Abstract
Ischemic brain injury causes tissue damage and neuronal death. The deficits can often be permanent because adult neurons fail to regenerate. One barrier to neuronal regeneration is the formation of the glial scar, a repair mechanism that is otherwise necessary to seal off necrotic areas. The process of gliosis has been well described, but the mechanisms regulating the robust production of scar components after injury remain poorly understood. Here we show that the early growth response 1 transcriptional factor (Egr-1, also called Krox24, Zif268, and NGFI-A) is expressed in astrocytes in the ventricular wall, corpus callosum, and striatum of normal mouse brain. After experimental stroke caused by permanent occlusion of the middle cerebral artery, Egr-1 was expressed long term in reactive astrocytes that accumulate around the injury site. Gain- and loss-of-function studies in primary astrocytes indicated that Egr-1 regulates the transcription of chondroitin sulfate proteoglycans genes, the main extracellular matrix proteins of the glial scar. Egr-1 bound to a site within the phosphacan promoter and transactivated its expression. Egr-1-deficient mice accumulated lower levels of phosphacan RNA and protein than wild-type mice after stroke, but there were no measurable differences in neurite outgrowth toward the infarct area between the two groups. Our findings suggest that Egr-1 is an important component of the transcriptional network regulating genes involved in gliosis after ischemic injury.
A critical event after acute brain ischemia is the accumulation of astrocytes in the border zone of the tissue injury. Reactive astrocytes produce neuroprotective factors that promote neuronal survival. They also build the glial scar that isolates necrotic areas from the surrounding tissue.1 This phenomenon, called gliosis, involves large production of extracellular matrix (ECM) proteins, the molecular components of the scar. The glial scar protects healthy brain areas from exposure to toxic elements and cellular debris in the injured tissue and prevents excessive damage from the strong inflammatory response associated with the infarct. In addition to the protective effects of the dense scar, it is postulated that it forms a biochemical and mechanical obstacle to neuronal regeneration beyond the injury site.2,3,4
Recent studies implicate astrocytes-derived ECM products, such as chondroitin and keratan sulfate proteoglycans (CSPGs and KSPGs), as negative barriers of axon regeneration after stroke or mechanical injury to the central nervous system. These inhibitory proteoglycans are secreted rapidly (within 24 hours) after injury and can persist in the affected sites for many months.2,5,6,7,8 The molecular basis for this process remains unknown.
The zinc finger transcription factor Egr-1 (also known as Krox24, Zif268, or NGFI-A) was originally identified as an immediate-early serum-inducible gene in quiescent fibroblasts.9 Egr-1 undergoes rapid, transient activation downstream of a number of growth factors and environmental stress signals including hypoxia, physical force, or vascular injury.10 Once activated, Egr-1 regulates the transcription of a diverse array of genes including cytokines (interleukin-1β, platelet-derived growth factor A), chemokines (MIP-2, MCP-1), intercellular adhesion molecules (ICAM-1), coagulation proteins (plasminogen activator inhibitor-1, tissue factor), ECM components (fibronectin), and metalloproteases (MT1-MMP).11,12,13,14,15,16,17 Egr-1 can act synergistically with the hypoxia-inducible factor HIF-1α in promoting expression of genes such as vascular endothelial growth factor and plasminogen activator inhibitor-1.14,18,19
Mice with inactive Egr-1 gene are viable,20 but show different responses to pathological conditions compared with wild-type animals. Analysis of Egr-1-null mice in a lung ischemia/reperfusion model reveal that Egr-1 coordinates the expression of genes with critical roles in inflammation, coagulation, and vascular hyperpermeability.14 Egr-1-deficient mice also show impaired prostate tumorigenesis21 and decreased vascular wall inflammation when crossed to apolipoprotein E knockout mice.22 In the adult nervous system, Egr-1 is involved in neuronal plasticity and neurite outgrowth, and gene microarray data detected an up-regulation of Egr-1 after cerebral ischemia.23,24 These observations suggest that Egr-1 plays a role in postischemic events in the brain after stroke. In this report, we demonstrate that after focal cerebral ischemia, reactive astrocytes within the glial scar express high levels of Egr-1, which is shown by histological and molecular analyses to be an important transcriptional regulator of glial scar components.
