Abstract
Osteopontin (Opn) contributes to diverse biological processes that include immune responses, vascularization, and bone formation. Until recently, studies describing the activities of Opn have focused on the cytokine-like properties of the secreted protein. Here, we show that alternative translation of a single Opn mRNA species generates a secreted and intracellular isoform. Utilization of a 5′ canonical translation start site generates a protein that includes an N-terminal signal sequence allowing targeting to secretory vesicles and cytokine secretion, whereas usage of a downstream start site generates a shortened protein that lacks the N-terminal signal sequence and localizes mainly to cytoplasm. The coordinated action of these Opn gene products regulates the functional phenotype of subsets of dendritic cells.
Keywords: signal sequence, podosome
The osteopontin (Opn) glycoprotein regulates diverse biological processes, including immune responses, vascularization, and cell regeneration, through interactions with mononuclear and endothelial cells. Virtually all of these activities have been attributed to an interaction between secreted Opn (Opn-s) and its receptors on target cells (1, 2). For example, an interaction between Opn-s with αvβ3 and CD44 receptors on macrophages that enhances IL-12 and inhibits IL-10 responses (3) promotes Th1 development (4) and cellular resistance to apoptosis (5–7).
However, a nonsecreted species of Opn has been implicated in a growing number of cellular processes, including migration, fusion, and motility (8–11). Recent studies have revealed that an intracellular form of Opn (Opn-i) in plasmacytoid dendritic cells (pDC) interacts with IRF7 to induce IFNα expression (12), whereas expression of Opn-i in conventional DC (cDC) promotes differentiation of IL-17-producing T helper cells (Th17 cells) (13).
These findings prompted us to define the genetic and molecular origin of Opn-i and Opn-s in DC. Possibly, a weakly hydrophobic signal sequence may allow a single Opn protein to migrate into both secretory vesicles and cytoplasm, secondary to ER leakage into cytosol, as demonstrated for prion proteins (14). Alternatively, Opn may specify two distinct protein isoforms with distinct cellular locations and biologic function.
Here, we demonstrate that Opn-i and Opn-s represent alternative translational products of a single full-length Opn mRNA. Translation of Opn-s is initiated from the 5′ canonical AUG start site, whereas translation of the Opn-i isoform is initiated from a downstream non-AUG codon. Downstream translation of Opn-i is accompanied by deletion of the N-terminal 16-aa signal sequence, allowing the shortened protein product to localize in cytoplasm but not secretory vesicles. The shortened Opn translation isoform activates ifna4 gene expression and podosome formation in pDC, thus contributing to the characteristic biological activities of this DC subset. These findings indicate that factors that alter the translational balance of Opn in favor of either Opn-i or Opn-s may contribute to the phenotype of activated DC.
Results
DCs Express Two Opn Isoforms.
Full-length Opn (Opn-FL) contains a signal peptide that targets nascent protein to secretory vesicles, however, the mechanism allowing intracellular retention of Opn is unknown. To determine whether both Opn isoforms were present in DC, we performed Western blot analysis of whole cell lysates. We detected two bands (75 kDa/70 kDa) in both pDC (Fig. 1A) and cDC (data not shown) using the mAb (O-17) specific for Opn sequence immediately downstream of 16-aa signal sequence (Fig. 1B). This observation opened the possibility that the difference in size (≈5 kDa) reflected deletion of the 16-aa signal sequence. We then analyzed proteins translated in vitro from full-length Opn mRNA and an Opn mRNA truncation mutant (Δ48Koz) that lacked the first 48 nucleotides encoding the Opn signal sequence (amino acid 1–16). An artificial AUG/Kozak sequence was introduced to initiate translation (Fig. 1 B and C). In vitro translation (IVT) of full-length Opn mRNA gave rise to a doublet of similar size to that identified by Western blot analysis of pDC (Fig. 1A), whereas the Δ48Koz construct generated a single band that migrated with the lower band of the doublet (Fig. 1C). These results suggest that the doublet represented Opn-FL protein (upper band) and a shortened protein (lower band) that may correspond to intracellular Opn, provisionally designated Opn-i.
Fig. 1.
