Abstract
We describe here an extraordinary purple-colored DNA ligase, LigFa, from the acidophilic ferrous iron-oxidizing archaeon Ferroplasma acidiphilum, a di-ferric enzyme with an extremely low pH activity optimum. Unlike any other DNA ligase studied to date, LigFa contains two Fe3+-tyrosinate centers and lacks any requirement for either Mg2+ or K+ for activity. DNA ligases from closest phylogenetic and ecophysiological relatives have normal pH optima (6.0–7.5), lack iron, and require Mg2+/K+ for activity. Ferric iron retention is pH-dependent, with release resulting in partial protein unfolding and loss of activity. Reduction of the Fe3+ to Fe2+ results in an 80% decrease in DNA substrate binding and an increase in the pH activity optimum to 5.0. DNA binding induces significant conformational change around the iron site(s), suggesting that the ferric irons of LigFa act both as structure organizing and stabilizing elements and as Lewis acids facilitating DNA binding at low pH.
Keywords: iron metalloenzyme, Picrophilus torridus, Sulfolobus acidocaldarius, Thermoplasma acidophilum, Ferroplasma acidiphilum
DNA ligases (EC 6.5.1.1/2) catalyze the formation of phosphodiester bonds between adjacent 3′-hydroxyl and 5′-phosphoryl groups at single-strand breaks in double-stranded DNA (1, 2); reestablish the integrity of the DNA backbone after events that introduce strand breaks, such as DNA replication (3), recombination (4), damage, and repair (5); and hence are pivotal enzymes in the central processes of cell division, genetic reassortment, and the maintenance of genome integrity in all organisms (2). Acidic environments might be expected to be particularly challenging for DNA ligase function and hence for these vital functions, because acidophiles tend to have cytoplasmic pH values <6.0, and available information indicates that DNA ligases function suboptimally at such pH levels (e.g., www.brenda-enzymes.info). However, there is currently little information on DNA ligases of acidophiles (6–9). We have therefore isolated and characterized DNA ligases of five acidophilic organisms, namely Ferroplasma acidiphilum, Thermoplasma acidophilum, and Picrophilus torridus (three extremely acidophilic archaea representing all three families of the order Thermoplasmatales, phylum Euryarchaeota); Sulfolobus acidocaldarius (an acidophilic member of phylum Crenarchaeota); and Acidithiobacillus ferrooxidans (an acidophilic iron-oxidizing bacterium). Interestingly, while DNA ligases of the latter four microbes were unremarkable in terms of their pH optima in the neutral range and requirement for Mg2+ or K+ ions for catalysis (but not enzyme formation or stability), that of F. acidiphilum is a purple enzyme containing two ferric ions that organize the 3D structure of the protein and play a role in substrate binding and having an in vitro pH optimum of 1.5–3.0.
Results
LigFa Requires Iron for Activity and Has a Uniquely Low pH Optimum.
We recently reported that F. acidiphilum, an extremely acidophilic archaeon of the order Thermoplasmatales, phylum Euryarchaeota (10), has a thus far unique iron-protein-dominated metabolic machinery (11). One such protein not expected from either current knowledge or its biochemical role to contain iron was the DNA ligase, LigFa. We therefore undertook characterization of this enzyme. Because biomass yields of Ferroplasma cultures are very low, and LigFa represents only 0.04% wt/wt of total soluble protein, we elected to isolate it from a hyperexpressing recombinant Escherichia coli strain. However, to confirm that the recombinant enzyme had properties similar to those of the native enzyme, we isolated a small quantity of LigFa (40 ng) from nondenaturing 2D gels of F. acidiphilum cell extracts (11) and analyzed some of its properties: it contains iron in a molar stoichiometry of 2Fe:1LigFa, has an exceptionally low pH activity optimum, and is purple in color [supporting information (SI) Fig. S1]. The ligFa gene of F. acidiphilum was cloned in the expression vector pET-3a, and the hybrid plasmid introduced into the ORIGAMI(DE3)pLysS strain of E.coli (SI Text). Although significant quantities of LigFa were expressed by the recombinant clone, no active enzyme was obtained. Because the native enzyme contains iron, we tried supplementation of the growth medium with ferrous chloride (up to 120 μM), but this did not yield active enzyme. However, incubation of acidified (optimum: pH 3.0) cell extracts with 0.01 M freshly prepared ferric nitroacetate Fe(NTA)2, solution followed by extensive dialysis against 100 mM sodium citrate buffer, pH 3.0, resulted in an increase in the iron content of the enzyme, and a gain in catalytic activity equal to the levels of the native enzyme (see below). Subsequent dialysis of the iron-reconstituted enzyme against iron-free buffer did not result in iron loss from the enzyme, so iron seemed to be tightly bound in the protein, which is consistent with its ability to be detected as a metal-containing protein after nondenaturing 2D gel electrophoresis (11). Purified LigFa (SI Text) is deep purple in color, a feature thus far unique in DNA ligases, and its similar relative electrophoretic mobilities on denaturing and nondenaturing polyacrylamide gels indicate it has a monomeric structure (Fig. S2).