Materials and Methods
Animal Breeding and Genotyping
C57BL/6J mice were purchased from Charles River, Sulzfeld, Germany. Egr-1-deficient mice (Egr-1−/−)20 and control littermates (wild type) came from Taconic (Hudson, NY). Heterozygous Egr-1+/− mice were bred to generate Egr-1+/−, Egr-1−/−, and wild-type siblings. Genomic DNA was isolated from tail biopsies and genotyped by polymerase chain reaction (PCR) analysis as previously described.20
Permanent Focal Cerebral Infarction in Mice and Rats
Animal care and experimental procedures were performed in accordance to the German and National Institutes of Health animal legislation guidelines and were approved by the local animal care and use committees. Animals were housed under standard conditions with free access to mice chow and tap water before and after surgery. Irreversible occlusion of the left middle cerebral artery (MCA) was performed as described previously in 8-week-old male mice weighing 20 to 26 g.25,26 Briefly, animals were anesthetized by intraperitoneal injection of ketamine (80 mg/kg; Apharmo, Arnhem, The Netherlands) and xylazine (10 mg/kg; Bayer, Leverkusen, Germany). Using an operating microscope, a U-shape incision was made between the left ear and the left eye. The top and back segments of the temporal muscle were transected, and the skull was exposed by retraction of the temporal muscle. A small opening (1 to 2 mm in diameter) was made in the region over the MCA with a handheld drill. The MCA was occluded by ligation with a 10-0 nylon thread (Ethylon, Norderstedt, Germany) and transected distally to the ligation point. Finally, retracted soft tissues were replaced, wounds were sutured, and mice were placed back into their cages. Sham-operated control animals were prepared in a similar manner except that the exposed MCA was not occluded. Body temperature was maintained at 37°C during surgery and until the animals regained consciousness by a rectal probe connected to a heating pad. Thereafter, rectal temperature was checked every 10 to 15 minutes during the following 2 hours and, if necessary, it was corrected to 37°C by placing a heating pad below the cage. After middle cerebral artery occlusion (MCAO), animals were analyzed at 12, 24, and 48 hours, 4 days, 1 week, 10 days, and 6 weeks after surgery (n = 5 plus 3 sham-operated mice per group). Male Long-Evans rats (Charles River), 250 to 280 g, were used in ischemia experiments. Permanent distal MCAO was performed according to the method described previously.27 Brain tissues were analyzed 4 days, 10 days, and 6 weeks after MCAO (n = 5 per group).
Immunohistological Analysis
Mice were perfused via the left ventricle of the heart with saline. Brain tissues were dissected and frozen in OCT embedding medium (Sakura Finetec, Torrance, CA) using dry-ice-cooled isopentane. Tissues were stored at −80°C until further processing. For staining with the neurocan (rat model)- and phosphacan KAF13 (mouse model)-recognizing antibodies, we perfused deeply anesthetized mice and rats with a phosphate-buffered 4% paraformaldehyde solution. Brains were removed, incubated in 4% paraformaldehyde, submerged in 30% sucrose for 24 hours, and frozen in OCT embedding medium as described above. Embedded frozen tissues were sectioned at 10 μm with a Leica cryostat (Wetzlar, Germany). Frozen sections were thawed on silanized (3-aminopropyltriethoxysilane; Fluka, Deisenhofen, Germany) glass slides and dried for at least 2 hours at 37°C. We performed serial sectioning of the infarcted area using every 10th section for antigen-specific antibody staining.
