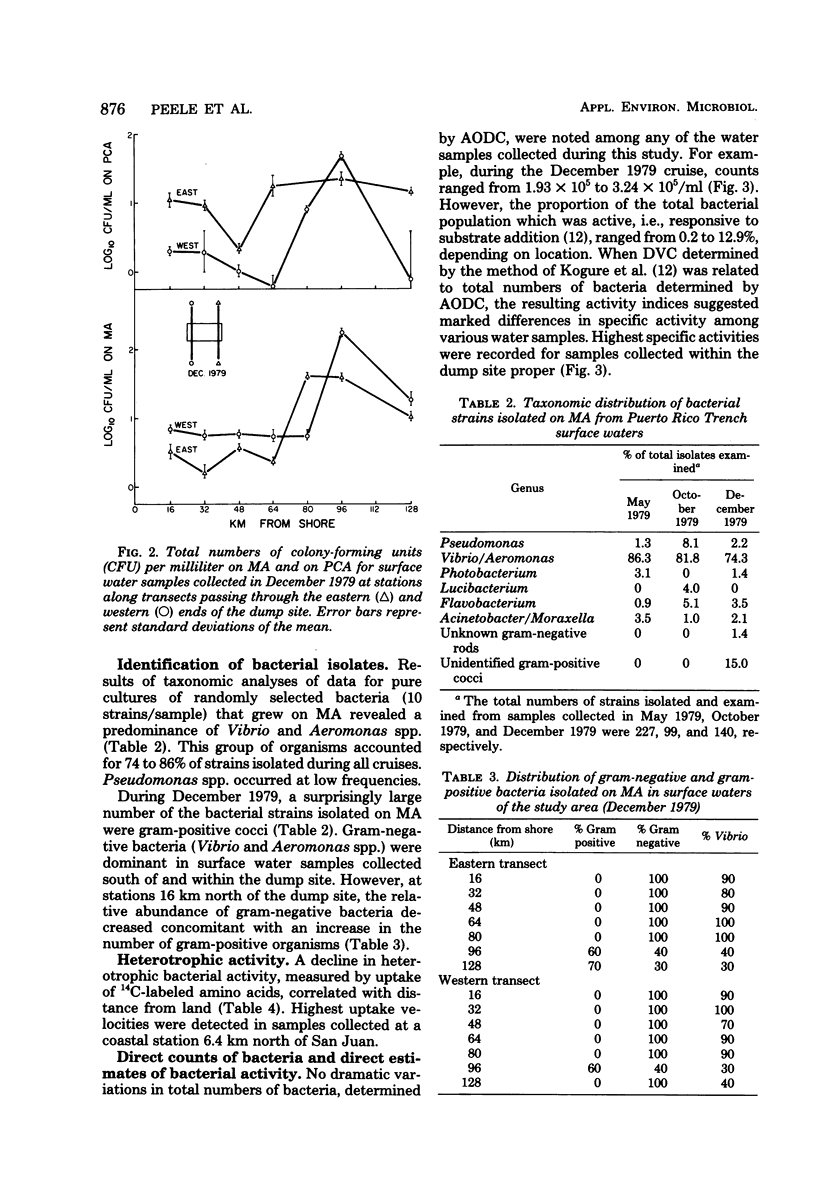
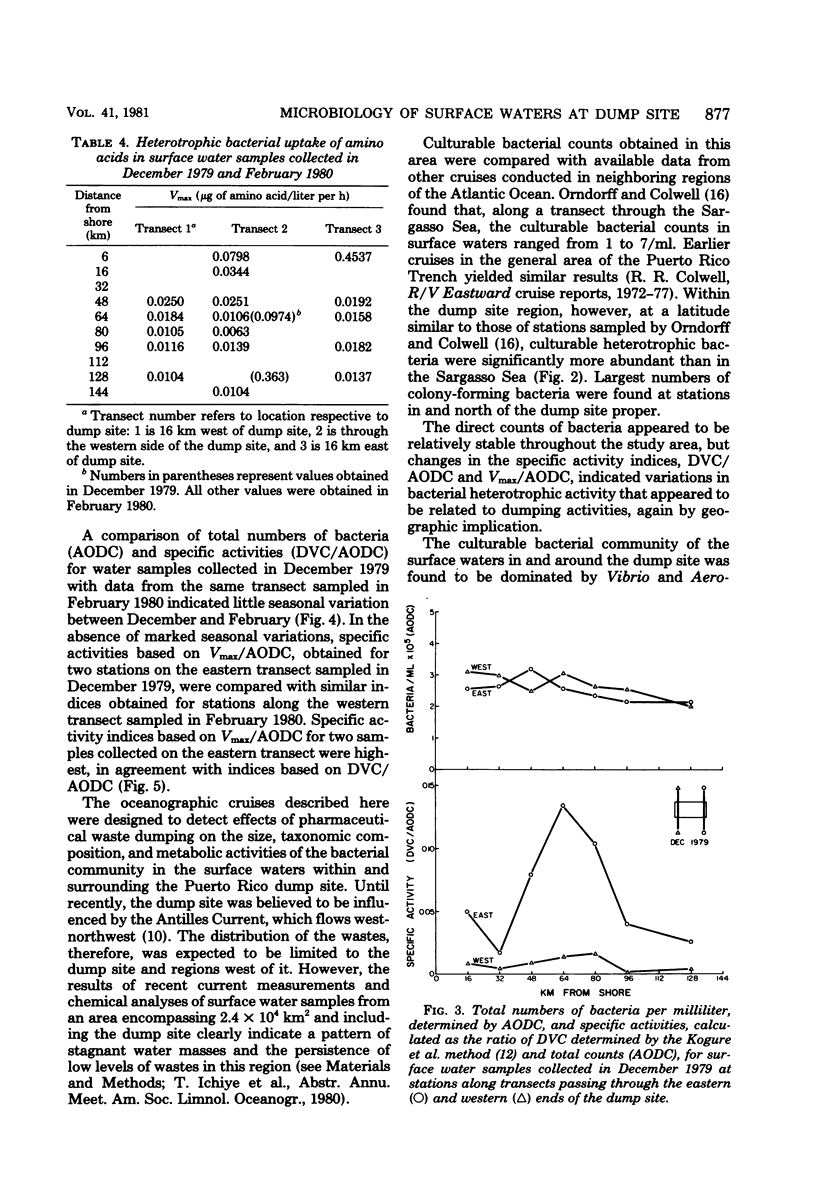
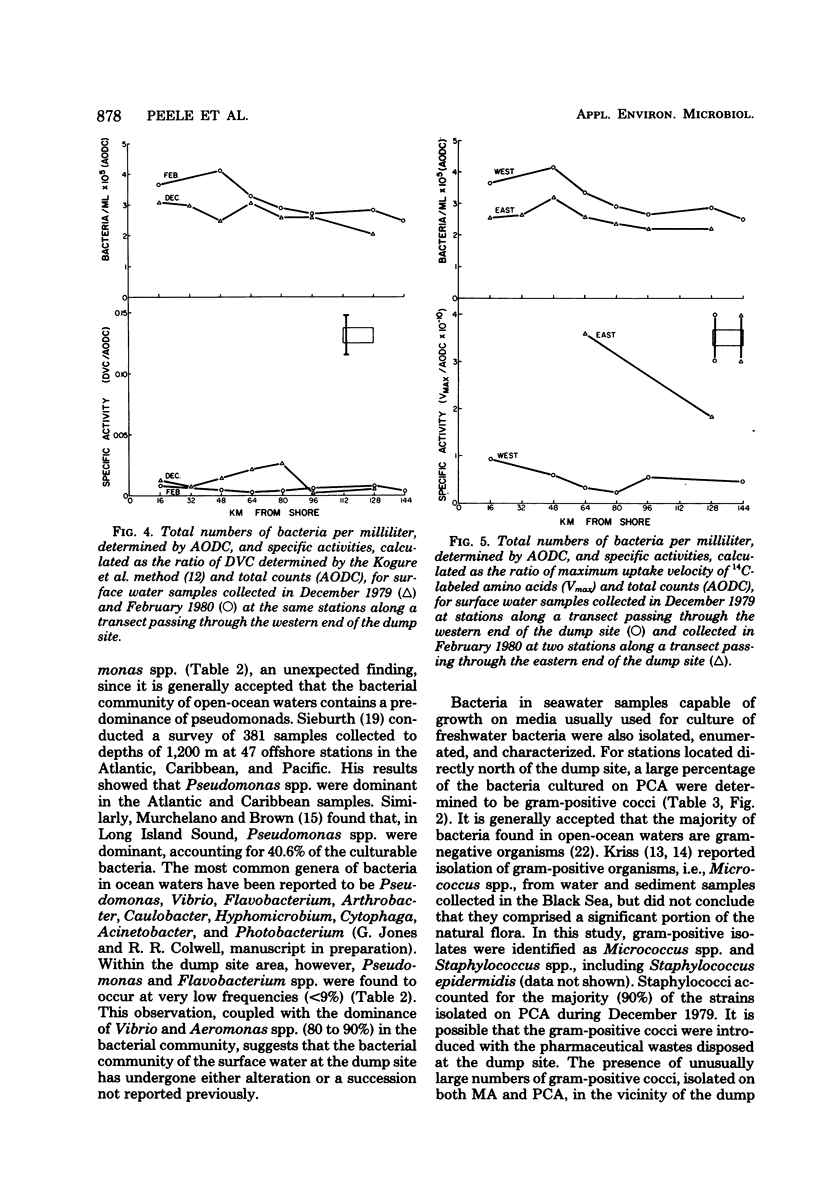
Abstract
A series of cruises during 1979 and 1980 to the pharmaceutical dump site located 64 km north of Arecibo, Puerto Rico, in the Atlantic Ocean, was carried out to evaluate effects of wastes on the ecology of the microflora of surface waters of the dump site. In addition to bacteriological monitoring of the waste plume created by the release of wastes from the disposal barge, stations along a series of transects, extending north from coastal waters through and beyond the dump site, were sampled. Largest numbers of culturable bacteria on marine agar were found at stations closest to shore and in the vicinity of the dump site. Bacteria recovered on marine agar were predominantly Vibrio and Aeromonas spp., with the relative abundance of these organisms decreasing as gram-positive organisms (staphylococci, micrococci, and bacilli) became dominant in areas immediately affected by waste dumping. Total numbers of bacteria (determined by acridine orange direct counts [AODC]), which were relatively stable throughout