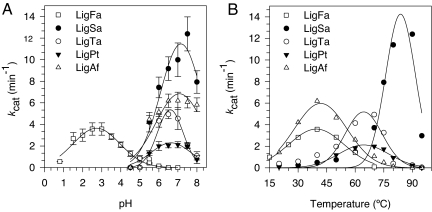
Purified LigFa catalyzed the ligation of two short (25- and 35-mer) contiguous synthetic oligonucleotides base-paired to a complementary strand (nick-joining reaction), sticky-ended Sau3A-generated λ DNA fragments, and blunt-ended SspI-generated bacteriophage φX174 DNA fragments over a rather narrow pH range of 1.5 to 4.0 (Fig. S3). As shown in Fig. 1, these ligation activities, at 40°C under conditions of substrate saturation, were highest at pH 2.5–3.0 (turnover rate up to 3.57 ± 0.57 min−1), only slightly lower at pH 1.5–2.0 (≈80%), but were practically undetectable above pH 5.0. LigFa is also acid-tolerant and retained >95% of its activity after incubation at pH 2.0–3.0 for 10 h but was unstable at higher pH levels (data not shown). Its catalytic activity neither depended on nor was stimulated by added Mg2+ or K+, obligatory cofactors of all other known DNA ligases. To our knowledge, LigFa is the only example of a ferric DNA ligase lacking any requirement for Mg2+/K+ cofactor; its in vitro pH activity optimum is by far the lowest among DNA ligases reported until now.
Fig. 1.
Recombinant LigFa has a uniquely low pH optimum. Activity vs. pH assays were performed by using a standard fluorimetric assay based on the ligation of a 5′-phosphorylated 35-meric and 5′-fluorescein-labeled 25-meric oligonucleotides annealed at the complementary 70-mer, as described in Materials and Methods, at the optimal temperature 40°C (LigFa and LigAf), 70°C (for LigTa and LigPt), or 80°C (for LigSa) in reaction buffers supplemented with 20 mM MgCl2 and 2 mM ATP, except for LigFa, where MgCl2 was not added. Plot of kcat against pH (A) and temperature (B) obtained by Eadie–Hofstee linearization for LigFa and DNA ligases from other acidophiles. Buffers (100 mM) used were sodium citrate for pH 0.5–3.0, sodium acetate for pH 4.0–5.0 and Mes for pH 6.0–7.0. All data were calculated from three independent assays ± SD and are not fitted to any model.
DNA ligases from other acidophiles are neither metalloenzymes, iron-dependent, nor active at low pH. Given the extraordinary properties of LigFa, the question arises: do DNA ligases of phylogenetic or ecophysiological relatives of F. acidiphilum exhibit characteristics similar to those of LigFa? F. acidiphilum belongs to the order Thermoplasmatales, which consists of three families: the Thermoplasmaceae, Picrophilaceae, and Ferroplasmaceae, each represented by the species T. acidophilum and Thermoplasma volcanium, Picrophilus oshimae and P. torridus, and F. acidiphilum, “Ferroplasma acidarmanus” and “Ferroplasma cupricumulans,” respectively (12). All of these archaea grow well at pH values between 0 and 2 and are the most acidophilic microorganisms, although they are reported to exhibit an intracellular pH of 4.6–5.6 (13–15). Other organisms inhabiting the same type of environment as F. acidiphilum are crenarchaea of the species Sulfolobus acidocaldarius (from sulfur- and metal-rich acidic high-temperature habitats) and the acidophilic mesophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans. We therefore cloned and hyperexpressed in E. coli the DNA ligase genes from T. acidophilum, P. torridus, S. acidocaldarius, and A. ferrooxidans and purified and characterized the corresponding enzymes LigTa, LigPt, LigSa, and LigAf (SI Text).
Unlike the situation with LigFa, no difficulties were encountered in obtaining active DNA ligases by normal procedures: neither iron nor low pH reconstitution was required for the activity. They also exhibited pH optima typical of all other DNA ligases characterized thus far, i.e., 6.0–7.5 (Fig. 1A), with turnover numbers ranging from 2.1 to 12.4 min−1 at optimal temperatures (which reflected optimal growth temperatures; Fig. 1B). No metal ions were detected in the purified proteins (SI Text). In contrast to LigFa, all these DNA ligases required for activity MgCl2 (optimal concentrations 15, 22, 20, and 4 mM, for LigTa, LigPt, LigSa, and LigAf, respectively) or KCl (optimal concentrations 0.8, 2, 8, and 7 mM, in the same order); no other mono- or divalent cations, including Fe2+/3+, influenced their activity. LigFa exhibited maximal activities when provided with either ATP or NAD+ as cofactors (Table S1). These nucleotides also served as cofactors for LigTa and LigPt, although ATP was preferred. In contrast, LigAf and LigSa could use only ATP. Interestingly, although ATP-dependent DNA ligases are ubiquitous in eukaryotes, bacteria, archaea, bacteriophages, and viruses (6, 7), NAD+-dependent ligases have been described so far only in bacteria and eukaryotic viruses (8). The ability of DNA ligases of representatives of all three families of the Thermoplasmatales to use either ATP or NAD+ in adenylation and nick-sealing reactions is therefore unexpected. Comparison of the protein sequences of LigFa, LigTa, LigPt, LigSa, and LigAf and of others available in public databases revealed family branch structures largely similar to those based on 16S rRNA gene sequences, although the relative positions of the family branches differ somewhat (Fig. S4). Thus, although LigFa exhibits phylogenetic relatedness to, and nucleotide cofactor requirement and catalytic parameter similarities with, DNA ligases of other acidophiles, it is thus far unique in its iron content and low pH activity optimum.
The Principal Role of Ferric Iron in LigFa Is Structural.
