [RNQ+] amyloid formation.
Fig. 5.
Rnq1 L94A toxicity and assembly status in [rnq−] yeast. (a) (Upper) Growth of 5-fold serial dilutions of [rnq−] strains in which WT or L94A Rnq1 was overexpressed from the GAL1 promoter. Sis1 was overexpressed from the GPD promoter. (Lower) Western blots of cell extracts. (b) Fluorescent foci formed by Rnq1-YFP and Rnq1-YFP L94A in [RNQ+] and [rnq−] cells. (c) (Upper) SDD-AGE analysis of aggregates formed by Rnq1-YFP and Rnq1-YFP L94A in [RNQ+] and [rnq−] cells. (Lower) Western blots of cell extracts. (d) Thioflavin-T staining of untagged Rnq1 and Rnq1 L94A in [RNQ+] and [rnq−] cells. Fixed yeast was decorated with α-Rnq1 sera that was detected with a fluorescent secondary antibody. The same cells were simultaneously stained with the amyloid indicator dye, thioflavin-T. (e) Cell extracts from [rnq−] cells overexpressing either Rnq1 or Rnq1 L94A were incubated with purified Rnq1-His or Rnq1-His L94A. The assembly status of purified Rnq1-His was determined by SDD-AGE. As a control, the assembly status of purified Rnq1-His incubated with [RNQ+] cell extract also was determined.
Sis1-dependent [RNQ+] amyloid formation appears to protect cells from toxicity caused by the overexpression of Rnq1. If amyloid formation is a critical aspect of Sis1's ability to suppress Rnq1 toxicity, then Sis1 overexpression should not protect [rnq−] cells from Rnq1 L94A-mediated death because these cells lack the [RNQ+] prion seeds required for amyloid assembly. Indeed, overexpression of Sis1, which binds Rnq1 L94A with reduced efficiency, protected [RNQ+], but not [rnq−], strains from Rnq1 L94A toxicity. This finding further confirms that Rnq1 toxicity is not caused by the sequestration of Sis1 into [RNQ+] prion complexes. Furthermore, the presence of the [RNQ+] prion assembly pathway and Sis1 overexpression are both required for the suppression of Rnq1 toxicity.
Rnq1 L94A Does Not Form Prion Amyloids in [rnq−] Cells.
To rule out the possibility that Rnq1 L94A assembled into [RNQ+] prions spontaneously in [rnq−] cells, we compared its assembly status to that of WT Rnq1 in [rnq−] strains (Fig. 5 b–e). In [rnq−] cells, Rnq1 L94A exhibited a higher propensity than WT Rnq1 to coalesce into foci (Fig. 5b). Gel-filtration chromatography showed that Rnq1 L94A formed high-molecular-weight aggregates in these cells (Fig. S4). Notably, these aggregates were not SDS-resistant and (Fig. 5c) were unable to bind the amyloid indicator thioflavin-T (Fig. 5d). Thus, Rnq1 toxicity is not related to the accumulation of excess pools of [RNQ+] amyloid and may be caused by a SDS-soluble Rnq1 species.
Because small prion seeds in the form of detergent-soluble prefibrillar species could have escaped detection by SDD-AGE and thioflavin-T staining, we applied another test for the existence of such forms of Rnq1 L94A in [rnq−] cell extracts (Fig. 5e). Prion seeds in cell extracts can be sensitively detected through their ability to catalyze the conversion of exogenously added native prion protein into SDS-resistant amyloids. Prion seeds were readily detected in lysates of [RNQ+] cells overexpressing Rnq1 L94A (Fig. 4b). However, extracts of [rnq−] cells that contained toxic levels of Rnq1 L94A failed to seed assembly of purified His-Rnq1 or His-Rnq1-L94A into SDS-resistant amyloids (Fig. 5e).
The Rnq1 L94A assemblies in [rnq−] cells fail to meet three classification standards of [RNQ+] amyloid. They are SDS-soluble, they do not stain with thioflavin-T, and they do not seed polymerization of soluble Rnq1 protein. Rnq1 L94A is more lethal than Rnq1 and can assume a toxic conformation in the absence of [RNQ+] templates. Rescue from Rnq1 L94A toxicity requires Sis1 overexpression and active propagation of the [RNQ+] prion. Therefore, it appears that the conversion of Rnq1 L94A to [RNQ+] amyloid prevents the accumulation of toxic Rnq1 conformer whose true nature remains obscure.
Discussion
Our data suggest a model in which efficient chaperone-dependent conversion of soluble Rnq1 into SDS-resistant [RNQ+] amyloid is critical to prevent the formation of other toxic Rnq1 conformers. We demonstrate that toxic Rnq1 conformers accumulate in a nonamyloid form when [RNQ+] assembly is made inefficient by multiple means. We propose that nonproductive templating of Rnq1 monomers by [RNQ+] seeds predisposes Rnq1 to leave the amyloid pathway and accumulate as a toxic species, whose exact nature is not yet clear. Templating of native proteins to form amyloid is a basic feature of amyloidogenesis, and we suggest that inefficiencies in this process contribute to the proteotoxicity associated with certain protein conformational diseases. This templating model explains how amyloid formation can serve a protective function, whereas the [RNQ+] prion state is a prerequisite for toxicity of the WT Rnq1 protein.