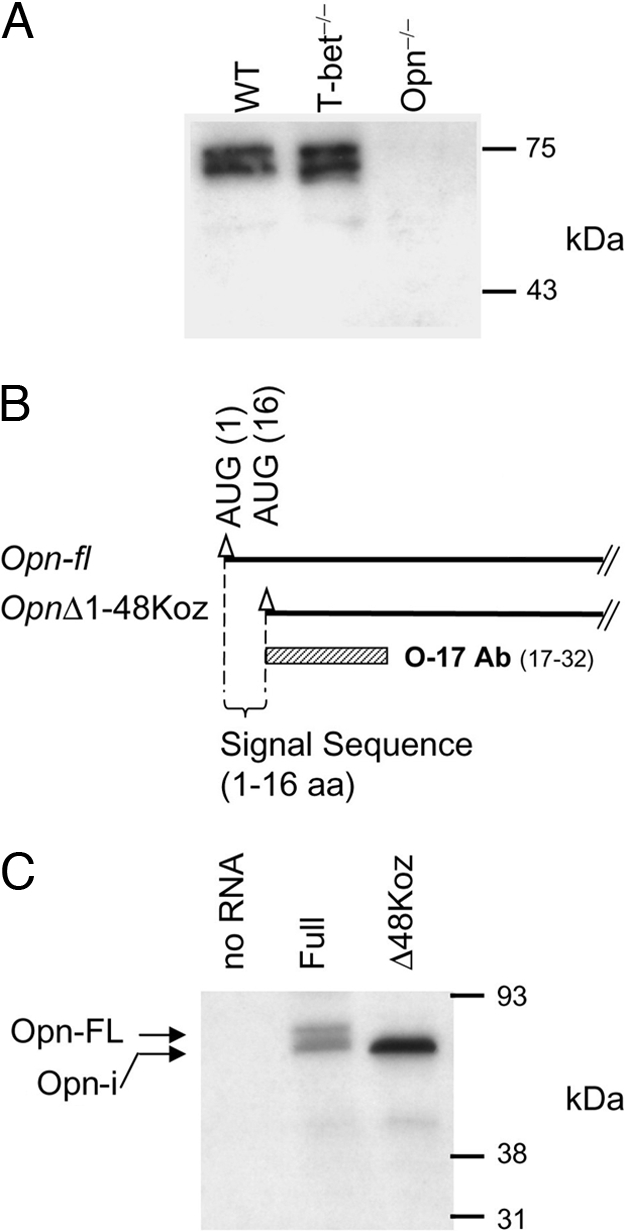
Two peptides of different size translated from a single Opn mRNA species. (A) Opn detection from pDC lysate by immunoblotting with O-17 Ab. (B) Transcripts from “Full” (Opn-fl) and “OpnΔ48Koz.” Triangles (▵) denote AUG codons, including the second endogenous AUG at position 84 aa. O-17 Ab recognizes amino acids spanning position 17–32 aa. (C) IVT of two Opn constructs shown in B. Peptide products were phosphatase-treated. Negative control of IVT was performed without RNA template (“no RNA”).
The Opn-i Isoform Is Not Generated from Alternative Opn Transcription Initiation or Splicing.
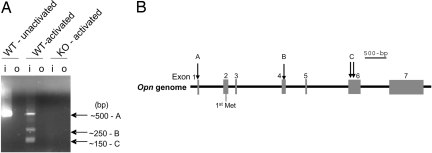
We identified transcription initiation sites using RLM-RACE (RNA ligase-mediated RACE) analysis (15). Although a single band was amplified in total cDNA from unactivated bone marrow (BM)-derived DC (BM-DC) (Fig. 2A, band A), multiple bands appeared after CpG activation of DC (Fig. 2A, bands A–C). Cloning and sequencing revealed that band A was transcribed from the adenine residue at position −90 in exon 1 (90 nucleotides upstream of the ATG translation start codon) [Fig. 2B and supporting information (SI) Fig. S1 and SI Text]. Alternative transcription start sites in exon 4 (band B) and exon 6 (band C) were too far downstream to account for the shorter Opn peptide shown in Fig. 1.
Fig. 2.
RLM-RACE of Opn mRNA and structure of the Opn gene. (A) Opn WT and KO DC were activated with or without CpG-1668 (0.2 μg/ml) × 3 h. Agarose gel shows nested PCR (“o,” outer PCR; “i,” inner). Bands corresponding to products from Opn transcripts generated by RLM-RACE are labeled A–C. The position of the Opn specific inner primer is shown in Fig. S1. (B) Schematic of transcription start sites A–C on Opn.