Immunohistochemistry and immunofluorescence were performed as described previously.26 Cryosections were washed thoroughly in phosphate-buffered saline (PBS) before and after each incubation step. Nonspecific binding sites were blocked by incubation in 20% normal goat serum (Sigma, Deisenhofen, Germany) and 5% bovine serum albumin in PBS (BSA Fraction V, Sigma). We used the following primary antibodies: mouse anti-glial fibrillary acidic protein (GFAP, 1:500 dilution; Chemicon-Millipore, Temecula, CA), rabbit anti-Egr-1 (catalogue number SC-110, 1:50 dilution; Santa Cruz, Heidelberg, Germany), rat anti-CD31 (1:100 dilution; BD Biosciences, Heidelberg, Germany), mouse anti-neuronal nuclei (NeuN) protein (1:100 dilution; Molecular Probes-Invitrogen Corporation, Carlsbad, CA), mouse anti-α-smooth muscle actin (1:400 dilution, BD Biosciences), rabbit anti-ephrin B1 (C-18, 1:50 dilution, recognizing also ephrin B2; Santa Cruz), anti-160-kDa neurofilament medium protein (1:400 dilution; Abcam, Cambridge, UK), mouse anti-neurocan (1:200 dilution, Chemicon), rabbit anti-NG2 chondroitin proteoglycan (1:400 dilution, Chemicon), and rabbit anti-laminin-α1 and rabbit anti-laminin-γ1 (1:500 dilution; a kind gift of the late Dr. R. Timpl, Munich, Germany). For detection of phosphacan, the following antibodies were used: rabbit KAF13 anti-phosphacan/RPTPβ (1:200 dilution) and rabbit PTP1 anti-RPTPβ28 (1:100 dilution; antibodies were a kind gift of Dr. A. Faissner, Bochum, Germany). To further evaluate phosphacan protein in brain tissue, we have also used mouse anti-phosphacan (1:200 dilution, Chemicon), mouse anti-phosphacan (1:200 dilution, clone 3F8; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), and rat monoclonal IgM 473 HD anti-DSD-128 (1:200 dilution) with comparable results. As secondary antibodies, we used goat anti-mouse Alexa 488- or 568-conjugated IgG (7.5 μg/ml), goat anti-rabbit Alexa 488- or 568-conjugated IgG (7.5 μg/ml), and mouse anti-rat Alexa 488- or 568-conjugated IgG (7.5 μg/ml) from Molecular Probes, as well as the biotinylated goat anti-rabbit and the horseradish peroxidase-coupled rabbit anti-rat IgM secondary antibodies from Dianova (1:200 dilution; Hamburg, Germany). Omission of primary antibodies served as controls. For co-stains with two rabbit polyclonal antibodies, we used the ZenonAlexa Fluor 488 rabbit IgG labeling kit (Invitrogen). Staining using mouse monoclonal antibodies on mouse tissue sections was performed with the M.O.M. immunodetection kit (Vector, Burlingame, CA) to reduce unspecific background. Slides for peroxidase staining were incubated for 30 minutes at room temperature with peroxidase-conjugated streptavidin (Vectastain KIT ABC, Vector). Thereafter, slides were rinsed four times with PBS and incubated with 3-amino-9-ethylcarbazole (Vector) and 0.006% H2O2. Color developed within 30 minutes. Sections were rinsed in distilled H2O, counterstained with hematoxylin, and mounted with elvanol.
For immunofluorescence analysis of cells transfected with the CMV-Egr-1-IRES-EGFP plasmid, cell growth medium was removed 24 hours after transfection and cells were washed two times with PBS and then fixed with 4% paraformaldehyde in PBS for 10 minutes at 4°C. Paraformaldehyde was removed and cells were washed five times with blocking buffer (1% bovine serum albumin, 0.05% saponin in PBS) and then kept in blocking buffer for 2 hours at 4°C. The blocking buffer was removed and the cells were incubated overnight with rabbit anti-green fluorescent protein Alexa Fluor 488 conjugate (Invitrogen) together with either anti-laminin α1 antibody, rabbit PTP1 anti-RPTPβ (transmembrane phosphacan isoform), rabbit anti-GFAP antibody (Abcam), or mouse anti-β-tubulin antibodies (Sigma). The next day cells were washed five times with PBS, incubated first with blocking buffer for 30 minutes and then with the secondary antibody (goat anti-rabbit Alexa 568, 1:600 dilution) at room temperature for 2 hours. Cells were washed in PBS and analyzed. Photographic documentation of stained tissue sections and cells was performed with a Zeiss Axiovert 135 or a Zeiss Axiovert 200M microscope (Oberkochen, Germany). Confocal images were recorded with a Leica TCS SP5 microscope. To measure neurite outgrowth and scar area size, images were digitalized and analyzed using ImageJ software (National Institutes of Health, Bethesda, MD).