One of Sis1's functions in [RNQ+] prion propagation appears to be promoting the shearing of [RNQ+] amyloid fibers into smaller pieces, thereby creating new surfaces to more efficiently seed the assembly of the prion (28). This reaction also requires Hsp104 and Hsp70. Hence, binding of Sis1 to the non-prion-forming domain of full-length Rnq1 may help facilitate this shearing process. However, because overexpression of Hsp70 and Hsp104 does not suppress Rnq1 toxicity, Sis1 may have additional functions in [RNQ+] prion propagation that do not overlap with those of other chaperones. Sis1 stably associates with assembled [RNQ+] conformers in a 1:1 molar ratio (29). Therefore, Sis1's binding to the non-prion-forming domain has the potential to stabilize [RNQ+] prions in a conformation that is optimal for efficient amyloid fibril growth.
Many neurodegenerative diseases involve the accumulation of intracellular and/or extracellular amyloid protein aggregates. In the past, these amyloid aggregates were thought to be the cytotoxic, disease-causing protein conformer. However, recent studies have begun to question this view. As one striking example in mice, deletion of the GPI anchor of the prion protein PrP leads to massive extracellular amyloid plaque formation in mice injected with infective scrapie, but causes no overt clinical manifestations of scrapie (32). Furthermore, it has been suggested that neurodegenerative diseases are caused by the ability of very different proteins to adopt common toxic nonamyloid conformers such as protofibrils or soluble oligomers (3, 5). Hence, amyloid formation may serve to convert oligomeric amyloid precursors into a highly stable, nontoxic form (33).
Although amyloid is not toxic, its interaction with soluble protein forms could give rise to pathogenic species via nonproductive templating. Case in point, when GPI-anchorless PrP was expressed together with WT PrP, it accelerated scrapie disease and resulted in increased deposits of both amyloid and nonamyloid proteinase K-resistant PrP (32). Similarly, in yeast, the toxicity of Huntington exon 1 depends on the [RNQ+] prion state (16, 22). This concept may even extend to the heterokaryon incompatibility mediated by the [Het-s] prion in Podospora anserina (34). HET-s in its prion form only leads to cell death when coexpressed with the HET-S allele that cannot form amyloid. Templating of the nonamyloidogenic HET-S protein by [Het-s] prion seeds could lead to the formation of a toxic species, whereas templating of HET-s protein would result in the nontoxic prion amyloid species.
The aggregation state and the toxicity of aggregation-prone proteins are strongly modulated by host factors such as Hsp70 and its associated cochaperones, but the mechanisms for chaperone function in this process are just being defined (9). Molecular chaperones generally act to antagonize protein aggregation. Yet our observations that the chaperone-dependent assembly of amyloid conformers can be cytoprotective provide a different view of the effects of chaperones in conformational disease. Thus, molecular chaperones can antagonize protein toxicity in conformational disorders by two different mechanisms: They can solubilize misfolded proteins or aid in sequestering them into benign, amyloid-like species. The most central aspect of antagonizing toxicity of misfolded proteins appears to be preventing accumulation of the detergent-soluble misfolded species, rather than preventing the formation of amyloid conformers.
Methods
Strains.
W303 and 74D-694 strains were used to take advantage of the different genetic markers or gene deletions. Unless noted, yeast harbored Rnq1 in its [RNQ+] prion form. The generation of isogenic [rnq−] strains was accomplished via sequential passage of cells on plates containing 3 mM guanidinium-HCl (35).
Analysis of Rnq1 Cytotoxicity.
Strains harboring pRS416-RNQ1 or pRS416-RNQ1-YFP were grown overnight in synthetic dropout media containing 2% raffinose before 5-fold serial dilutions were spotted on either glucose or galactose plates. Alternatively, strains that harbored pRS316-RNQ1-GFP or pRS315-RNQ1-GFP were cultured overnight in synthetic media containing glucose before dilutions were spotted on plates that contained 500 μM CuSO4. Plates were photographed after 3–5 days of incubation at 30°C.
Screening of an Rnq1 Peptide Array.
A 25mer Rnq1 cellulose-bound peptide array was prepared by automated spot synthesis and screened essentially as described (31) (see also SI Methods).
Pulse–Chase Analysis of [RNQ+] Prion Formation.
Pulse labeling of yeast to analyze the kinetics of Rnq1 assembly in 74D-694 cultures was performed essentially as described (30) (see also SI Methods). Time points were taken after the addition of 50 μM CuS04 to growing cells.
[RNQ+] Prion Formation by Fluorescence Microscopy.
Assembly of Rnq1-GFP into fluorescent foci that represent prion amyloids was preformed essentially as described (26) (see also SI Methods).
SDD-AGE.