We next amplified by PCR the region between exons 1 and 4 of Opn cDNA to ask whether alternative splicing might produce the shorter Opn-i product using DC and T cells. Although no shorter bands were detected, some samples showed a larger band than expected, i.e., alternative splicing might provide an additional mRNA sequence rather than reducing the size of mRNA (Fig. S2A). Although sequencing of the additional band revealed that alternative splice added a 52-nt extension at the 3′-end of exon 1 (Fig. S1), this did not account for Opn-i, because Opn mRNA containing the 52-nt extension failed to generate an additional Opn peptide product compared to canonical Opn mRNA (Fig. S2 B and C).
Identification of a Nucleotide Sequence Required for Alternative Translation Initiation of Opn-i.
Although the GUG codon is preferred for non-AUG translation initiation (16), GUG codons immediately downstream of the signal peptide-encoding sequence did not initiate translation: GUG→ GUC mutations at positions +55 and +61 (Fig. S3A) did not impair generation of Opn-FL and Opn-i peptides (Fig. S3B)§.
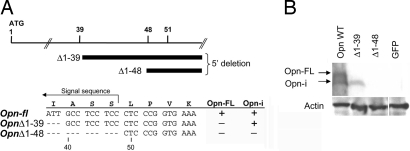
We next tested series of Opn deletion mutants with no AUG codon to define sequences essential for production of the Opn-i isoform. Expression of full-length Opn cDNA construct was compared to Opn nucleotide deletion constructs in transfected 293FT cells by immunoblotting (Fig. 3A). Opn plasmid Δ1–39, which lacks the first 39 nucleotides (Fig. 3A), generated the lower but not the upper Opn band (Fig. 3B), suggesting that the Opn-i protein did not require the canonical AUG codon but was initiated by a non-AUG codon downstream of position +39. Additional deletion to position +48 (Δ1–48) completely eliminated alternative translation of the shortened protein (Fig. 3B), suggesting that the 9-nt sequence between positions +40 and +48 (GCC TCC TCC) encoding Asp-Ser-Ser was essential for translation of the Opn-i isoform.
Fig. 3.
Codons encoding aa sequence Ala-Ser-Ser participate in Opn alternative translation. (A) Schematic of Opn deletion mutants to assess alternative translation. (B) 293FT cells were transfected with Opn expression constructs: Opn-fl, OpnΔ1–39, OpnΔ1–48, GFP (negative control); Opn-FL and Opn-i were detected by immunoblotting with O-17 Ab in 293FT cell lysates.
The Opn-i Isoform Localizes Outside Secretory Vesicles.
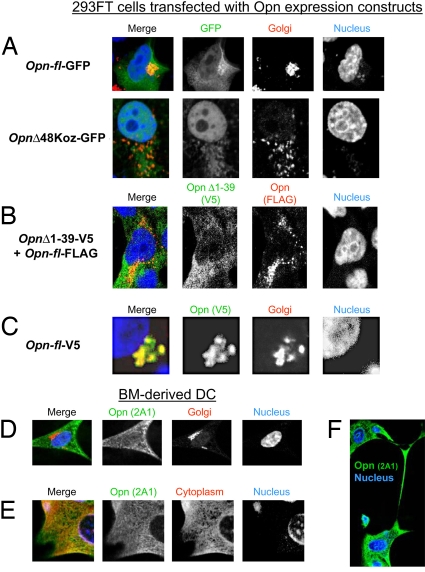
We examined subcellular localization of Opn in 293FT cells transfected with GFP-fusion constructs of full-length Opn (Opn-fl) or a Δ48Koz Opn deletion mutant. Opn-fl-GFP protein colocalized with Golgi and cytoplasm, whereas OpnΔ48Koz-GFP appeared to reside mainly in cytoplasm (Fig. 4A). To further examine distribution of Opn isoforms expressed without an added Kozak sequence, we cotransfected 293FT cells with Opn-fl and OpnΔ1–39 (see Fig. 3A) tagged with FLAG and V5 epitopes, respectively. Proteins generated from deletion mutant OpnΔ1–39 appeared to localize in cytoplasm, whereas proteins encoded by the Opn-fl construct localized to perinuclear secretory vesicles (Fig. 4B). The latter observation was confirmed using V5-tagged Opn-fl constructs that displayed colocalization of Opn-FL with Golgi marker GOLPH4 (Fig. 4C). Occasional colocalization of Opn-FL FLAG-tagged products with OpnΔ39 V5-tagged products was consistent with alternative translation of Opn-fl mRNA (Fig. 4B).
