Abstract
Spliceosomal introns, a hallmark of eukaryotic gene organization, were an unexpected discovery. After three decades, crucial issues such as when and how introns first appeared in evolution remain unsettled. An issue yet to be answered is how intron positions arise de novo. Phylogenetic investigations concur that intron positions continue to emerge, at least in some lineages. Yet genomic scans for the sources of introns occupying new positions have been fruitless. Two alternative solutions to this paradox are: (i) formation of new intron positions halted before the recent past and (ii) it continues to occur, but through processes different from those generally assumed. One process generally dismissed is intron sliding—the relocation of a preexisting intron over short distances—because of supposed associated deleterious effects. The puzzle of intron gain arises owing to a pervasive operational definition of introns, which sees them as precisely demarcated segments of the genome separated from the neighboring nonintronic DNA by unmovable limits. Intron homology is defined as position homology. Recent studies of pre-mRNA processing indicate that this assumption needs to be revised. We incorporate recent advances on the evolutionarily frequent process of alternative splicing, by which exons of primary transcripts are spliced in different patterns, into a new model of intron sliding that accounts for the diversity of intron positions. We posit that intron positional diversity is driven by two overlapping processes: (i) background process of continuous relocation of preexisting introns by sliding and (ii) spurts of extensive gain/loss of new intron sequences.
Keywords: intron drift, intron migration, intron movement, intron sliding, intron slippage
Eukaryotes traveled disparate trajectories of intron gain and/or loss since they split from their last common ancestor. Most of what is known about newly originated intron positions has been obtained from phylogenetic reconstructions of ancestral character states.
Background
Intron Positions Arise de Novo in Evolution.
Approaches to the evolution of intron positions have become increasingly sophisticated since the early comparisons of GenBank data (1). Yet the prevalence with which new intron positions arise in evolution continues to be debated (2–5). At the root of the controversy are differences in methodological postulates, phylogenetic sampling scopes, and criteria for deciding intron positions.
Ancestral intron positions are inferred from a matrix of intron presence/absence built by projecting present positions onto automated multiple sequence alignments of genome scale sets of orthologous proteins. Rogozin et al. (6) compiled 684 clusters of orthologous genes (KOGs) from eight model eukaryotes, including one vertebrate (human), two arthropods (Drosophila melanogaster and Anopheles gambiae), one nematode (Caenorhabditis elegans), two fungi (Saccharomyces cerevisiae and Schizosaccharomyces pombe), one plant (Arabidopsis thaliana), and one protist (Plasmodium falciparum). The resulting 16,577 unique intron positions were condensed into 7,236 (≈43%) by retaining only those located within well conserved tracts of alignment. The full and conserved matrices were analyzed by Dollo parsimony (6). The conserved matrix was subsequently reanalyzed by other authors. Roy and Gilbert (7) devised a local maximum-likelihood (ML) approach that corrects for the known bias of Dollo parsimony toward the overestimation of intron gain at peripheral branches, owing to a failure to detect intron losses that are not directly observed. However, when the number of target sites (i.e., observed plus unobserved intron positions) is taken into account explicitly in ML simultaneous comparison of all species (8–10), the numbers of ancestral intron positions are fewer than those obtained previously (7). The reason could be that the method of ref. 7 does not allow for homoplastic gains (i.e., introns arising more than once at the same homologous position) (8, 9, 11), but it also could be that homoplastic gains are overestimated by ML methods (e.g., due to sparseness of phylogenetic sampling). Homoplastic gains seem to have been extremely overestimated by Qiu et al. (12), who claim that the vast majority of intron positions are new apparently because, in their Bayesian analysis of 10 gene families, the number of target sites is bounded to be equal to the number of observed intron positions (8, 9).
The dataset shown previously (6) has been expanded from 8 to 18 eukaryotic species using a new criterion to determine intron positional homology (10). The 10 added species split long branches of the tree near the tips. The result is a 30% reduction of KOGs (from 684 to 483), but not of intron positions in the matrix, which increases by 10% (from 7,236 to 8,044), almost twice the value that obtains (4,136) by extrapolating from the corresponding numbers in the conserved dataset of ref. 6. A factor contributing to the increase in intron positions may be that ref. 10 rewards matching of intron positions to help align the amino acids, which relaxes the minimum of protein conservation required for identifying intron positions. The ref. 6 dataset also has been expanded by ref. 11 by adding 11 species, 6 of which are not included in ref. 10. Previous models allow for variation of the rates of intron gain and loss, among either lineages (7–10) or genes (12), and the Carmel et al. (11) model accommodates both, plus rate variation among sites within a gene, thus avoiding the difficulty of having to estimate the number of target sites separately (8–10). Five of the 11 new species involve intron positions in deuterostomia. The other five species (except for Oryza sativa, which is closely related to Arabidopsis) belong to new long peripheral branches. The increase in the number of species results in a 40% reduction of KOGs (from 684–391) and a 20% reduction in the number of analyzed intron positions (from 7,236 to 5,755).
Most of the studies cited above agree that the last eukaryotic common ancestor (LECA) had a high intron density. A fraction (10–40%) (1, 3, 6) of the ancestral introns has persisted to the present time, although the degree of ancestral intron retention varies among species owing to vast differences in rates of intron loss. But the inferred and/or observed intron positions at many nodes cannot be explained without also invoking differences in rates of gain. Patterns of gain appear to be due to episodic bursts superimposed on much lower background rates. Except for those spurts, intron losses dominate over intron gains in most lineages (11). Apparently, both gain and loss rates have decreased during the last tens to hundreds of million years, but at a rate decrease much greater for intron gain than for intron loss (10, 11).
Mechanisms for the Origin of Novel Intron Positions.