Isolation of Total RNA and Reverse Transcriptase (RT)-PCR Analysis
Total RNA was extracted and prepared from cultured cells as well as brain hemispheres (ipsilateral and contralateral) 10 days after permanent occlusion of the MCA using the RNA lipid tissue mini kit (Qiagen, Hilden, Germany). Residual genomic DNA was removed by the RNase-Free DNase Set (Qiagen) and RNA was reverse-transcribed into cDNA as follows: 1 μg of RNA was mixed with 100 ng of oligo(dT)15 and incubated for 5 minutes at 65°C. One mmol/L dNTPs, 60 mmol/L KCl, 15 mmol/L Tris-Cl, pH 8.4, 3 mmol/L MgCl2, 0.3% Tween 20, 10 mmol/L β-mercaptoethanol, 10 U RNasin (Promega, Madison, WI), and 100 U Mo-MLVRT (Invitrogen) were added, and the mix was incubated for 55 minutes at 37°C followed by enzyme inactivation for 5 minutes at 95°C. PCR was performed with 20 ng of cDNA using TaqDNA polymerase (Promega) for 24 to 35 cycles (1 minute at 95°C, 1 minute at 60 to 65°C, and 1 minute at 72°C). Primer sequences are listed in Table 1. Real-Time PCR was performed with Lightcycler in a reaction volume of 20 μl using SYBR Green I (LC-FastStart DNA MasterPlus; Roche, Mannheim, Germany) according to the manufacturer’s protocol. Primers (Table 1) were used at a final concentration of 0.5 μmol/L each. An initial denaturation step at 95°C for 10 minutes was followed by 45 cycles of denaturation (95°C, 1 second), annealing (65°C, 10 seconds), and extension (72°C, 20 seconds).
Table 1.
List of Primer Sequences Used for Gene Expression Analysis and Molecular Cloning
| Gene | Forward primer | Reverse primer | Product (bp) |
|---|---|---|---|
| Primers used for RT-PCR and Q-PCR analysis
| |||
| Aldolase | 5′-AGCTGTCTGACATCGCTCACCG-3′ | 5′-CACATACTGGCAGCGCTTCAAG-3′ | 571 |
| m_β-actin | 5′-CTACGAGGGCTATGCTCTCCC-3′ | 5′-CCGGACTCATCGTACTCCTGC-3′ | 602 |
| CSPG2 | 5′-GCCTTCAGTTCAGTAGACAGACTTC-3′ | 5′-AGTAGCATTTGCACACTCTTCTTCT-3′ | 573 |
| CSPG3 | 5′-TGCAACTACAACCTACCCTATGTCT-3′ | 5′-ACTCTCTTGTTTCTCTCTGGGTTCT-3′ | 460 |
| CSPG4 | 5′-GGGTCTGAGGATCTGGTCTACA-3′ | 5′-CTCATACAGAATATTCCCAGCGTAG-3′ | 465 |
| h_EGR-1 | 5′-CGGCAGAAGGACAAGAAAGCAGAC-3′ | 5′-GGGGAAGTGGGCAGAAAGGATTG-3′ | 561 |
| m_Egr-1 | 5′-CGAACAACCCTATGAGCACCTG-3′ | 5′-CAGAGGAAGACGATGAAGCAGC-3′ | 276 |
| m_Egr-1 genotype | 5′-AGGCTCTTAATACCACCTACCAATC-3′ | 5′-TTTCTTGTCCTTCTGTCTTAAATGG-3′ | 362 |