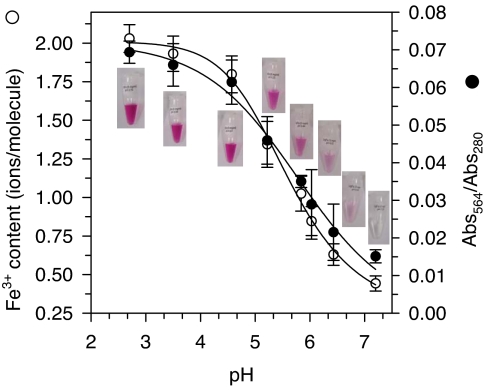
LigFa contains 2.03 ± 0.09 iron ions per molecule (SI Text), both in the ferric state, as shown by Mössbauer spectroscopy (see below), and exhibits an absorption maximum (λmax) at 564 nm (Fig. S5), which is unusual in biomolecules but characteristic of interactions between tyrosine ligand(s) and a bound ferric iron (16, 17). As seen in Fig. 2, the iron content and the purple color of LigFa are also pH-dependent, with loss of absorption at 564 nm beginning at pH 4.0 and continuing to decrease as the pH increases (apparent pKa of 5.09 ± 0.94; rate constant of iron release is 4.7 × 10−4 min−1 at this pH). Colorless inactive LigFa obtained by exposure to high pH, followed by restoration of low pH, regains both its purple color and enzymatic activity upon addition of ferric iron. Thus, purple color and enzymatic function of LigFa are obligatorily coupled to its ferric iron content.
Fig. 2.
Iron (III) content, purple color, and low pH activity optimum of LigFa are coupled characteristics. Iron content (left y axis) and Abs564/280 (right y axis) vs. pH dependence was determined by equilibration of LigFa [(E) 0.3 mM], prepared by dissolving freeze-dried LigFa in 100 mM buffer at each of different pHs, followed by 24-h incubation at 25°C and extensive dialysis against the same buffer through a Centricon YM-10 membrane. The metal ion content was measured, after dilution of the enzyme solution with 5 ml of 0.5% (vol/vol) HNO3 to digest the protein and release metal ions (SI Text). The LigFa concentration was determined spectrophotometrically (ε280 = 52,322 M−1 ·cm−1, calculated from the amino acid sequence at www.expasy.org/tools/protparam.html). The characteristic color phenotype of protein solutions at each pH is shown.
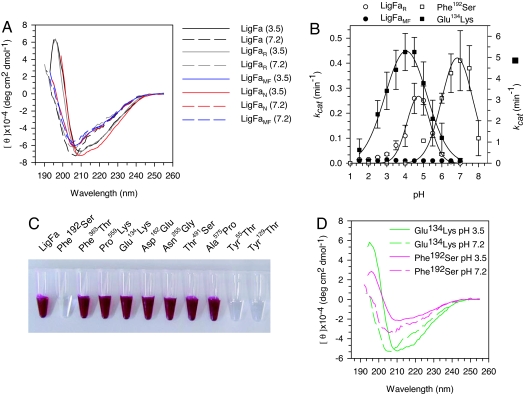
The influence of high and low pH on the secondary structure of LigFa was assessed by CD (18). The CD spectra of the ferric recombinant (LigFa) and native (LigFaN) enzymes at pH 3.5 were similar with a maximum at 196 nm, a minimum at 210 nm, and a shoulder at 218 nm (Fig. 3A). At pH 7.2, only a minimum at 206 nm, indicative of the loss of ordered secondary structure, was observed. The secondary structure content of LigFa at pH 3.5 estimated by the best-fitting CD analysis method (Fig. S6 and SI Text) was 16% α-helix, 30% β-sheet, 21% turns, and 32% unordered structure. The average of eight methods was coincident: 15 ± 5% helix, 30 ± 10% sheet, 24 ± 7% turns, and 31 ± 9% unordered. More interestingly, the estimated changes in secondary structure upon transferring the protein from pH 3.5 to 7.2 were −10% helix, 0% sheet, −5% turns, and +15% unordered. Thus, iron retention by isolated LigFa, its A564 signal/purple color, and its ordered secondary structure are correlated with low pH values.
Fig. 3.
Properties of LigFa variants. (A) pH dependence of CD spectra of LigFa, reduced/ferrous iron-containing enzyme, LigFaR, and metal-free enzyme, LigFaMF. CD spectra of LigFa variants at acidic and neutral pHs were acquired as described in SI Text. (B) Influence of iron status and single point mutations on LigFa ligation activity (kcat) at each of different pHs. Iron variants were prepared and analyzed as described in SI Text. Assay conditions and buffers were as in Fig. 1. All data were calculated from three independent assays ± SD at 40°C and are not fitted to any model. Note the scale differences on the y axes (left axis: LigFaR, LigFaMF, and Phe192Ser; right, Glu134Lys). (C) Color phenotypes of LigFa variants (10 mg/ml) at pH 3.0. (D) CD spectra of Glu134→Lys134 and Phe192→Ser192 LigFa mutants at acidic and neutral pHs. Spectra were acquired as described in SI Text.