There are at least three global mechanisms for the de novo origin of intron positions: (i) transposition, which would include duplication of preexisting introns; (ii) insertion of intron-like transposons; and (iii) tandem duplication of exon sequences that happen to include splice sites (4, 13). These mechanisms assume that (i) every new intron position originates from a “formative” intron, (ii) formative introns derive from intron donors elsewhere in the genome (including introns, transposons, and exons), and (iii) formation of a novel intron position is instantaneous. Formative introns are at first identical to their donors and are expected to remain detectably similar for millions of years. A straightforward approach to show that intron positions arise by any of the proposed mechanisms is finding the donors of formative introns, which should not be difficult provided recent intron positions arise in sufficient numbers.
Results
Recent Rates of Origination of New Intron Positions.
Rates of intron gain are inferred to have strongly declined during the last tens to hundreds of million years (6, 8–11). Table 1 (“Long-term”) shows the rates of intron gain for long peripheral branches, not appropriate for evaluating recent gains, but given for comparison. The rates tend to decrease as the complexity of the evolutionary model increases. The lowest values are attained by allowing for gain rate variation among lineages, genes, and sites within a gene (11), but the models' relative performance in capturing the evolution of intron numbers has not been evaluated statistically. Refs. 8 and 10 use the same ML approach, but the latter produces smaller estimates perhaps because the alignment strategy minimizes intron positional discordance. Some of the long branches were divided by refs. 10 and 11 to provide ML estimates of recent intron gain (Table 1, “Recent”). Accordingly, the human lineage gained as a minimum 0.53 introns per gene per billion years (By) since the split from rodents. The corresponding minimum rates for C. elegans (× C. briggsae), A. thaliana (× O. sativa), and O. sativa (× A. thaliana) are, respectively, 0.8, 0.56, and 0.95 per gene/By.
Table 1.
Estimates of long-term and recent rates of intron gain (per gene per 109 years) for some better studied lineages
| Ref. | Dataset* (sps; KOGs) | Method† | Lineage‡ (subtree; calibration time in My) |
|||
|---|---|---|---|---|---|---|
| H. sapiens | D. melanogaster | C. elegans | A. thaliana | |||
| Long-term | ((Ag,Dm),Ce,Hs); 1,130 | (Ag,Dm); 470 | ((Ag,Dm),Ce,Hs); 1,130 | ((An,Fu),At); 1,670 | ||
| 6 | 8;684 | Dollo parsimony | 2.39 | 0.45 | 1.04 | 1.75 |
| 7 | 8;684 | Dollo parsimony + local ML + AL | 1.10 | 0.28 | 0.93 | 1.51 |
| 8 | 8;684 | global ML + AL | 1.50 | 0.36 | 1.11 | 1.89 |
| 9 | 8;684 | global ML + AL | 1.52 | 0.36 | 1.09 | 1.90 |
| 10 | 18;483 | global ML + AL | 0.99 | 0.28 | 2.12 | 1.80 |
| 11 | 19;391 | global ML + AL + AG + AS | 1.09 | 0.15 | 0.89 | 1.46 |
| Recent | ((Mm,Rn),Hs); 95 | — | (Cb,Ce); 100 | (Os,At); 250 | ||
| 10 | 18;483 | global ML + AL | 0.53 | — | 0.80 | 0.56 |
| 11 | 19;391 | global ML + AL + AG + AS | 0.63 | — | — | 0.68 |
*Number of species (sps) and number of eukaryotic clusters of orthologous genes (KOGs).
†Allowance for rate variation among lineages, genes, or sites is denoted as AL, AG, and AS, respectively.
‡Subtrees are subsets of the trees used in the referenced studies and are given in Nexus format. The number next to each subtree is the duration (My; from ref. 11) of the branch over which rates are calculated (underlined). Ag, Anopheles gambiae; An, animals; At, Arabidopsis thaliana; Cb, Caenorhabditis briggsae; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Fu, fungi; Hs, Homo sapiens; Mm, Mus musculus; Os, Oryza sativa; Rn, Rattus norvegicus.
ML estimates of recent intron gain are almost always larger than corresponding estimates obtained with parsimony using closely related species (14–24). Excluding distantly related species allows for more efficient exploitation of whole-genome data, including longer and better alignments, because it allows the use of synteny and gene order and orientation to establish orthology (e.g., refs. 17 and 21). The approach begins by identifying discordant intron positions between closely related homologs. The discordant positions are then compared to an outgroup. The discordances that match an intron in the outgroup are attributed to intron loss; otherwise they are attributed to intron gain. Roy et al. (14) found no evidence of intron gain from 1,560 human-mouse and 360 mouse-rat orthologs (using the fish Fugu and human as outgroups, respectively).
No case of gain was reported in a mapping of annotated intron–exon boundaries of either 17,242 human or 16,068 mouse genes in alignments of human, mouse, rat, and dog genomic sequences (17) (this result appears to be at variance with that obtained by ref. 25, which reported many novel introns in humans, although the new intron-containing genes are either unannotated or in copy-number variant regions). D. melanogaster (subgenus Sophophora) is inferred to have gained ≈0.45 introns/gene/By during the ≈40 My elapsed since it split from the Drosophila subgenus (18). Table 2 gives parsimony estimates of intron gain from closely related species/lineages.
Table 2.
Parsimony estimates of recent rates of intron gain (per gene per 109 years) for some better studied lineages
| Ref(s). | Gene set | Rate (intron/gene/By) | Lineage* (tree; calibration time in My) |
|---|---|---|---|
| 15, 22 | 16,590 h | 0.0034–0.0127 | C. elegans ((((((Cb,Cr),Csp4),Ce),Bm),Hs),Sp); 100 |
| 24 | 4,690 h | 0.0023 | C. neoformans ((Cnn,Cng),(CgR,CgW)); 37 |
| 16 | 1,447 h | 0.66 | Three ascomycete fungi (((Mg,Nc),Fg),An); 630 |
| 19, 23 | 2,563 p | 0.30–0.90 | A. thaliana (((At1,At2),Le),Os); 20–60 |
| 20 | 3,101 p | 0.15 | O. sativa ((Os1,Os2),At); 70 |
| 21, 26 | 3,479 p | 0.0014–0.0115 | P. falciparum (((((Pk,Pv),Py),Pf),Pg),(Ta,Tp)); 100 |
The number next to each tree is the duration (My; from references in the leftmost column) of the branch over which rates are calculated (underlined). h, homolog; p, paralog.