| m_Egr-2 | 5′-CCCAGTAACTCTCAGTGGTTTTATG-3′ | 5′-ATTGATGATACCTTCTGGATAGCAG-3′ | 313 |
| m_Egr-3 | 5′-CCATTACAATCAGATGGCTACAG-3′ | 5′-TAGTCAGGAATCATGGGGAAAAGAT-3′ | 450 |
| m_Egr-4 | 5′-GTATCCTGGAGGCGACTTCTT-3′ | 5′-CCTCTACTCCCCAGATCTGAGTT-3′ | 314 |
| Ephrin B2 | 5′-GTTGATAAAGACCAAGCAGACAGAT-3′ | 5′-GATGATGATGACGATGAAGATGAT-3′ | 489 |
| Fibronectin | 5′-GTCTATTCGCCATCAGTAGAAGGTA-3′ | 5′-CCAGGACAGTAGAATCAGTTTCATT-3′ | 514 |
| GAPDH | 5′-AGAACATCATCCCTGCCTCTACTG-3′ | 5′-TGTCGCTGTTGAAGTCAGAGGAGA-3′ | 258 |
| m_GFAP | 5′-AGGAGATCCAGTTCTTAAGGAAGAT-3′ | 5′-GCTTAACGTTGAGTAGATCCTGGTA-3′ | 486 |
| Laminin α1 | 5′-AGAGTGAGACAGGAACAAGAAGTAG-3′ | 5′-TAGCCAGAAGTACACACATCACAAT-3′ | 475 |
| Laminin α2 | 5′-GACCAGATGATTAAAGAACTGAGGA-3′ | 5′-CTCAGACATAGGTGGCAATTTAGTT-3′ | 441 |
| Laminin β1 | 5′-TGATTCTCGATATTCTGACATTGAA-3′ | 5′-CACGTACATCGTTCACAGAAATTAG-3′ | 630 |
| Phosphacan | 5′-AAAGGGAAGTTAAGAGCTTTATCCA-3′ | 5′-GAATCTCTTCCTTTCCAGTGTATGA-3′ | 403 |
| m_Phosphacan
|
5′-CATATACTGGAAAGGAAGAGATCCA-3′
|
5′-GTTCCTTTTTCCTTATTTGGTTTGT-3′
|
480
|
| T7 promoter-containing human EGR-1 PCR primers for siRNA preparation*
| |||
| EGR-1
|
5′-taatacgactcactatagggGGTCAGTGGCCTAGTGAGCATG-3′
|
5′-taatacgactcactataggggTCTGCTTTCTTGTCCTTCTGCCG-3′
|
559
|
| Primers used for mouse Egr-1 full length cloning†
| |||
| m_Egr1
|
5′-ggaattcCGCTGGTCCGGGATGGCAGCGGCC-3′
|
5′-gggatcccGCTTTCTTTTATTCCCTTTAGCAAAT-3′
|
1739
|
| OGNs used in the electrophoresis mobility shift assays‡
| |||
| Phos-1 | 5′-CTGCGCCCCCGCCCCCTCCAG-3′ | 5′-CTGGAGGGGGCGGGGGCGCAG-3′ | |
| GCE | 5′-GATCTCTCTCCTCCCCCGCGCCCCGGGG-3′ | 5′-CCCCGGGGCGCGGGGGAGGAGAGAGATC-3′ | |
| mGCE | 5′-GATCTCTCTCCTATACCGCGCCCCGGGG-3′ | 5′-CCCCGGGGCGCGGTATAGGAGAGAGATC-3′ | |
| OCT-1
|
5′-TGTCGAATGCAAATCACTAGAA-3′
|
5′-TTCTAGTGATTTGCATTCGACA-3′
|
|
| Primers used for human phosphacan promoter constructs§
| |||
| PTPRZ2_402 | 5′-CTGCGCCCCCGCCCCCTCCAG-3′ | 5′-CACTCTGAGAAGCAGAGGAGCCGC-3′ | |
| PTPRZ2_402mut | 5′-CTGCGCATATGCCCCCTCCAG-3′ | Same as above | |
| PTPRZ2_390
|
5′-CCCCTCCAGGAGCCGCGGCGC-3′
|
Same as above
|
|
| Primers used in the ChIP analysis
| |||
| Egr-1 site | 5′-GATTGTCGGTGTGTGAATTG-3′ | 5′-CTGTGCGCGCCGCGGCTCCTG-3′ | 149 |
| Distal 4.5 kb | 5′-CCTCCTACTTGTTTTATGCCTGATA-3′ | 5′-TTAAAAAGCAGTTCATTCCTACCTG-3′ | 109 |
T7 recognition sequences are underlined and in italics.