Because high pH results in expulsion of ferric iron from LigFa, polypeptide chain unfolding at high pH revealed by CD spectra could reflect either iron expulsion or the influence of pH per se. To uncouple these two possibilities and assess the consequence of reduction of the ferric ions to ferrous, we obtained reduced (LigFaR) and metal-free (LigFaMF) forms of the enzyme by appropriate treatments at low pH (SI Text). Dithionite treatment produced Fe2+–Fe2+-LigFa complexes (>99%) having the same stoichiometry of two mol of iron per molecule, as confirmed by high-resolution inductively coupled plasma mass spectrometry, UV-Vis, and Mössbauer analysis (SI Text and Fig. S7), and a CD spectrum similar to that of LigFa (Fig. 3A). However, LigFaR had only ≈2% of the activity of LigFa (Fig. 3B) and lacked absorbance at 564 nm (Fig. S7), which strongly suggests that the ferric form is the principal iron valency in the enzyme. Interestingly, reduction of Fe3+ to Fe2+ resulted in not only a depression of enzymatic activity but also a marked upward shift of two units in the pH activity optimum (Fig. 3B). Metal-free enzyme, LigFaMF (obtained by treatment of the reduced enzyme with chelating agents, such as EDTA or EGTA, which quantitatively abstracted iron from the enzyme), was essentially inactive (Fig. 3B), and its CD spectrum was similar to that obtained with LigFa at pH 7.2, with a single minimum at 206 nm, which did not change significantly when the pH was increased from 3.5 to 7.2 (Fig. 3A). Secondary structure content calculations gave similar changes for LigFaMF as for LigFa at pH 7.2 (−10% helix, +3% sheet, −5% turns, and +12% unordered). These results clearly demonstrate that it is the ferric content per se that determines the structure, activity, and low pH optimum of LigFa.
To ascertain whether reduced/loss of activity of the LigFa variants reflects changes in substrate binding or reaction rate, their ability to bind substrate DNA was analyzed by EMSA. EMSA was carried out with a double-stranded DNA substrate consisting of a 32P-labeled 35-mer oligonucleotide (5′-TAAGCTCCGGATTGTCCGGGAGGTAAAGCCCTGAT-3′) and its nonphosphorylated complementary strand. Quantitation of the formation of the protein–DNA complex was followed by densitometric scanning of autoradiographs of the gels (see SI Text). As shown in Fig. S8 A–C, reduction of Fe3+ to Fe2+ lowered enzyme binding to the oligonucleotide substrate by a factor of ≈320-fold (from 0.026 ± 0.008 to 8.4 ± 0.3 μM, measured at 40°C and pH 3.0), whereas removal of iron lowered it 1,600-fold (to 43.0 ± 2.6 μM). To confirm that the migrating complex did indeed consist of LigFa bound to the DNA, and that no nonspecific binding occurred, EMSA was also carried out with denatured protein (see SI Text) under identical conditions; in this case, no migrating complex was formed, which confirms that the native LigFa indeed complexes the DNA specifically (see lanes 1 and 2 in Fig. S8D). Although it was expected that secondary structure-disordered LigFaMF would fail to bind the DNA substrate, the reduced substrate binding of the ferrous-containing variant indicates that the ferric ions may play a dual role, namely in organizing the polypeptide secondary structure and in substrate binding.
Assessment of the ability of other divalent cations to replace iron revealed that Al(III), W(IV), and Cr(III) could substitute (albeit poorly) for iron, giving variants with 7%, 36%, and 14% activities, respectively, relative to the ferric-containing form (Fig. S9).
Substrate DNA Does Not Bind to Iron But Induces Major Conformational Changes of the Iron Centers.
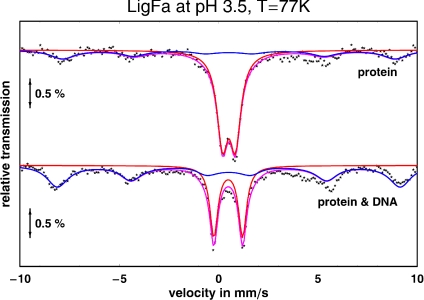
Mössbauer spectroscopy of LigFa was carried out to further characterize the iron:protein interactions. Spectra for freshly reconstituted 57Fe-LigFa were obtained at 77, 140, and 190 K at pH 3.5 and 5.5 (SI Text). As shown in Fig. 4, the spectrum of a sample of recombinant 57Fe-LigFa in 0.1 M citrate buffer pH 3.5 at 77 K exhibits a doublet [quadrupole splitting ΔEQ = 0.65 (1) mms−1, isomer shift δ = 0.52 (1) mms−1, line width Γ = 0.67 (1) mms−1, relative area A = 70 (2) %), representing Fe(III) in paramagnetic (S = 5/2) state, and a sextet (ΔEQ = 0 mms−1, δ = 0.51 (1) mms−1, Γ = 1.0 (1) mms−1, A = 30 (2) %, internal hyperfine field Bhf = 51.5 (5)T], representing iron-containing clusters of considerable size (see below). The doublet represents the iron-binding site of the protein. Its overall effect of ≈2% indicates that the sample at pH 3.5 was highly (1–2 mM) enriched in 57Fe. The relatively large line width (0.67 mms−1) could be an indication of two structurally slightly different iron sites in the protein. At pH 5.5, iron binding was reduced by ≈40% (data not shown), which is consistent with the UV-Vis spectra. When the 57Fe-LigFa was exposed to DNA substrate (Sau3A-digested bacteriophage λ DNA fragments) in 0.1 M sodium citrate buffer pH 3.5 (see details in SI Text), its spectrum recorded at 77 K again exhibited a doublet representing the paramagnetic ferric high spin state of iron bound to the protein [ΔEQ = 1.44 (1) mms−1, δ = 0.48 (1) mms−1, Γ = 0.49 (1) mms−1], but, in this case, the quadrupole splitting was more than two times larger: it changed from 0.65 mm/s to 1.44 mms−1. It should be noted that in a control experiment involving 57Fe metal ions and DNA substrate but no LigFa (see details in SI Text), performed under identical conditions, no resonance absorption was detected (data not shown), so any strong direct binding of 57Fe metal ions to DNA substrate can be excluded. Therefore, DNA substrate binding to the protein induces major conformational changes in the iron coordination sphere. We cannot exclude the possibility that iron may directly participate in binding to the phosphodiester backbone of the DNA molecule. To explore the possibility of a functional role of iron cations as a redox element in LigFa, we monitored spectral changes associated with the reduction of LigFa at different applied potentials in the presence of the electrochemical mediator, methyl viologen. Initially, at −300 mV, only an absorbance associated with Fe3+ was observed, and this signal was used as background (Fig. S10). A decrease in the applied potential to more negative values initiated reduction of the ferric ion and an increase in the proportion of ferrous-containing LigFa. However, a subsequent switch in the applied potential to more positive values, i.e., +120 mV, did not result in reoxidation of the ferrous ion, which we interpret to mean that, at least in vitro, the ferric cation does not act as a redox element. Whatever the case, the source of the observed iron-containing clusters (sextet in Fig. 4) is as yet unknown: their Mössbauer parameters (see above) were very similar to those observed for iron-oxide (hydroxide) clusters (19) and, from the superparamagnetic behavior of the sextet measured at 77, 140, and 190 K, we conclude that the diameter of these clusters is >10 nm.