*Species not given in Table 1 are Cr and Csp4, Caenorhabditis remanei and sp. 4., respectively; Bm, Brugia malayi; Sp, Schizosaccharomyces pombe; Cnn and Cng, Cryptococcus neoformans var. neoformans and C. neoformans var. grubii, respectively; CgR and CgW, Cryptococcus gattii strains R265 and WM276, respectively; Mg, Magnaporthe grisea; Nc, Neurospora crassa; Fg, Fusarium graminearum; An, Aspergillus nidulans; At1 and At2, Arabidopsis thaliana duplicates 1 and 2, respectively; Le, Lycopersicon esculentum; Os1 and Os2, Oryza sativa duplicates 1 and 2, respectively; Pk, Pv, Py, Pf, and Pg, Plasmodium knowlesi, P. vivax, P. yoelii, P. falciparum, and P. gallinaceum, respectively; Ta and Tp, Theileria annulata and T. parva, respectively.
The higher ML rates of recent intron gain, compared with those obtained with parsimony, cannot be accounted for by systematic differences in calibration dates between the two optimality criteria. Under a range of models, parsimony is an ML estimator, but not for the model that allows multiple changes (gains or losses) at a position (26). Intron gain/loss has only two alternative states and, thus, is more vulnerable to homoplasy. Homoplastic gains represent 5–20% of shared intron positions (8, 9, 27, 28). Although the potential for homoplastic gain decreases with the divergence in the sample, closely related sequences are prone to it by virtue of their high similarity (provided gains do not occur at random) (29). Studies of closely related species that use distantly related outgroups (e.g., 15, 17, 21, 22) have enhanced likelihood of parallel gain. However, both the ML and parsimony estimates would be downwardly biased if newly gained intron positions tend to be excluded by data filtering.
To avoid database errors in intron–exon boundaries and annotation, analyses of intron gain are typically confined to positions in windows of protein alignment that are highly conserved and often do not contain gaps (6, 8–11, 14–18, 20, 21, 24). In addition, slight discordances (<6 nt) are either excluded (14, 16, 21) or treated as orthologous (6, 8–11). Consequently, conclusions about the incidence of intron gain are based on small subsets (≤50%) of the positional discordances in the unfiltered data (6, 8–11, 14, 17, 20, 30). But the subsets will be impoverished in gained intron positions if the mechanisms that create new positions can cause indels and/or changes in the amino acid composition of flanking exons (see below).
An indication that filtering methods may underestimate the rates of intron gain comes from ref. 6, where the ratio between gains and losses in the unfiltered dataset (5,377/14,341 = 2.67) is ≈50% greater than in the conserved dataset (3,306/5,951 = 1.80). The proportion of counted gains at peripheral branches also is greater for the full dataset (50% vs. 40% for the full and conserved datasets respectively, excluding deep-branching fungi, Arabidopsis, and Plasmodium). It seems reasonable to assume that the rates of recent intron gain are somewhere between the ML and the parsimony estimates. Some parsimony rates would still imply substantial gains of introns in the recent past when extrapolated to a genomic gene number scale. In particular, the number of introns gained/My would be ≈6.1 in the lineage of D. melanogaster (assuming 13,600 genes), between 7.6 and 22.9 in A. thaliana (25, 500), 7.5 in rice (50,000), and 6.6 in Ascomycete fungi (10,000).
The Puzzle of the Origin of Novel Intron Positions.
Genome scans carried out in human, D. melanogaster, C. elegans, and A. thaliana could not detect a single case of homologous introns in nonhomologous genes (31). Subsequent studies in Drosophila, Caenorhabditis, and rice searching specifically for donors of introns supposed to be novel because of restricted phylogenetic distribution (15, 18, 20) also have been fruitless (7, 18, 22, 23). This may not be surprising for humans and nematode if one assumes that the rates of recent intron gain in those species are closer to the low-rate parsimony (14, 17, 22) than to the ML estimates (10, 11). But the outcome is most unexpected for Drosophila, Arabidopsis, and rice even if we assume parsimony estimates. In Drosophila, a 100-bp-long formative intron evolving at 1.5 × 10−5 substitutions per site/My (32) should remain 80% identical to its donor after 6.6 My of divergence. During that time, Drosophila should have acquired >40 novel intron positions. In the case of Arabidopsis and rice, assuming a rate of 1 × 10−5 substitutions per site/My (33), the corresponding outcome would be 10 My, during which each should have gained ≈75 novel positions (≈150 with the ML rates). Drosophila and the two plants meet the conditions for the proposed mechanisms of creation of new intron positions, namely, the occurrence of reverse transcription, as well as transposon and tandem duplication activities. The failure at identifying intron donors is puzzling. Either intron gain is an ancient process, no longer active (31), which would be at variance with phylogenetic studies, or novel intron positions originate by additional mechanisms other than the postulated mechanisms of intron gain, which is the alternative we favor.
Discussion
What Is an Intron?
Models of what introns are constrain our understanding of how intron positions arise in evolution. What has become popularly known as the “mystery of intron gain” (4, 28, 31) may be a corollary of the operational definition of introns that has pervaded evolutionary approaches to the origin of new intron positions. The term “intron” was introduced (34) with the electron micrographs in mind of the ssDNA loop configuration that obtains when a mature transcript is hybridized with its encoding genomic DNA (35, 36). Within the dominant “one gene-one enzyme” hypothesis and the “central dogma” paradigm, it was natural to map the loop's ends “top down” from that specific mRNA molecule to a precise location in the genomic DNA. The genome became invested with a property that primarily pertains to the transcriptome/proteome realm. How intronic information is incorporated at the DNA level became identified with how it is effected through splicing of a particular mRNA. This formal identification fostered a categorization of introns as precisely demarcated segments of the genome separated from the nonintronic DNA by fixed, unmovable ends. Recent advances in the understanding of pre-mRNA processing suggest that this received notion of introns, used to approach the question of the evolutionary origin of new intron positions, needs to be revised.