Fig. 4.
Subcellular localization of Opn-FL and Opn-i. (A) Two Opn-GFP fusion constructs were transfected into 293FT cells and counterstained with Golgi marker GM130. “Opn-fl-GFP” includes WT Opn with the canonical Opn ATG translation start codon. “OpnΔ48Koz-GFP” lacks a signal sequence and an artificial ATG codon with the Kozak sequence. (B) Cotransfection of “Opn-fl-FLAG” and “OpnΔ1–39-V5” constructs into 293FT. (C) Using a system similar to (B), Opn-fl-V5 construct was transfected into 293FT and detected with V5 Ab and counterstained with GOLPH4 Ab. BM-DC stained with Opn 2A1 Ab and counterstained with GOLPH-4 Ab (D) or GAPDH Ab (E). (F) Dendrites of BM-DC stained with Opn 2A1 Ab. Nucleus staining was carried out with DRAQ5 (A) or TOPRO3 (B–E).
We analyzed subcellular localization of endogenous Opn in BM-derived DC using Opn 2A1 Ab that detects both Opn-FL and Opn-i isoforms. Although there was almost no detectable colocalization of Opn with the GOLPH4 Golgi marker (Fig. 4D), Opn localized extensively in the cytoplasm (Fig. 4E) and was also detected in dendritic processes (Fig. 4F).
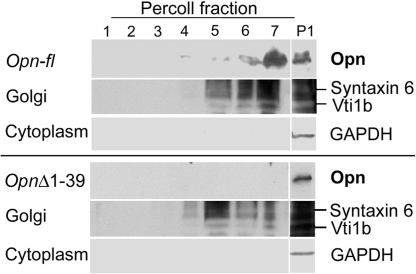
To confirm localization of Opn-FL, we fractionated 293FT lysates by density gradient. Golgi content (detected by Syntaxin 6 and Vti1b Ab) was found at the top of the gradient (fractions 5–7) (Fig. 5). A single band was detected mainly in fraction 7 of the Opn-fl transfectants (Fig. 5 Upper), whereas no band was detected in any Percoll fraction derived from the OpnΔ1–39 transfectants (Fig. 5 Lower), suggesting that Opn-FL, but not Opn-i, localized in Golgi. Because these gradient fractions did not include cytoplasmic material, as judged from the absence of GAPDH markers, a fraction (P1) containing total cell lysate was analyzed. Opn-fl transfectants displayed two bands in P1, whereas OpnΔ1–39 transfectants gave rise to a single band in this fraction. These findings indicate that the short isoform of Opn is found mainly in cytoplasm, rather than secretory vesicles and Golgi.
Fig. 5.
Biochemical detection of Opn-FL targeted to Golgi vesicles. Percoll gradient analysis of Opn in 293FT cells transfected with Opn-fl or OpnΔ1–39 expression constructs. Opn was detected with O-17 Ab.
An Opn Isoform Lacking the Signal Sequence Restores Biological Activity of DC.