Fig. 4.
Substrate DNA induces major conformational changes of the iron centers. Mössbauer spectra of 57Fe-LigFa and 57Fe-DNA-LigFa (1 mM) were recorded at pH 3.5 (100 mM sodium citrate) and 77 K by using a spectrometer in the constant acceleration mode. See SI Text for experimental details and sample preparation.
The Exceptional Features of LigFa Are Genetically Determined: Identification of Key Residues.
The data presented relate to the in vitro properties of isolated LigFa enzyme and variants created by physical-chemical treatments, so it is not possible thus far to exclude an environmental/technical basis of the enzyme phenotype. We have therefore investigated the possible genetic basis of this phenotype.
Phe192.
The ligFa gene of this study was obtained by PCR amplification from genomic DNA of F. acidiphilum with primers based on the sequence of an ORF coding for a putative ATP-dependent DNA ligase in the genome sequence of the Ferroplasma relative “F. acidarmanus” (20). The deduced protein sequence specified by the ligFa gene was, as expected, closely related (99.5% identity) to the translation product of the corresponding ORF in “F. acidarmanus.” This latter enzyme was recently isolated and characterized by Jackson et al. (21); perhaps surprisingly, its activity optimum is pH 6–7, and it requires either Mg2+ or Mn2+ as a cofactor, just like other typical DNA ligases. Inspection of the deduced amino acid sequence revealed three amino acid differences, namely Phe192, Phe363, and Pro559 in LigFa, which are substituted by Ser, Thr, and Lys in “F. acidarmanus” ligase. To investigate the role of these residues in the major differences in properties of the two enzymes, the single mutant variants Phe192→Ser192, Phe363→Thr363, and Pro559→Lys559 of LigFa, and combinations thereof, were generated by site-directed mutagenesis, and the variant enzymes were hyperexpressed and purified to homogeneity (SI Text). All variants showed the typical deep-purple color characteristic of the wild-type protein, with the exception of Ser192-containing variants, which were colorless (Fig. 3C). Moreover, whereas all non-Phe192 variants exhibited features identical to those of the wild-type enzyme (data not shown), all Phe192 variants exhibited very similar iron contents, pH optima, and catalytic activities, namely almost complete iron depletion (average: 0.07 ± 0.02 mol iron/mol protein), a major change in CD spectra at both low and neutral pH (Fig. 3D), a shift in the pH optimum from ≈3.0 to 7.0 at 40°C (Fig. 3C), a 10-fold drop in turnover rate from 3.57 ± 0.57 to 0.41 ± 0.11 min−1, a loss of NAD+ and iron dependency, and a gain in Mg2+-cofactor requirement (Table S1). Thus, replacement of the single Phe192 residue of LigFa by Ser192 transformed the unique biferric DNA ligase with acidic pH optimum to a typical nonpurple DNA ligase, albeit with greatly reduced activity, and suggests that the Phe192 residue plays a significant role in the ferric iron cluster:low pH optimum characteristic of the enzyme.
Glu134.