Intron Sliding in a World Lacking Alternative Splicing (AS).
AS, and the notion that there is not necessarily a one-to-one correspondence between intronic DNA and splicing products at the RNA/protein level, was implicit in the experimental observations that led to the discovery of introns (36). The fact that the AS products were of the cassette type (i.e., exons that are alternatively included/skipped from the mature transcript) did not question the emerging conceptualization of introns as definite DNA segments. Yet seeking to identify mechanisms for the rapid evolution of protein-coding sequences (34, 37–39), records were cited of so-called cryptic donor/acceptor splice sites (40, 41), and speculations were advanced that splicing-altering mutations could cause extensions/contractions of exons at intron junctions.
The discovery of increasing examples of lineage-specific introns (e.g., 42–44) launched the debate on the origins of new intron positions. The hypothesis of intron sliding (IS), also named “intron drift,” “intron migration,” or “intron slippage,” holds that new intron positions arise by the relocation of preexisting introns (45, 46). Relocation events would take place through the reassignment of an intron's donor and acceptor splice junctions to nearby positions, both offset in the same direction by the same distance (47, 48). But owing to its likely stepwise mechanism (see below), IS may account for alignment gaps frequently observed lying adjacent to exon–intron junctions (29, 37), as well as for discordant intron positions close to each other in homologous genes. With IS, the sequences of formative introns and intron donors overlap each other, which may explain why looking for the donors anywhere else within a genome has been unsuccessful (1, 15, 18, 20). A ML model of intron loss plus IS provides a highly significant better fit to the intron–exon structure of aldehyde dehydrogenase genes than other models of intron gain (47). IS would increase the diversity of intron positions without increasing the number of introns. Hence, IS would not be a valid explanation for introns in intron-bearing genes that were previously intronless, such as processed pseudogenes (although initial intron positions may slide later).
Interest in IS models diminished on the belief that IS could not be a frequent phenomenon (4, 6, 47, 48). Under the notion of introns as fixed genomic segments, IS is perceived as uncommon because it calls for the simultaneous occurrence of two mutations. Other paths, by a series of two or more short-range extension/contraction events of intron–exon boundaries, were deemed likely to be deleterious at the protein level (47, 48). Such events would be feasible when the aberrant mRNAs contained premature stop codons that could be targeted by nonsense mediated decay (NMD) (49). Provided the locus is haplosufficient, degradation of the transcript would turn out the mutant allele completely recessive, which would enhance its persistence in the population and, thus, the likelihood of a compensatory mutation. But the requirement of haplosufficiency requires a second, physically distinct genomic copy of the gene for expression of the correct function (see below) (49).
IS is thought to exhibit low potential for intron relocation because standing formulations neglect that AS can facilitate the process. Moreover, phylogenetic approaches, which provide the evidence for the incidence of IS, have overlooked AS as a fundamental consideration in deciding the positional homology of introns. One reason for this neglect is that homologous intron positions have largely been established by extrapolation from unannotated or poorly annotated genes with respect to AS (1, 6, 10, 11, 21, 22, 47, 48, 50, 51). At the time that the hypothesis of IS was launched, AS was still thought to be a minor processing pathway (52).
An AS-Driven Model of IS.
New splice sites can arise by point mutation because donor and acceptor splice sites are short and imprecise (53). Any gene region likely includes many more donor and acceptor splice sites than those implied by the exon junctions of mature transcript molecules (54–56). There is not a one-to-one correspondence between donor and acceptor splice sites. One donor may pair with more than one of several acceptors and the other way around, giving rise to a profile of AS products or transcript isoforms, which can differ in the exons they contain, but also in the location of exon junctions (56, 57). Alternative mRNA isoforms evince that fixed intron locations are not suitable for determining positional homology at the genome (DNA) level.
AS has been reported in animals, fungi, plants, and various protists and was probably present in the intron-rich LECA (58). Many AS events, especially those involving weak splice sites, are idiosyncratic across species (38, 59–62). Most AS events can be classified into four basic patterns, including exon skipping, alternative 3′ and 5′ splice site selection, and intron retention. The patterns required for IS, namely, alternative 3′ and 5′ splice site selection, are the most or the second most prevalent type of AS event, accounting for at least one third of all AS events in invertebrates, vertebrates, and Arabidopsis (55–57). A typical human gene may yield 2.53 splicing isoforms translatable to protein (63). Such a diversity of mRNAs and proteins may, in part, be redundant and carry out new functions and may not be “visible” to natural selection (38, 39, 63, 64). However, a substantial fraction will involve changes unlikely to be tolerated (63, 65–67).
Donor-acceptor splice pairs can be strong or weak variants according to frequency of use. Strong splice pairs yield major isoforms, present in >50% of the transcripts of an allele, whereas weak splice pairs yield minor isoforms, which are a small fraction of the normally spliced, mature mRNA (38, 39). Differential production/processing of transcript isoforms may be at the core of organismal robustness to the diversity of AS products (38, 39, 63). It has been proposed that newly arising, potentially deleterious AS products convey only weak splice signals and, hence, are minor isoforms (38, 60). Because of their low abundance, minor isoforms would not often have a major impact on physiology; thereby, they would evolve relatively unconstrained, provided the major fraction of transcripts upholds the gene's function (38). So-called “tunneling” of aberrant AS forms enhances their retention in a population, which increases the likelihood of compensatory mutations to a restored or novel function if they happen to be disclosed to selection (38, 64, 68). Unlike the standing model of IS via NMD (49), in IS via AS, a second genomic copy of the gene would not be required to maintain the original function because AS would furnish internal paralogs of the gene. This hypothesis is supported by a study showing that (i) minor-form AS relaxes selection pressure against premature termination codons (PTCs) that are likely targets of NMD (to the same degree as having two copies of the gene), and (ii) the combined effects of AS and diploidy yield a >9-fold increase in tolerance for PTCs (69). By enhancing the rate of compensatory mutation, AS expands the potential paths to IS over those under NMD. The threshold of approximately four codons above which IS is considered to be unviable (47, 70) is most likely an underestimate.