Rnq1 assembly into SDS-resistant [RNQ+] prions was monitored by SDD-AGE as described (23) (see also SI Methods).
Rnq1 Coimmunoprecipitation with Sis1.
Expression of Rnq1-GFP in log-phase cells harboring the indicated form of pRS316-Rnq1-GFP was induced by supplementation with 50 μM CuSO4. Native cell extracts were prepared 1 h later under the conditions described (26), and α-Sis1 was added to cell lysates. Rnq1-GFP was coimmunoprecipitated with Sis1 and detected with α-GFP by Western blot.
Seeded Polymerization of Purified Rnq1.
Purified Rnq1-His was added to lysates of the indicated strains. Assembly was monitored by SDD-AGE (see also SI Methods).
Size-Exclusion Chromatography.
[RNQ+] or [rnq−] cells were grown overnight in synthetic media at 30°C. Rnq1-YFP expression was induced by the addition of 2% galactose for 4 h before the collection of 100 OD units of cells. Proteins in extracts created with a nondenaturing lysis buffer were resolved on a Superose 12S sizing column (Amersham Pharmacia). Indicated proteins in column fractions were detected by Western blot.
Supplementary Material
Acknowledgments.
We thank Jessica Brown, Daniel Summers, and James Shorter for helpful discussions; Karen Allendoerfer for critical reading; and Elizabeth Craig (University of Wisconsin, Madison, WI) for providing the Sis1 ΔG/F construct and Sis1 antisera. This work was supported by the National Institutes of Health (D.M.C. and S.L.), a Howard Hughes Medical Institute Investigatorship (to S.L.), an American Heart Association Predoctoral Fellowship (to P.M.D.), and a National Science Foundation predoctoral training grant (to R.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802593105/DCSupplemental.
References
- 1.Carrell RW, Lomas DA. Conformational disease. Lancet. 1997;350:134–138. doi: 10.1016/S0140-6736(97)02073-4. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 4.Nelson R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 6.Cheng IH, et al. Accelerating amyloid-beta fibrillization reduces oligomer levels and functional deficits in Alzheimer disease mouse models. J Biol Chem. 2007;282:23818–23828. doi: 10.1074/jbc.M701078200. [DOI] [PubMed] [Google Scholar]
- 7.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 8.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 9.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 10.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 11.Lindquist S. Mad cows meet psi-chotic yeast: The expansion of the prion hypothesis. Cell. 1997;89:495–498. doi: 10.1016/s0092-8674(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 12.Wickner RB. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 13.Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 14.Derkatch IL, et al. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? EMBO J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sondheimer N, Lindquist S. Rnq1: An epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 16.Meriin AB, Zhang X, He X, Newnam GP, Sherman MY. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 18.Taneja V, Maddelein M, Talarek N, Saupe S, Liebman SW. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol Cell. 2007;27:67–77. doi: 10.1016/j.molcel.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: The story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 20.Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci USA. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper AA, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duennwald ML, Jagadish S, Giorgini F, Muchowski PJ, Lindquist S. A network of protein interactions determines polyglutamine toxicity. Proc Natl Acad Sci USA. 2006;103:11051–11056. doi: 10.1073/pnas.0604548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 24.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan CY, Lee S, Ren HY, Cyr DM. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol Biol Cell. 2004;15:761–773. doi: 10.1091/mbc.E03-03-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong T, Luke MM, Arndt KT. Transcriptional regulation of the yeast DnaJ homologue SIS1. J Biol Chem. 1996;271:1349–1356. doi: 10.1074/jbc.271.3.1349. [DOI] [PubMed] [Google Scholar]
- 28.Aron R, Higurashi T, Sahi C, Craig EA. J-protein co-chaperone Sis1 required for generation of [RNQ(+)] seeds necessary for prion propagation. EMBO J. 2007;26:3794–3803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez N, Aron R, Craig EA. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ(+)] Mol Biol Cell. 2003;14:1172–1181. doi: 10.1091/mbc.E02-09-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luke MM, Sutton A, Arndt KT. Characterization of SIS1, a Saccharomyces cerevisiae homologue of bacterial dnaJ proteins. J Cell Biol. 1991;114:623–638. doi: 10.1083/jcb.114.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chesebro B, et al. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 2005;308:1435–1439. doi: 10.1126/science.1110837. [DOI] [PubMed] [Google Scholar]
- 33.Piccardo P, Manson JC, King D, Ghetti B, Barron RM. Accumulation of prion protein in the brain that is not associated with transmissible disease. Proc Natl Acad Sci USA. 2007;104:4712–4717. doi: 10.1073/pnas.0609241104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coustou-Linares V, Maddelein ML, Begueret J, Saupe SJ. In vivo aggregation of the HET-s prion protein of the fungus Podospora anserina. Mol Microbiol. 2001;42:1325–1335. doi: 10.1046/j.1365-2958.2001.02707.x. [DOI] [PubMed] [Google Scholar]
- 35.Eaglestone SS, Ruddock LW, Cox BS, Tuite MF. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:240–244. doi: 10.1073/pnas.97.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.