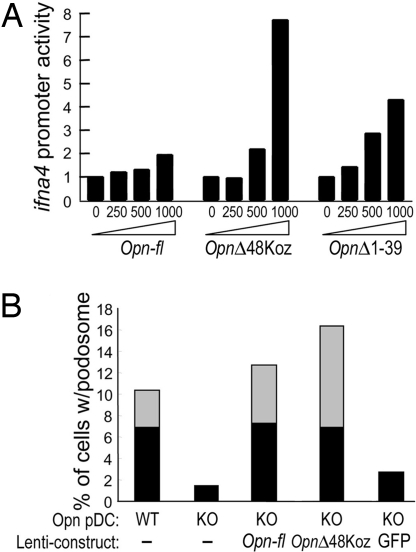
We measured Opn-mediated IFN-α production using a reporter assay (12). Transfection of 293FT cells with the OpnΔ1–39 or OpnΔ48Koz construct gave 4–8-fold more ifna4 promoter activity than transfection of the full Opn construct (Fig. 6A). Expression of Opn-i by the OpnΔ48Koz construct also induced a 15-fold increase in formation of podosomes (17) by pDC (Fig. 6B). Without stimulation, both Opn WT and Opn-deficient pDC were spherical (Fig. S4 A and B). After stimulation with CpG, Opn WT but not Opn-deficient pDC displayed efficient podosome formation (Fig. S4 C and D), and the Opn-deficient defect was particularly evident in their failure to form long podosomes (Fig. 6B). Expression of lenti-Opn-fl or lenti-OpnΔ48Koz in Opn-deficient DC rescued podosome formation (Fig. S4 E and F), whereas lentiviral expression of GFP did not (Fig. S4G). Results are quantitatively analyzed in Fig. 6B. These data suggest that Opn-i is required for IFN-α production and podosome formation and that it is functionally distinct from the Opn-s isoform.
Fig. 6.
Functional analyses of the Opn-i isoform: ifna4 promoter activation and podosome formation. (A) Opn expression and ifna4 activation in 293FT cells transiently transfected with increasing concentrations of Opn expression vectors along with IRF7 and MyD88, as described (12). (B) Opn WT and KO pDC were not infected or were infected with the indicated Opn lentiviral constructs. Proportions of pDC with long (length of podosome > cell diameter; gray) and short (length of podosome < cell diameter; black) podosomes were calculated from total 120 cells per sample.
Discussion
Although previous studies have suggested that an intracellular form of Opn (Opn-i) may mediate some of its biological activities, its genetic origin has not been established. Here, we show that Opn-i is generated from alternative translation of a non-AUG site downstream of the canonical AUG sequence. This mechanism, which does not involve alternative mRNA transcription initiation or splicing, generates a full-length secreted Opn protein (Opn-s) and a smaller intracellular product (Opn-i) from a single full-length mRNA species.
Analysis of the mammalian genome revealed that unexpectedly large numbers of proteins are produced from a relatively small number of genes using several posttranscriptional mechanisms. Alternative translation, which allows generation of multiple proteins from a single mRNA species, is used by Opn to expand the biological range of the products through a translational mechanism that allows differential expression of the N-terminal signal sequence. The two proteins generated by this mechanism –Opn-s and Opn-i– localize to characteristic intracellular sites and mediate distinct functions in DC and T cells.
More than a dozen instances of mammalian genes that produce isoforms by in-frame alternative translation initiation have been reported. A striking feature of these genes is that most encode regulatory proteins, including transcription factors, protooncogenes, kinases, and growth factors (reviewed in ref. 16). Alternative translation initiation of non-AUG codons can also regulate the localization of proteins, including Fgf2 (18, 19), Fgf3 (Int-2) (20), hck (21), and VEGF (22). In the case reported here, the alternative Opn translation initiation site is downstream of the canonical AUG start codon and therefore differs from previously reported instances of alternative translation in which codons locate upstream of the first in-frame AUG codon. In this case, the downstream location of the alternative translation site allows generation of either full-length or truncated protein products that are secreted or retained by the cell.
Selection of the downstream initiation codon can occur by leaky scanning of ribosome, internal ribosome entry or ribosomal shunting (16, 23, 24). In the case of Opn alternative translation, leaky scanning of ribosome is most likely. Entry through an internal ribosome entry site (IRES) is unlikely because the 40-nt stretch between the AUG codon, and the alternative translation region is probably not sufficiently long to form an IRES. Ribosomal shunting is also unlikely, because ribosomes are not likely to skip the canonical AUG, which is surrounded by a sequence (ACGACCAUGA) that closely resembles the Kozak translation initiation consensus sequence (GCCRCCAUGG: R = purine) and probably accounts for the consistent generation of Opn-FL from Opn-fl mRNA templates.
Ribosomes that escape the canonical Opn AUG start codon may continue to scan downstream mRNA sequences until the ribosome encounters another Kozak-like sequence and/or a stable mRNA hairpin secondary structure (16, 24, 25) that stalls the ribosome and allows recognition of an alternative translation initiation site (26, 27). Folding prediction analysis of Opn mRNA suggests that extremely stable secondary structures (kcal/mol) can be formed, and their hairpin stability is greater (−14.7 and −11.4 kcal/ml) than the hairpin formed immediately 3′ to the canonical AUG codon (−11.1 kcal) (Fig. S5).