Sequences for the restriction enzymes EcoRI and Bam HI, with an extra g added to improve digestion efficiency, are underlined and in italics.
Mutated nucleotides appear in bold and italics. The Egr-1 consensus site is underlined.
The Egr-1 consensus site is underlined. Mutated nucleotides appear in bold and italics.
Plasmids and Transfection
Mouse Egr-1 full-length cDNA was generated by PCR amplification of cDNA from mouse brain tissue using primers listed in Table 1. The PCR product was digested with the indicated restriction enzymes and cloned into the pBluescriptII SK(+/−) vector (Stratagene, La Jolla, CA) and then into the pIRES2-EGFP (enhanced green fluorescent protein) expression vector (BD Biosciences) to generate CMV-Egr-1-IRES-EGFP. The inserts of the resulting plasmids were sequenced to confirm the identity and orientation of the constructs.
For transfection experiments, we used human nontransformed, nonimmortalized, primary astrocytes from Cambrex (East Rutherford, NJ) maintained in AGM, ie, astrocyte medium including serum-free basal medium and SingleQuot growth factors (Cambrex). HeLa cells were cultured under standard conditions. For transient transfection of the CMV-Egr-1-IRES-EGFP construct, astrocytes were grown to 80% confluence in 10-cm tissue culture dishes, medium was exchanged, and cells received fresh serum-free medium without antibiotics for the next 24 hours. Approximately 8 μg of plasmid were used for transfection with the Lipofectamine 2000 reagent following the manufacturer’s specifications (Invitrogen). Transfection with the empty CMV-pIRES2-EGFP vector served as control. Cells were harvested 24 hours later for protein and RNA analysis.
To silence Egr-1, we used the Silencer siRNA cocktail kit protocol according to the manufacturer’s instructions (Ambion, Austin, TX). The PCR primers from the human Egr-1 gene with appended T7 promoter sequences to the 5′ end are included in Table 1. The primers were annealed to Egr-1 cDNA and the two complementary RNA transcripts were synthesized by T7 polymerase. The resulting dsRNA was cleaved with RNase III to produce siRNA (12 to 30 bp long) containing a mixture of siRNA target sites. Twenty-four hours before transfection, cells received fresh medium without antibiotics. To transfect one six-well plate, 2.5 μg of siRNA were mixed with 100 μl of Opti-MEM1 (Invitrogen). In a separate tube, 6 μl of oligofectamine were added to 24 μl of Opti-MEM1, gently mixed, and incubated for 10 minutes at room temperature. After incubation, both solutions were mixed together and incubated for another 25 minutes at room temperature. Finally, liposome complexes were added to the culture wells and carefully mixed for 30 seconds. Cells were incubated for 70 hours at 37°C before harvesting. Control cells received oligofectamine alone, or oligofectamine containing siRNAs against GAPDH (Ambion) or control (scrambled) siRNA (Qiagen). To ensure the specificity of the Egr-1 siRNA results, the knockdown experiments were repeated with the TranSilent human Egr-1 siRNA vector mix from Panomics (Redwood City, CA) with the same results. In this case, we used Lipofectamine 2000 as the transfection agent.