The LigFa sequence was also closely related to those of the DNA ligases of the archaeal species P. torridus, T. acidophilum, Desulfurolobus ambivalens, Pyrobaculum aerophilum Thermococcus kodakaerensis and Pyrococcus abyssi (65, 56, 57, 42, 39 and 36% aa identity, respectively; Fig. S11). It differs, however, at several positions, such as Glu134, Asp162, Asn255, Thr491 and Ala575, which are otherwise highly conserved in archaeal ATP-dependent DNA ligases (corresponding LigFa residues: Lys134, Glu162, Gly255, Ser491 and Pro575). To determine whether one or more of these differences may account for the differences in properties of LigFa and the other archaeal DNA ligases, we created and purified single mutant variants of LigFa carrying the conserved residues (SI Text). The single mutations Asp162→Glu162 (kcat 3.00 ± 0.72 min−1), Asn255→Gly255 (kcat 3.07 ± 0.92 min−1), Thr491→Ser491 (kcat 2.90 ± 0.96 min−1) and Ala576→Pro576 (kcat 3.04 ± 0.73 min−1) hardly affected ligation activity at pH 3.0, whereas the Glu134→Lys134 mutation caused a 60% reduction at this pH, with no net change in iron content and purple color (Fig. 3C): its kcat for nicked DNA fell from 3.57 ± 0.53 (wild type) to 1.5 ± 0.31 min−1 (Fig. 3B). The mutant Glu134→Lys134 LigFa variant had a similar CD spectrum to that of wild type LigFa (Fig. 3D). Determination of the pH optima of the mutant ligases revealed no changes except for the Glu134→Lys134 mutant, which had an activity optimum of pH ≈5.0, two pH units above that of LigFa, and at its pH optimum, a 1.5-fold higher turnover number (5.3 vs. 3.6 min−1) (Fig. 3B) than the wild-type enzyme. The Glu134→Lys134 and the ferrous LigFa forms are therefore phenotypically similar in regard to their pH activity optima, although not in regard to their catalytic rates. This suggests that Glu134 contributes to the acidic phenotype of LigFa, although further analysis is required to define the functional role of this residue in the enzyme and its relationship to the iron clusters in LigFa.
Tyr55 and Tyr129.
The color/spectroscopic characteristics of LigFa indicate Fe (III)-tyrosinate species involvement in iron coordination. To gain insights into the possible identity of such residues, all 23 tyrosine residues in the protein were individually replaced by threonines through site-directed mutagenesis and the resulting variants purified and characterized (SI Text). Only 2 of 23 of the enzyme variants, Tyr55→Thr55 and Tyr129→Thr129, exhibited properties significantly different from those of the wild-type enzyme; both lacked iron, catalytic activity, absorbance at 564 nm (Table S2), and purple color (Fig. 3C). This is consistent with Fe-tyrosinate complexes being mediators of iron coordination in LigFa and points to Tyr55 and Tyr129 as iron ligands that play a key role in the assembly and/or maintenance of iron atoms in LigFa. That neither of the two iron atoms of LigFa was retained in the Tyr55 and Tyr129 mutants suggests loss of both atoms may occur under circumstances where retention of one is hampered.
These results demonstrate that the unusual properties of LigFa are genetically determined and identify Phe192 and Glu134 as residues implicated in the acidophilic phenotype of the enzyme and Tyr55 and Tyr129 as potential ferric iron ligands.
Discussion
We report here the finding of a purple DNA ligase, LigFa, a ferric iron-containing enzyme with an exceptionally low pH activity optimum in vitro. Ferric iron content, low pH optimum, and high catalytic activity appear to be obligatorily coupled properties, because chemical (reduced) or genetic (Phe192 and Glu143 mutants) variants having either ferrous iron or no iron are characterized by low activities and pH activity optima 2–3 units higher than the parental enzyme. Because removal of the iron results in structural rearrangement and partial or total loss of enzyme activity, a key role of the iron is clearly the organization and/or maintenance of protein secondary structure, a function we have termed the “iron rivet” (11). However, that other DNA ligases require added Mg2+ or K+ for DNA binding, and that these metals do not stimulate LigFa activity, suggest the possibility that the ferric cations may play a similar role in substrate DNA binding. In this context, it is generally accepted that the “two-magnesium-ion” functional mechanism of “common” DNA ligases involves the positioning of catalytic groups, neutralizing the negative charges of the environment, stabilizing the active site structure, catalyzing the nucleotide insertion and transferring the AMP moiety from the ligase to the 5′-phosphate at the ligation junction, through coordination of the α-phosphate of nucleotide cofactor and the 3′-O of the DNA molecule. All of these roles may be assumed by the iron (III) ions in LigFa (see details in Fig. S12). Indeed, it may be recalled that reduction of the ferric ions to ferrous produced a LigFa with much lower DNA-binding activity, and that Mössbauer spectroscopy detected major changes in the ligand symmetry around the iron centers in response to DNA substrate binding. This suggests perhaps that Fe3+ modifies pKa values or the positioning of amino acid residues involved in DNA binding and/or acts as a Lewis acid without changing its oxidation state (as expected for a DNA ligation reaction where no electron flux is involved) and directly participates in enzyme binding to the phosphodiester backbone of DNA substrate molecules (see details in Fig. S12). The finding that other metals with Lewis acid character can substitute (albeit less effectively) ferric iron in LigFa is consistent with this notion. A functional role for Fe3+ as a Lewis acid has been proposed for purple acid phosphatases, nitrile hydratase, acotinase, and intradiol dioxygenase (16). Other potential roles of iron include pH- and/or metal-dependent folding effects, or alternatively, the formation of metal-ion hydroxides acting as a general base, similar to that characterizing some ribozyme-mediated RNA cleavage reactions (22).
LigFa is a remarkable protein, not only because it is thus far unique and even unlike DNA ligases of close phylogenetic and ecophysiological neighbors, but also because its features seem physiologically counterintuitive. DNA ligases are central enzymes in the replication and repair of DNA and thus play a pervasive role in genome duplication and maintenance of genome integrity. LigFa properties seem to be counterintuitive because low pH promotes depurination, a mutagenic activity, and Fe efficiently mediates redox reactions that may generate mutagenic oxygen radicals, two features that would tend to counteract the DNA integrity maintenance function of a DNA ligase (23). These considerations might suggest that Ferroplasma could be a highly mutable organism with an unstable genome, but this is unlikely to be the case for a small genome chemoautotroph. The tendency of iron to mediate potentially harmful redox reactions might, however, be minimized through restrictive coordination in the protein interior (23). Consistent with this is that LigFa neither showed any appreciable capacity to be reoxidized after electrochemical reduction in vitro nor released the reduced iron. Therefore, the chemical/ionic environments of the ferric centers in LigFa may restrict redox activity.