The relative use of a given donor–acceptor splice pair depends on the interactions between trans-acting factors and the splicing code. The splicing code is made up of an extensive and complex array of cis-acting elements featuring two layers of information. The first layer comprises the splice site sequences that define potential intron–exon junctions on the target pre-mRNA. The second layer consists of splicing enhancers and silencers distributed all over the introns and exons of the target pre-mRNA. This second informational layer determines which and with what frequency splice sites of the first layer will become targets of the trans-acting factors (71).
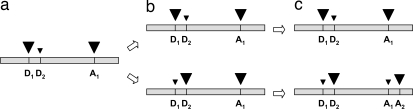
The interactions between trans- and cis-acting splicing elements are highly context-dependent. Every site of a pre-mRNA molecule can potentially influence the production of a transcript isoform (55, 56), which implies that there is an extensive genomic target for mutations that can affect AS profiles. This conclusion is supported by the large and growing number of inherited human diseases found to be caused by AS-altering mutations (56, 71, 72). Likely, those mutations represent only extreme cases of an abundant class of genetic polymorphisms that generate quantitative variation in the ratios of isoforms among individuals (73–75). The mutations responsible for this variation may spread and become fixed or lost under the forces of population genetics, just like any genetic variant. Minor splice isoforms would evolve into major isoforms, replacing preexisting predominant gene products, which would then become minor isoforms and be lost over time. The discovery of ancient human pseudogenes, originated by reverse transcription of AS products not presently expressed by the parent gene (76), suggests that the strength of a splice site is dynamic during evolution. This idea is further supported by observations that AS profiles tend to diverge rapidly after gene duplication (77) or speciation events (61, 78). If a preexisting major isoform is superseded by another isoform bearing expansions/contractions of exon limits or slid exon junctions, the replacement would cause a change in the distribution of intron positions of the gene (see Fig. 1).
Fig. 1.
Sliding of an intron position to a nearby position via AS. (a) A sequence with two donors (D1 and D2) and one acceptor (A1) splice sites, with relative strengths proportional to the size of the arrowheads, undergoes a branching event (e.g., duplication or speciation). (b) In the upper branch, the AS profile remains unchanged; in the lower branch, D2 evolves into a strong splice site, such that D2–A1 becomes the major isoform. Note that, at this stage, alignment of the cDNAs corresponding to the major isoforms of the two branches (D1–A1 and D2–A1) contain a gap adjacent to the position of the intron. (c) In the lower branch, a mutation creates a new acceptor site (A2) at the same distance from A1 as D2 is from D1, which evolves into a strong splice site. Alignment of the major isoforms of the two branches (D1–A1 and D2–A2, for the upper and lower branches, respectively) will show two intron positions. At the time the process is completed, the original intron sequence may have drifted away completely, such that the IS event will look the same as an intron gain that happened long ago. In the figure, it is assumed that new major isoforms are functional at the time they become abundant (either because they were always so or because they underwent function-restoring mutations along the process) and that their splicing profiles spread in the population.
De Novo Origin of Intron Positions: Intron Sliding Versus Intron Gain.
The arguments given suggest that AS could provide a major avenue for the occurrence of IS, one that may have been seriously underestimated as a source of intron positional diversity. A reason that IS has been disfavored over gain of new introns in accounts of intron positional diversity is the assumption that IS must involve large deleterious effects (47, 48). However, increasing understanding of the complexity of splicing codes suggests otherwise. Optimal splicing codes must require time to evolve (79). It seems unlikely that a de novo intron-formation event, regardless of whether it derives from another intron, a transposon, or an exon donor, can lead to an immediately efficiently spliced product. If splicing of a formative intron is inefficient, then the unspliced, intron-retaining, and, hence, unlikely to be functional transcript will set off as a major isoform, hence exposed to negative selection. Thus, the creation of intron positions from new introns may have larger fitness costs than IS of preexisting introns over short distances because the latter would take place through the readjustment of preexisting splicing codes via changes in minor isoforms.
IS events are not expected to occur instantaneously. After the emergence of a novel donor/acceptor splice site, millions of years might be necessary until the fixation of the mutation(s), as well as the occurrence of changes in splicing code allowing for the replacement of preexisting major isoforms. At the process completion, little may be left of the original intron sequence. IS events may be more easily detected by retracing phylogenetically the AS events that led to the intron relocation than by interspecific comparison of intron sequences. Comparing closely related genomes, such as those of 12 Drosophila species (80), may help identify such footprints. The persistence of alternative isoforms for long periods of evolutionary time would provide a natural path to parallel gain of intron positions if, after a duplication/speciation event, the same isoform replacement takes place in more than one descendant lineage.
IS may help explain the observed preference of introns to be located at mAG|Gt contexts (where “m” can be A or C, uppercase letters indicate a stronger preference, and “|” denotes the placement of the intron), termed “protosplice sites.” In actin genes, elimination of normal splice sites in a gene triggers AS of the mutant transcripts via use of cryptic splice sites, which happen to coincide in location with functional splice sites in other orthologs of that gene (51). Newly activated donor (GT) and acceptor (AG) splice sites exhibit a bias to be flanked, respectively, by AG and GT dinucleotides at the exonic side. However, IS may be instrumental to understand reported correlations between intron positions and structural/functional features of the encoded proteins if less harmful AS events have a greater associated likelihood of compensatory mutation.