For phosphacan promoter analysis, we scanned the genomic sequences upstream of the human phosphacan gene for the presence of putative Egr-1 sites. Egr-1 binds preferentially, but not exclusively, to GC-rich elements with a consensus sequence of 5′-GCG(T/G)GGGCG-3′.17 We identified a consensus Egr-1 binding sequence (GCGGGGGCG) 283 nucleotides upstream from the initiation of transcription site (or 430 nucleotides upstream from the initiation of translation ATG). We created a phosphacan promoter construct (402 bp long) containing the potential Egr-1 binding site and including most of the 5′UTR, and two additional constructs, one introducing mutations to abolish the Egr-1 site, the other with a 12-bp deletion to cut out the Egr-1 site (390-bp construct). The three fragments were amplified using human genomic DNA as template (prepared with the Wizard genomic DNA purification kit, Promega) and inserted into the pGL3 promoter vector (Promega). Cloning primers are listed in Table 1. Constructs were verified by sequence analysis.
For luciferase assays, HeLa cells and primary human astrocytes were grown on six-well plates at a density of 2 × 105 cells/well and treated with Lipofectamine 2000 in the presence of 2 μg of reporter constructs alone or combined with 2 μg of Egr-1 expression plasmids. Cells were co-transfected with the SV40 β-galactosidase expression vector (pSV-β-Gal, Promega) to normalize results for transfection efficiency using the β-galactosidase enzyme assay system (Promega). Cells were maintained in the presence of Lipofectamine 2000 complexes for 6 hours and then grown in Dulbecco’s modified Eagle’s Medium containing 5% fetal bovine serum. After 24 hours of incubation, cells were harvested and luciferase activity was measured with an assay kit (Promega).
Protein Extraction and Western Blotting
For analysis of glial scar proteins, the scar was dissected from the left cortical area 10 days after MCAO, whereas tissue from the corresponding contralateral site served as control. Tissue samples were mechanically homogenized in 50 mmol/L Tris, 50 mmol/L Na-acetate, and 60 mmol/L n-octyl-β-d-glycopyranoside, pH 8.5, supplemented with a protease inhibitor cocktail (20 μl/ml, Sigma) and lysed for 10 minutes on ice. The homogenates were centrifuged at 13,000 rpm for 5 minutes and supernatants were collected. After a second lysis step, the supernatants were pulled together and the protein content was determined using the BCA protein assay kit (Bio-Rad, Munich, Germany). For the analysis of phosphacan expression in astrocytes or brain tissue, a total of 100 μg of protein of each sample was incubated for 6 hours at 37°C with chondroitinase ABC (Sigma) at 20 mU per 100 μg of extracted protein. Samples were boiled in reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer and separated on 4 to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Western blot analysis was performed using standard techniques.29 After antibody staining and color development, blots were digitalized and analyzed using ImageJ software.
Oligonucleotides (OGNs) and Electrophoresis Mobility Shift Assay (EMSA)
Nuclear protein extracts were prepared from whole brain tissues of wild-type or Egr-1-deficient mice and from human astrocytes using reagents from Pierce (Rockford, IL) according to the protocol of the supplier. A double-stranded OGN containing the putative Egr-1 site found ∼283 bp upstream from the transcription initiation site of the human phosphacan promoter, termed Phos-1, was synthesized and end-labeled with biotin. Two additional unlabeled OGNs carrying the previously characterized Egr-1 binding site from the human fibronectin promoter (with and without mutations in the Egr-1 site, named GCE and mGCE, respectively) were used in competition experiments.17 To control for nuclear extract quality, we used the OCT-1 OGN that contains the octamer site of the AHSP gene promoter, which binds the widely expressed Oct-1 transcription factor.30 EMSAs were performed according to the manufacturer’s protocol (LightShift chemiluminescent EMSA kit, Pierce). Briefly, nuclear extracts (20 μg of protein) were incubated with biotin end-labeled OGNs for 15 minutes at 4°C, separated by electrophoresis on native polyacrylamide gels, transferred to a positive nylon membrane, UV cross-linked, probed with streptavidin-horseradish peroxidase conjugates, and incubated with the chemiluminescent substrate.