Also counterintuitive is the finding that enzyme exhibits a pH activity optimum 2 pH units lower than that assumed for the cytoplasm of the Ferroplasma cell, the so-called “pH anomaly” that characterizes several enzymes of this organism (24). This uniquely (for DNA ligases) low pH activity optimum of isolated LigFa is, however, an in vitro value. Moreover, the enzyme may not (need to) operate at optimal efficiency under physiological conditions. In this context, it may be recalled that the DNA ligase of “Ferroplasma acidarmanus” exhibits an in vitro optimal activity of only 10% of that of LigFa, and that LigFa exhibits a similar activity in the pH range thought to characterize the cytoplasm of Ferroplasma. However, pH measurements for cytoplasm are average values, and there may exist different cellular compartments characterized by distinct pH values. Furthermore, LigFa may operate in vivo as one element of a multicomponent cellular machinery that either functions as an entity at normal cytoplasmic pH values (in this case, the low pH activity optimum of LigFa in vitro would reflect an inherent property of the isolated ligase but not of its physiological form) or that provides a local physical-chemical environment for the LigFa element characterized by low pH. Another factor may be a highly positively charged cytoplasmic environment in Ferroplasma, resulting from a repertoire of proteins with high densities of positively charged amino acids on their surfaces, a strategy known to be exploited by acidophiles to compensate for high extracellular proton concentrations (13–15) and an environment proxied in vitro by low pH. Indeed, the reason for the pH anomaly of Ferroplasma proteins in general, and of LigFa in particular, may well be a combination of these factors. LigFa of F. acidiphilum is the pioneer protein of a thus far unique iron-protein-dominated metabolic machinery, the investigation of which promises to reveal entirely new biochemical and evolutionary features of proteins.
Materials and Methods
A full description of materials and methods is available in SI Text.
Protein Samples.
All recombinant DNA ligases used in the present study were PCR-amplified from genomic DNA, cloned, expressed, and purified as described in SI Text. Details of the preparation of reduced, metal-free, and LigFa variants containing metals other than iron and protein variants containing 57Fe and 57Fe-DNA are given in SI Text.
Nick-Joining DNA Ligase Assay (Standard Assay).
For kinetic studies of DNA ligases, we modified a fluorimetric assay based on the ligation of a 5′-phosphorylated 35-mer (5′-TAA GCT CCG GAT TGT CCG GGA GGT AAA GCC CTG AT-3′), to a 5′-fluorescein-labeled 25-mer (5′-CAC AGG AAG CTC TAC AGG TAC TCC G-3′), annealed to the complementary 70-mer (5′-TGG TCA TCA GGG CTT TAC CTC CCG GAC AAT CCG GAG CTT ACG GAG TAC CTG TAG AGC TTC CTG TGC AAG C-3′). In this reaction, the 3′-OH of the 25-mer is ligated to the phosphorylated 5′-PO4 of the 35-mer to produce a 60-mer, which is detected after denaturing from the 70-mer. Assays were performed in a final volume of 20 μl of buffer containing 10 μM of the 35-and 25-mer, 5 μM of the 70-mer, 5 mM DTT, 0.1 mM ATP, 25 μg/ml BSA, and 20 nM DNA ligase. The reaction medium was supplemented with appropriate amount of MgCl2 when analyzing DNA ligases other than LigFa. For kinetic studies, 75–100 pmol of 5′-fluorescein labeled “substrate template” were used. Samples were incubated for 30 min at 40°C for LigFa and LigAf, 70°C for LigTa and LigPt, or 80°C for LigSa, and the reactions stopped with 8 μl of 98% (vol/vol) formamide, 10 mM EDTA, and 0.25 mg/ml bromophenol blue. After heating for 3 min at 95°C, reaction mixtures consisting of substrates (25- and 35-meric oligonucleotides), annealing substrate (70-mer), and the ligation product (60-mer) were fractionated by electrophoresis on a 16% denaturing PAGE, which was subsequently scanned with a Molecular Dynamics densitometer and the intensity of bands quantified by volume integration, using ImageQuant software (Bio-Rad). The system was calibrated with a 20-pmol to 500-fmol range of the fluorescein-labeled 25-nt substrate. One unit of DNA ligase is defined as the amount of protein that ligates 1 pmol of substrate-template in 30 min at the corresponding temperature.
Supplementary Material
Acknowledgments.
We thank Rita Getzlaff (HZI) for protein sequence analyses. A.B. thanks the Spanish Ministry of Science and Education for a Formacion de Profesorado Universitario (FPU) fellowship. K.N.T. thanks the Fonds der Chemischen Industrie for generous support. Work carried out at HZI was supported by Grant 0313751K from the Federal Ministry for Science and Education (BMBF) within the GenoMikPlus initiative.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. AJ850915).
This article contains supporting information online at www.pnas.org/cgi/content/full/0800071105/DCSupplemental.