Gain of an intron position by IS implies loss of the previous position of that intron. IS should generate a strong and positive correlation between the rates of intron gain and loss. Such a correlation has been reported in a recent ML reconstruction of intron evolution in 19 model eukaryote species (11). The study partitioned intron evolution into three modes: balanced mode, characterized by proportional gain and loss rates, and elevated loss or gain modes. Rates of gain and loss were found to be positively correlated only for the balanced mode, as expected of IS, which cannot either create or remove intron sequences. These results suggest that the diversity of intron positions may be dominated by two main effects: a background effect due to the continuous relocation of introns by IS, superimposed by episodes of active addition/removal of new intron sequences by intron gain/loss mechanisms. In this respect, it is important to pinpoint that AS may contribute to the evolution of the diversity of intron positions not only as a catalyst of IS, but also as a potentially powerful mechanism of intron gain. Indeed, a large-scale analysis of the role of AS in exon creation and loss during vertebrate evolution (81, 82) found that new alternative exons set off as minor splice forms in most cases. These minor splice forms originate via mutations that introduce new splice sites inside preexisting intron sequences. Exonization of an intron's partial sequence effectively splits the original intron sequence in two, thus increasing the initial intron number by one. Because this process creates new introns from separate parts of preexisting introns, it cannot be identified by intragenomic similarity searches. AS-driven exonization of intron partial sequences complements our current knowledge of molecular mechanisms of intron gain. The efficiency of this mechanism should increase with intron length.
If IS is an important determinant of the diversity of intron positions, then it might be expected that the rate of intron position evolution would be positively correlated with the rate of sequence evolution. The two types of evolution depend on the same set of mutations (i.e., point mutations that by changing the sequence would influence the rates of splicing code evolution). The issue has not been investigated in depth, but there are some indications that such a correlation may occur. The sea anemone Nematostella vectensis, the marine annelid Platynereis dummerilii, and humans evolve more slowly than Caenorhabditis and Drosophila at the sequence level. Apparently, they also share larger numbers of ancestral intron positions (83, 84), although anemones and annelids are more distantly related to humans than nematodes and flies. Sverdlov et al. (28) reported a shortage of conserved intron positions in ancient eukaryotic paralogs compared with the higher rate of conservation of intron positions in more recent paralogs. This finding would be consistent with an effect of IS, taking into account that widespread AS appears to be an ancient feature (58), as well as the tendency of AS patterns to diverge after duplication (69, 77).
Acknowledgments.
We thank Dr. David Penny and Dr. Christopher Lee for helpful suggestions for improving the manuscript, and Dr. Miklós Csűrös for making data available. This work was supported by Centro de Investigación Biomédica en Red de Enfermedades Raras and Ramón y Cajal from the Spanish Ministerios de Sanidad y Consumo and Educación y Ciencia (R.T. and F.R.-T.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Fedorov A, Merican AF, Gilbert W. Large-scale comparison of intron positions among animal, plant, and fungal genes. Proc Natl Acad Sci USA. 2002;99:16128–16133. doi: 10.1073/pnas.242624899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogozin IB, Sverdlov AV, Babenko VN, Koonin EV. Analysis of evolution of exon–intron structure of eukaryotic genes. Brief Bioinform. 2005;6:118–134. doi: 10.1093/bib/6.2.118. [DOI] [PubMed] [Google Scholar]
- 3.Jeffares DC, Mourier T, Penny D. The biology of intron gain and loss. Trends Genet. 2006;22:16–22. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Roy SW, Gilbert W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat Rev Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 5.Rodríguez-Trelles F, Tarrío R, Ayala FJ. Origins and evolution of spliceosomal introns. Annu Rev Genet. 2006;40:47–76. doi: 10.1146/annurev.genet.40.110405.090625. [DOI] [PubMed] [Google Scholar]
- 6.Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol. 2003;13:1512–1517. doi: 10.1016/s0960-9822(03)00558-x. [DOI] [PubMed] [Google Scholar]
- 7.Roy SW, Gilbert W. Rates of intron loss and gain: Implications for early eukaryotic evolution. Proc Natl Acad Sci USA. 2005;102:5773–5778. doi: 10.1073/pnas.0500383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Csűrös M. Likely scenarios of intron evolution. Lect Notes Comput Sci. 2005;3678:47–60. [Google Scholar]
- 9.Nguyen HD, Yoshihama M, Kenmochi N. New maximum likelihood estimators for eukaryotic intron evolution. PLos Comp Biol. 2005;1:7e79. doi: 10.1371/journal.pcbi.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Csűrös M, Holey JA, Rogozin IB. In search of lost introns. Bioinformatics. 2007;23:i87–i96. doi: 10.1093/bioinformatics/btm190. [DOI] [PubMed] [Google Scholar]
- 11.Carmel L, Wolf YI, Rogozin IB, Koonin EV. Three distinct modes of intron dynamics in the evolution of eukaryotes. Genome Res. 2007;17:1034–1044. doi: 10.1101/gr.6438607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu WG, Schisler N, Stoltzfus A. The evolutionary gain of spliceosomal introns: sequence and phase preferences. Mol Biol Evol. 2004;21:1252–1263. doi: 10.1093/molbev/msh120. [DOI] [PubMed] [Google Scholar]