The size of Opn-i generated in 293FT cells from the 5′ deletion mutant OpnΔ1–39 was similar to the isoform generated from a non-AUG codon residing downstream of the nucleotide sequence that encodes the signal peptide. Analysis of subcellular localization of Opn-i by confocal microscopy and biochemical analyses revealed that alternatively translated Opn-i localized mainly to the cytoplasm. We did not detect Opn-i in Golgi/secretory granules, presumably reflecting the absence of a signal sequence and direct entry of the nascent protein into cytoplasm after synthesis. These observations explain the contribution of Opn-i to intracellular signaling pathways, including IRF7 activation after association with MyD88 in pDC (12), and its association with ezrin (8) and the cytoplasmic portion of CD44 (9, 10). The impact of Opn-i on podosome formation may reflect its contribution to actin polymerization at the leading edge of migration, consistent with its association with the ERM (ezrin/radixin/moesin) complex that couples cell surface adhesion molecules with actin filaments (8). Ongoing studies are intended to define the non-AUG codon within positions 40–51 that initiates alternative translation and to delineate the mechanism of this process.
We have shown that, although APC, including DC and macrophages, are rich in Opn-i, T cells tend to secrete rather than retain Opn (4). Cell type-specific translation of Opn from the canonical and alternative sites defined here may account for the dominance of Opn-s expression in activated T cells compared with the dominance of Opn-i in DC (4). Tissue specific regulation of translation of Fgf2 results in kidney and skeletal muscle expression of the AUG-initiated isoform, whereas production of the alternative CUG-initiated translational isoforms predominate in brain and liver. Cell type-specific factors expressed by T cells and DC that determine the relative expression levels of Opn-i and Opn-s after cellular activation may account for the impact of Opn gene expression on the functional phenotype of these cells.
Methods
Cells and Reagents.
DC and T cells were prepared as described (12). Rabbit polyclonal mouse Opn O-17 Ab was raised against a synthetic peptide (LPVKVTDSGSSEEKLY) (IBL America) immediately downstream of the Opn signal peptide (29). Protein samples for IVT and Western blot analysis were treated with calf intestine alkaline phosphatase (CIP, New England Biolabs). Mouse Opn 2A1 Ab (not signal sequence; Santa Cruz) was used for immunofluorescence. Additional Abs include: mouse anti-GM130, mouse anti-syntaxin 6, mouse anti-Vti1b (BD PharMingen), and rabbit anti-GOLPH4 (Abcam) (Golgi markers), rabbit anti-GAPDH (Abcam) (cytoplasmic marker), mouse anti-V5 (Invitrogen), rabbit anti-FLAG (Sigma), Alexa 488-conjugated anti-mouse IgG, and Alexa 546-conjugated anti-rabbit IgG (Molecular Probes).
Opn Expression Constructs.
Lentiviral expression constructs pMLS3 (Opn-fl) and pMLS5 (no signal sequence, OpnΔ49Koz) were described (12). Constructs for IVT and Opn expression in 293FT cells contain backbone expression vector pcDNA5/FRT/V5-His TOPO TA (Invitrogen). Point mutations in the Opn-fl sequence were introduced by site-directed PCR mutagenesis using Opn-fl constructs as templates with QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene). N-terminal FLAG-tagged Opn-FL constructs were produced with pCMV-Tag4 vector (Stratagene). Opn-GFP fusion expression vector was produced by inserting EGFP downstream of Opn cDNA in-frame.
5′ RLM-RACE to Determine Transcription Initiation Sites.
RLM-RACE was performed using FirstChoice RLM-RACE kit (Ambion). Outer primer annealing site was in Exon 7, inner primer binding region is shown in Fig. S1. Amplified bands were excised from an agarose gel, cloned into vectors, and the 5′-end of Opn transcripts determined by sequencing.
IVT.
Transcription was carried out with T7 RNA polymerase (Promega) from linearized Opn expression constructs. IVT was performed in rabbit reticulocyte lysates (Promega) with [35S] Met at 30°C for 90 min. Samples were phosphatase-treated.