References
- 1.Martin IV, MacNeill SA. ATP-dependent DNA ligases. Genome Biol. 2002;3:1–7. doi: 10.1186/gb-2002-3-4-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson A, O'Donnell M. DNA ligase: Getting a grip to seal the deal. Curr Biol. 2005;15:90–92. doi: 10.1016/j.cub.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 3.Li JJ, Kelly TJ. Simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1984;81:6973–6977. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jessberger R, Berg P. Repair of deletions and double-strand gaps by homologous recombination in a mammalian in vitro system. Mol Cell Biol. 1991;11:445–457. doi: 10.1128/mcb.11.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood RD, Robins P, Lindahl T. Complementation of the xeroderma pigmentosum DNA repair defect in cell-free extracts. Cell. 1988;53:97–106. doi: 10.1016/0092-8674(88)90491-6. [DOI] [PubMed] [Google Scholar]
- 6.Nakatani M, Ezaki S, Atomi H, Imanaka T. DNA ligase from a hyperthermophilic archaeon with unique cofactor specificity. J Bacteriol. 2002;182:6424–6433. doi: 10.1128/jb.182.22.6424-6433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rolland JL, Gueguen Y, Persillon C, Masson JM, Dietrich J. Characterization of a thermophilic DNA ligase from the archaeon Thermococcus fumicolans. FEMS Microbiol Lett. 2004;236:267–273. doi: 10.1016/j.femsle.2004.05.045. [DOI] [PubMed] [Google Scholar]
- 8.Keppetipola N, Shuman S. Characterization of a thermophilic ATP-dependent DNA ligase from the euryarchaeon Pyrococcus horikishii. J Bacteriol. 2005;187:6902–6908. doi: 10.1128/JB.187.20.6902-6908.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishida H, Tsuchiya D, Ishino Y, Morikawa K. Overexpression, purification and crystallization of an archaeal DNA ligase from Pyrococcus furiosus. Acta Crystallogr F. 2005;61:1100–1102. doi: 10.1107/S1744309105038649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golyshina OV, et al. Ferroplasma acidiphilum gen nov, sp nov, an acidophilic, autotrophic, ferrous-iron-oxidizing, cell-wall-lacking, mesophilic member of the Ferroplasmaceae fam nov, comprising a distinct lineage of the Archaea. Int J Syst Evol Microbiol. 2000;3:997–1006. doi: 10.1099/00207713-50-3-997. [DOI] [PubMed] [Google Scholar]
- 11.Ferrer M, Golyshina OV, Beloqui A, Golyshin PN, Timmis KN. The cellular machinery of Ferroplasma acidiphilum is iron-protein dominated. Nature. 2007;445:91–94. doi: 10.1038/nature05362. [DOI] [PubMed] [Google Scholar]
- 12.Golyshina OV, Timmis KN. Ferroplasma and relatives, recently discovered cell wall-lacking archaea making a living in extremely acid, heavy metal-rich environments. Environ Microbiol. 2005;7:1277–1288. doi: 10.1111/j.1462-2920.2005.00861.x. [DOI] [PubMed] [Google Scholar]
- 13.Searcy DG. Thermoplasma acidophilum: intracellular pH and potassium concentration. Biochim Biophys Acta. 1976;451:278–286. doi: 10.1016/0304-4165(76)90278-6. [DOI] [PubMed] [Google Scholar]
- 14.Van de Vossenberg JL, Driessen AJ, Zillig W, Konings WN. Bioenergetics and cytoplasmic membrane stability of the extremely acidophilic, thermophilic archaeon Picrophilus oshimae. Extremophiles. 1998;2:67–74. doi: 10.1007/s007920050044. [DOI] [PubMed] [Google Scholar]
- 15.Macalady JL, et al. Tetraether-linked membrane monolayers in Ferroplasma spp: A key to survival in acid. Extremophiles. 2004;8:411–419. doi: 10.1007/s00792-004-0404-5. [DOI] [PubMed] [Google Scholar]
- 16.Merkx M, Averill BA. The activity of oxidized bovine spleen purple acid phosphatase is due to an Fe(III)Zn(II) “impurity”. Biochemistry. 1998;37:11223–11231. doi: 10.1021/bi980389r. [DOI] [PubMed] [Google Scholar]
- 17.Zambonelli C, Roberts MF. An iron-dependent bacterial phospholipase D reminiscent of purple acid phosphatases. J Biol Chem. 2003;278:13706–13711. doi: 10.1074/jbc.M210363200. [DOI] [PubMed] [Google Scholar]
- 18.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nature Protocols. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlach M, Trautwein AX, Zecca L, Youdim MB, Riederer P. Mössbauer spectroscopic studies of purified human neuromelanin isolated from the substantia nigra. J Neurochem. 2007;65:923–926. doi: 10.1046/j.1471-4159.1995.65020923.x. [DOI] [PubMed] [Google Scholar]
- 20.Allen EE, et al. Genome dynamics in a natural archaeal population. Proc Natl Acad Sci USA. 2007;104:1883–1888. doi: 10.1073/pnas.0604851104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson BR, Noble C, Lavesa-Curto M, Bond PL, Bowater RP. Characterization of an ATP-dependent DNA ligase from the acidophilic archaeon “Ferroplasma acidarmanus” Fer1. Extremophiles. 2007;11:315–327. doi: 10.1007/s00792-006-0041-2. [DOI] [PubMed] [Google Scholar]
- 22.Pyle AM. Ribozymes: A distinct class of metalloenzymes. Science. 2003;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- 23.Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 24.Golyshina OV, et al. The “pH optimum anomaly” of intracellular enzymes of Ferroplasma acidiphilum. Environ Microbiol. 2006;8:416–425. doi: 10.1111/j.1462-2920.2005.00907.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.