the region, and a direct estimate of viable cells (DVC), i.e., those cells responsive to additions of yeast extract and nalidixic acid, were determined by acridine orange staining and epifluorescence microscopy. Heterotrophic bacterial activity, measured by the uptake (Vmax) of 14C-labeled amino acids, declined relative to distance from land. Increases in specific activity indices (DVC/AODC and Vmax/AODC) were observed near the dump site. The composite results of this study, i.e., increased specific activities (determined by two methods), increased numbers of culturable marine bacteria, and marked alteration of the taxonomic composition of the culturable bacterial community in waters within and surrounding the Puerto Rico dump site, indicate demonstrable changes in the marine microbial community in the region used for waste disposal.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Colwell R. R. Ecological aspects of microbial degradation of petroleum in the marine environment. CRC Crit Rev Microbiol. 1977 Sep;5(4):423–445. doi: 10.3109/10408417709102813. [DOI] [PubMed] [Google Scholar]
- Francisco D. E., Mah R. A., Rabin A. C. Acridine orange-epifluorescence technique for counting bacteria in natural waters. Trans Am Microsc Soc. 1973 Jul;92(3):416–421. [PubMed] [Google Scholar]
- Hobbie J. E., Daley R. J., Jasper S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 May;33(5):1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure K., Simidu U., Taga N. A tentative direct microscopic method for counting living marine bacteria. Can J Microbiol. 1979 Mar;25(3):415–420. doi: 10.1139/m79-063. [DOI] [PubMed] [Google Scholar]
- Kriss A. E. Okeanicheskaia mikrobiologiia: ékologiia i geografiia mikroorganizmov. Mikrobiologiia. 1971 Sep-Oct;40(5):904–911. [PubMed] [Google Scholar]
- Orndorff S. A., Colwell R. R. Distribution and identification of luminous bacteria from the sargasso sea. Appl Environ Microbiol. 1980 May;39(5):983–987. doi: 10.1128/aem.39.5.983-987.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. T. Measurement and significance of specific activity in the heterotrophic bacteria of natural waters. Appl Environ Microbiol. 1978 Aug;36(2):297–305. doi: 10.1128/aem.36.2.297-305.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]