Immunofluorescence Analysis by Confocal Microscopy.
Opn-expressing 293FT cells and BM-DC were harvested 24 h after transfection and replating, respectively. Images were obtained with a Nikon TE2000-U inverted Microscope equipped with C1 Plus Confocal Laser Scanning system.
Ifna4 Promoter Reporter Assay.
The assay was performed as described (12) by using QUANTI-Blue system (InvivoGen) with a slight modification using backbone vector pCMV4 for Opn expression. Total amounts of DNA were kept constant by supplementation with empty pCMV4 control vector.
Isolation of Secretory Vesicles.
Secretory granules of 293FT cells were isolated by Percoll density gradient centrifugation as described (28, 31). Homogenates of Opn-transfected cells were centrifuged at 150 × g for 5 min (P1 = the pellet), and supernatant was centrifuged at 1,500 × g for 10 min. The resulting pellet was suspended in homogenizing buffer containing 40% Percoll and centrifuged at 20,000 × g for 20 min to obtain seven fractions.
Supplementary Material
Acknowledgments.
We thank C. D. Novina and B. Wang for expertise and critical discussions and D. Laznik for technical assistance. This work was supported by the National Institutes of Health (Grants AI12184 and AI48125, to H.C.) and by National Research Service Award Fellowship T32CA70083 (to M.L.S.).
Footnotes
The authors declare no conflict of interest.
Examination of CUG codons [another common non-AUG translation initiation codon (16)] further downstream (at position +100 and +127; Fig. S3A) by mutagenesis did not change the pattern from Opn WT mRNA (“Full”) (Fig. S3B) nor did mutating GUG (Val) to GAC (Asp) affect translation (Fig. S3C).
This article contains supporting information online at www.pnas.org/cgi/content/full/0802301105/DCSupplemental.
References
- 1.Cantor H. T-cell receptor crossreactivity and autoimmune disease. Adv Immunol. 2000;75:209–233. doi: 10.1016/s0065-2776(00)75005-x. [DOI] [PubMed] [Google Scholar]
- 2.Denhardt DT, Noda M, O'Regan AW, Pavlin D, Berman JS. Osteopontin as a means to cope with environmental insults: regulation of inflammation, tissue remodeling, and cell survival. J Clin Invest. 2001;107:1055–1061. doi: 10.1172/JCI12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashkar S, et al. Eta-1 (osteopontin): an early component of Type 1(cell-mediated) immunity. Science. 2000;287:860–864. doi: 10.1126/science.287.5454.860. [DOI] [PubMed] [Google Scholar]
- 4.Shinohara ML, et al. T-bet-dependent expression of osteopontin contributes to T cell polarization. Proc Natl Acad Sci USA. 2005;102:17101–17106. doi: 10.1073/pnas.0508666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scatena M, et al. NF-kappaB mediates alphavbeta3 integrin-induced endothelial cell survival. J Cell Biol. 1998;141:1083–1093. doi: 10.1083/jcb.141.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan SA, et al. Soluble osteopontin inhibits apoptosis of adherent endothelial cells deprived of growth factors. J Cell Biochem. 2002;85:728–736. doi: 10.1002/jcb.10170. [DOI] [PubMed] [Google Scholar]
- 7.Hur EM, et al. Osteopontin-induced relapse and progression of autoimmune brain disease through enhanced survival of activated T cells. Nat Immunol. 2007;8:74–83. doi: 10.1038/ni1415. [DOI] [PubMed] [Google Scholar]
- 8.Zohar R, et al. Intracellular osteopontin is an integral component of the CD44-ERM complex involved in cell migration. J Cell Physiol. 2000;184:118–130. doi: 10.1002/(SICI)1097-4652(200007)184:1<118::AID-JCP13>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki K, et al. Colocalization of intracellular osteopontin with CD44 is associated with migration, cell fusion, and resorption in osteoclasts. J Bone Miner Res. 2002;17:1486–1497. doi: 10.1359/jbmr.2002.17.8.1486. [DOI] [PubMed] [Google Scholar]
- 10.Zhu B, et al. Osteopontin modulates CD44-dependent chemotaxis of peritoneal macrophages through G-protein-coupled receptors: evidence of a role for an intracellular form of osteopontin. J Cell Physiol. 2004;198:155–167. doi: 10.1002/jcp.10394. [DOI] [PubMed] [Google Scholar]