- 13.Rodríguez-Trelles F, Tarrío R, Ayala FJ. Models of spliceosomal intron proliferation in the face of widespread ectopic expression. Gene. 2006;366:201–208. doi: 10.1016/j.gene.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Roy SW, Fedorov A, Gilbert W. Large-scale comparison of intron positions in mammalian genes shows intron loss but not gain. Proc Natl Acad Sci USA. 2003;100:7158–7162. doi: 10.1073/pnas.1232297100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coghlan A, Wolfe KH. Origins of recently gained introns in Caenorhabditis. Proc Natl Acad Sci USA. 2004;101:11362–11367. doi: 10.1073/pnas.0308192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen CB, Friedman B, Birren B, Burge CB, Galagan JE. Patterns of intron gain and loss in fungi. PLoS Biol. 2004;2:e422. doi: 10.1371/journal.pbio.0020422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coulombe-Huntington J, Majewski J. Characterization of intron loss events in mammals. Genome Res. 2007;17:23–32. doi: 10.1101/gr.5703406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coulombe-Huntington J, Majewski J. Intron loss and gain in Drosophila. Mol Biol Evol. 2007;24:2842–2850. doi: 10.1093/molbev/msm235. [DOI] [PubMed] [Google Scholar]
- 19.Knowles DG, McLysaght A. High rate of ancient intron gain and loss in simultaneously duplicated Arabidopsis genes. Mol Biol Evol. 2006;23:1548–1557. doi: 10.1093/molbev/msl017. [DOI] [PubMed] [Google Scholar]
- 20.Lin H, Zhu W, Silva J, Gu X, Buell CR. Intron gain and loss in segmentally duplicated genes in rice. Genome Biol. 2006;7:R41. doi: 10.1186/gb-2006-7-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy SW, Penny D. Large-scale intron conservation and order-of-magnitude variation in intron loss/gain rates in apicomplexan evolution. Genome Res. 2006;16:1270–1275. doi: 10.1101/gr.5410606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy SW, Penny D. Smoke without fire: Most reported cases of intron gain in nematodes instead reflect intron losses. Mol Biol Evol. 2006;23:2259–2262. doi: 10.1093/molbev/msl098. [DOI] [PubMed] [Google Scholar]
- 23.Roy SW, Penny D. Patterns of intron loss and gain in plants: Intron loss-dominated evolution and genome-wide comparison of O. sativa and A. thaliana. Mol Biol Evol. 2007;24:171–181. doi: 10.1093/molbev/msl159. [DOI] [PubMed] [Google Scholar]
- 24.Stajich JE, Dietrich FS. Evidence of mRNA-mediated intron loss in the human-pathogenic fungus Cryptococcus neoformans. Eukaryot Cell. 2006;5:789–793. doi: 10.1128/EC.5.5.789-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuo D, Madden R, Elela SA, Chabot B. Modern origin of numerous alternatively spliced human introns from tandem arrays. Proc Natl Acad Sci USA. 2007;104:882–886. doi: 10.1073/pnas.0604777104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steel M, Penny D. Two further links between MP and ML under the Poisson model. Appl Math Lett. 2004;17:785–790. [Google Scholar]
- 27.Sverdlov AV, Rogozin IB, Babenko VN, Koonin EV. Conservation versus parallel gains in intron evolution. Nucleic Acids Res. 2005;33:1741–1748. doi: 10.1093/nar/gki316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy SW, Penny D. A very high fraction of unique intron positions in the intron-rich diatom Thalassiosira pseudonana indicates widespread intron gain. Mol Biol Evol. 2007;24:1447–1457. doi: 10.1093/molbev/msm048. [DOI] [PubMed] [Google Scholar]
- 29.Tarrío R, Rodríguez-Trelles F, Ayala FJ. A new Drosophila spliceosomal intron position is common in plants. Proc Natl Acad Sci USA. 2003;100:6580–6583. doi: 10.1073/pnas.0731952100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy SW, Hartl DL. Very little intron loss/gain in Plasmodium: Intron loss/gain mutation rates and intron number. Genome Res. 2006;16:750–760. doi: 10.1101/gr.4845406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fedorov A, Roy S, Fedorova L, Gilbert W. Mystery of intron gain. Genome Res. 2003;13:2236–2241. doi: 10.1101/gr.1029803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blumenstiel JP, Hartl DL, Lozovsky ER. Patterns of insertion and deletion in contrasting chromatin domains. Mol Biol Evol. 2002;19:2211–2225. doi: 10.1093/oxfordjournals.molbev.a004045. [DOI] [PubMed] [Google Scholar]
- 33.Wright SI, Lauga B, Charlesworth D. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol Biol Evol. 2002;19:1407–1420. doi: 10.1093/oxfordjournals.molbev.a004204. [DOI] [PubMed] [Google Scholar]
- 34.Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 35.Berget SM, Moore C, Sharp PA. Spliced segments at 5′ terminus of adenovirus 2 late messenger-RNA. Proc Natl Acad Sci USA. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chow LT, Gelinas RE, Broker TR, Roberts RJ. Amazing sequence arrangement at 5′ ends of adenovirus-2 messenger-RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 37.Craik CS, Rutter WJ, Fletterick R. Splice junctions: Association with variation in protein-structure. Science. 1983;220:1125–1129. doi: 10.1126/science.6344214. [DOI] [PubMed] [Google Scholar]
- 38.Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 39.Xing Y, Lee C. Alternative splicing and RNA selection pressure: Evolutionary consequences for eukaryotic genomes. Nat Rev Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- 40.Early P, et al. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980;20:313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- 41.DeNoto FM, Moore DD, Goodman HM. Human growth hormone DNA sequence and mRNA structure: Possible alternative splicing. Nucleic Acids Res. 1981;9:3719–3730. doi: 10.1093/nar/9.15.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Logsdon JM, et al. 7 newly discovered intron positions in the triose-phosphate isomerase gene: Evidence for the introns-late theory. Proc Natl Acad Sci USA. 1995;92:8507–8511. doi: 10.1073/pnas.92.18.8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tarrío R, Rodríguez-Trelles F, Ayala FJ. New Drosophila introns originate by duplication. Proc Natl Acad Sci USA. 1998;95:1658–1662. doi: 10.1073/pnas.95.4.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venkatesh B, Ning Y, Brenner S. Late changes in spliceosomal introns define clades in vertebrate evolution. Proc Natl Acad Sci USA. 1999;96:10267–10271. doi: 10.1073/pnas.96.18.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers J. Introns between protein domains: Selective insertion or frameshifting? Trends Genet. 1986;12:223. [Google Scholar]