- 11.Junaid A, Moon MC, Harding GE, Zahradka P. Osteopontin localizes to the nucleus of 293 cells and associates with polo-like kinase-1. Am J Physiol. 2007;292:C919–C9126. doi: 10.1152/ajpcell.00477.2006. [DOI] [PubMed] [Google Scholar]
- 12.Shinohara ML, et al. Osteopontin expression is essential for IFN-α production by plasmacytoid dendritic cells. Nat Immunol. 2006;7:498–506. doi: 10.1038/ni1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinohara ML, Kim JH, Garcia VA, Cantor H. Engagement of the Type-I interferon receptor on dendritic cells inhibits Th17 cell development: Central role of intracellular Osteopontin. Immunity. 2008 doi: 10.1016/j.immuni.2008.05.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rane NS, Yonkovich JL, Hegde RS. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. 2004;23:4550–4559. doi: 10.1038/sj.emboj.7600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maruyama K, Sugano S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 16.Touriol C, et al. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell. 2003;95:169–178. doi: 10.1016/s0248-4900(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 17.Carman CV, et al. Transcellular diapedesis is initiated by invasive podosomes. Immunity. 2007;26:784–797. doi: 10.1016/j.immuni.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bugler B, Amalric F, Prats H. Alternative initiation of translation determines cytoplasmic or nuclear localization of basic fibroblast growth factor. Mol Cell Biol. 1991;11:573–577. doi: 10.1128/mcb.11.1.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnaud E, et al. A new 34-kilodalton isoform of human fibroblast growth factor 2 is cap dependently synthesized by using a non-AUG start codon and behaves as a survival factor. Mol Cell Biol. 1999;19:505–514. doi: 10.1128/mcb.19.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acland P, Dixon M, Peters G, Dickson C. Subcellular fate of the int-2 oncoprotein is determined by choice of initiation codon. Nature. 1990;343:662–665. doi: 10.1038/343662a0. [DOI] [PubMed] [Google Scholar]
- 21.Lock P, et al. Two isoforms of murine hck, generated by utilization of alternative translational initiation codons, exhibit different patterns of subcellular localization. Mol Cell Biol. 1991;11:4363–4370. doi: 10.1128/mcb.11.9.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huez I, Bornes S, Bresson D, Creancier L, Prats H. New vascular endothelial growth factor isoform generated by internal ribosome entry site-driven CUG translation initiation. Mol Endocrinol. 2001;15:2197–2210. doi: 10.1210/mend.15.12.0738. [DOI] [PubMed] [Google Scholar]
- 23.Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 24.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kochetov AV, et al. AUG_hairpin: prediction of a downstream secondary structure influencing the recognition of a translation start site. BMC Bioinformatics. 2007;8:318. doi: 10.1186/1471-2105-8-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prats AC, Vagner S, Prats H, Amalric F. cis-acting elements involved in the alternative translation initiation process of human basic fibroblast growth factor mRNA. Mol Cell Biol. 1992;12:4796–4805. doi: 10.1128/mcb.12.10.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coffin JD, et al. Abnormal bone growth and selective translational regulation in basic fibroblast growth factor (FGF-2) transgenic mice. Mol Biol Cell. 1995;6:1861–1873. doi: 10.1091/mbc.6.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kon S, et al. Antibodies to different peptides in osteopontin reveal complexities in the various secreted forms 1. J Cell Biochem. 2000;77:487–498. [PubMed] [Google Scholar]
- 30.Fujita-Yoshigaki J, et al. Vesicle-associated membrane protein 2 is essential for cAMP-regulated exocytosis in rat parotid acinar cells. The inhibition of cAMP-dependent amylase release by botulinum neurotoxin B. J Biol Chem. 1996;271:13130–13134. doi: 10.1074/jbc.271.22.13130. [DOI] [PubMed] [Google Scholar]
- 31.Hara-Kuge S, Seko A, Shimada O, Tosaka-Shimada H, Yamashita K. The binding of VIP36 and alpha-amylase in the secretory vesicles via high-mannose type glycans. Glycobiology. 2004;14:739–744. doi: 10.1093/glycob/cwh082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.