- 46.Gilbert W, deSouza SJ, Long MY. Origin of genes. Proc Natl Acad Sci USA. 1997;94:7698–7703. doi: 10.1073/pnas.94.15.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rzhetsky A, Ayala FJ, Hsu LC, Chang C, Yoshida A. Exon/intron structure of aldehyde dehydrogenase genes supports the “introns-late” theory. Proc Natl Acad Sci USA. 1997;94:6820–6825. doi: 10.1073/pnas.94.13.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoltzfus A, Logsdon JM, Palmer JD, Doolittle WF. Intron “sliding” and the diversity of intron positions. Proc Natl Acad Sci USA. 1997;94:10739–10744. doi: 10.1073/pnas.94.20.10739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lynch M. Intron evolution as a population-genetic process. Proc Natl Acad Sci USA. 2002;99:6118–6123. doi: 10.1073/pnas.092595699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogozin IB, Lyons-Weiler J, Koonin EV. Intron sliding in conserved gene families. Trends Genet. 2000;16:430–432. doi: 10.1016/s0168-9525(00)02096-5. [DOI] [PubMed] [Google Scholar]
- 51.Sadusky T, Newman AJ, Dibb NJ. Exon junction sequences as cryptic splice sites: Implications for intron origin. Curr Biol. 2004;14:505–509. doi: 10.1016/j.cub.2004.02.063. [DOI] [PubMed] [Google Scholar]
- 52.Sharp PA. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 53.Irimia M, Penny D, Roy SW. Coevolution of genomic intron number and splice sites. Trends Genet. 2007;23:321–325. doi: 10.1016/j.tig.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 54.Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 55.Blencowe BJ. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 56.Buratti E, Baralle M, Baralle FE. Defective splicing, disease and therapy: Searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zavolan M, Nimwegen E. The types and prevalence of alternative splice forms. Curr Opin Struct Biol. 2006;16:362–367. doi: 10.1016/j.sbi.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 58.Irimia M, Rukov JL, Penny D, Roy SW. Functional and evolutionary analysis of alternatively spliced genes is consistent with an early eukaryotic origin of alternative splicing. BMC Evol Biol. 2007;7:188. doi: 10.1186/1471-2148-7-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nurtdinov RN, Artamonova II, Mironov AA, Gelfand MS. Low conservation of alternative splicing patterns in the human and mouse genomes. Hum Mol Genet. 2003;12:1313–1320. doi: 10.1093/hmg/ddg137. [DOI] [PubMed] [Google Scholar]
- 60.Zhang XHF, Chasin LA. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc Natl Acad Sci USA. 2006;103:13427–13432. doi: 10.1073/pnas.0603042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malko DB, Makeev VJ, Mironov AA, Gelfand MS. Evolution of exon–intron structure and alternative splicing in fruit flies and malarial mosquito genomes. Genome Res. 2006;16:505–509. doi: 10.1101/gr.4236606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang BB, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tress ML, et al. The implications of alternative splicing in the ENCODE protein complement. Proc Natl Acad Sci USA. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Masel J. Cryptic genetic variation is enriched for potential adaptations. Genetics. 2005;172:1985–1991. doi: 10.1534/genetics.105.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 66.Stetefeld J, Ruegg MA. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem Sci. 2005;30:515–521. doi: 10.1016/j.tibs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 67.Yura K, et al. Alternative splicing in human transcriptome: Functional and structural influence on proteins. Gene. 2006;380:63–71. doi: 10.1016/j.gene.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 68.Kan Z, States D, Gish W. Selecting for functional alternative splice sites. Genome Res. 2002;12:1837–1845. doi: 10.1101/gr.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xing Y, Lee CJ. Negative selection pressure against premature protein truncation is reduced by alternative splicing and diploidy. Trends Genet. 2004;20:472–475. doi: 10.1016/j.tig.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 70.Cerff R. The chimeric nature of nuclear genomes and the antiquity of introns as demonstrated by the GAPDH gene system. In: Gō M, Schimmel P, editors. Tracing Biological Evolution in Protein and Gene Structures. New York: Elsevier; 1995. pp. 205–227. [Google Scholar]
- 71.Wang G, Cooper TA. Splicing and disease: Disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 72.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 73.Marden JH. Quantitative and evolutionary biology of alternative splicing: How changing the mix of alternative transcripts affects phenotypic plasticity and reaction norms. Heredity. 2008;100:111–120. doi: 10.1038/sj.hdy.6800904. [DOI] [PubMed] [Google Scholar]
- 74.Hull J, et al. Identification of common genetic variation that modulates alternative splicing. PLoS Genet. 2007;3:e99. doi: 10.1371/journal.pgen.0030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kwan T, et al. Heritability of alternative splicing in the human genome. Genome Res. 2007;17:1210–1218. doi: 10.1101/gr.6281007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shemesh R, Novik A, Edelheit S, Sorek R. Genomic fossils as a snapshot of the human transcriptome. Proc Natl Acad Sci USA. 2006;103:1364–1369. doi: 10.1073/pnas.0509330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Su ZX, Wa JM, Yu J, Huang XQ, Gu X. Evolution of alternative splicing after gene duplication. Genome Res. 2006;16:182–189. doi: 10.1101/gr.4197006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cusack BP, Wolfe KH. Changes in alternative splicing of human and mouse genes are accompanied by faster evolution of constitutive exons. Mol Biol Evol. 2005;22:2198–2208. doi: 10.1093/molbev/msi218. [DOI] [PubMed] [Google Scholar]
- 79.Krull M, Brosius J, Schmitz J. Alu-SINE exonization: En route to protein-coding function. Mol Biol Evol. 2005;22:1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- 80.Drosophila Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 81.Alekseyenko AV, Kim N, Lee CJ. Global analysis of exon creation versus loss and the role of alternative splicing in 17 vertebrate genomes. RNA. 2007;13:661–670. doi: 10.1261/rna.325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sorek R. The birth of new exons: Mechanisms and evolutionary consequences. RNA. 2007;13:1603–1608. doi: 10.1261/rna.682507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raible F, et al. Vertebrate-type intron-rich genes in the marine annelid Platynereis dumerilii. Science. 2005;310:1325–1326. doi: 10.1126/science.1119089. [DOI] [PubMed] [Google Scholar]
- 84.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]