For competition of Egr-1 binding experiments, unlabeled OGN GCE or mutated mGCE were preincubated with the reaction mixture for 15 minutes at 4°C before addition of biotin-labeled Phos-1 OGNs. The specificity of the Egr-1 binding to its putative site was further evaluated in DNA-protein interaction assays in the presence of 2 μg of antibodies against Egr-1, Egr-2, Egr-3, and Egr-4 (Santa Cruz Biotechnology) that were added during the incubation time before gel electrophoresis. OGN sequences are listed in Table 1.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were performed in HeLa cells using the EZ-Zyme Chromatin Prep and the EZ ChIP Chromatin Immunoprecipitation kits from Upstate Biotechnology (Lake Placid, NY) following the manufacturer’s directions. Because HeLa cells express low Egr-1 levels under normal conditions, we increased Egr-1 expression in two different ways. First, before chromatin preparations, HeLa cells were transiently transfected with Egr-1 full-length human cDNA clones. Second, 4 × 106 HeLa cells per immunoprecipitation sample were stimulated for 2 hours at 37°C with PMA (25 ng/ml, Sigma) and ionomycin (1.5 μmol/L, Sigma) to induce endogenous Egr-1. ChIP was performed with the anti-Egr-1 antibody (catalogue number SC-189, Santa Cruz Biotechnology) or a rabbit IgG isotype control (Santa Cruz Biotechnology). Primers were designed to amplify a 149-bp fragment containing the putative Egr-1 binding site of the human phosphacan promoter and a 109-bp fragment ∼4.5 kb upstream from the translation initiation site of the human phosphacan promoter as a control (Table 1). Equal volumes of the immunoprecipitated DNA were analyzed by conventional PCR and real time PCR using the LightCycler (Roche). Fold differences between specific antibody and isotype control immunoprecipitations were calculated using the formula: 2−ΔCt, where ΔCt = Ctsample − Ctcontrol.
Results
Egr-1 Expression Pattern in Adult Brain
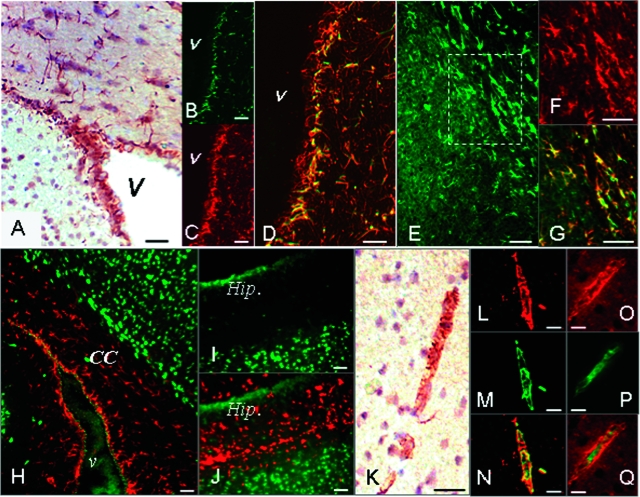
To appreciate changes in Egr-1 activity after stroke, we first analyzed Egr-1 expression patterns in normal adult mouse brain. Using anti-Egr-1 antibodies22 in immunohistochemistry and immunofluorescence experiments, we detected constitutive expression of Egr-1 in cells within and surrounding the brain ventricles (Figure 1, A–D), the corpus callosum (Figure 1, E–G), the external capsule, and the striatum (not shown). Double staining with anti-Egr-1 antibody and antibodies against astrocytic (GFAP) and neuronal (NeuN) markers identified the strongly positive Egr-1 cells primarily as astrocytes (Figure 1, B–G), in contrast to the weak anti-Egr-1 staining of neurons (Figure 1, H–J; and Supplemental Figure S1 at http://ajp.amjpathol.org).
Figure 1.
Egr-1 expression in the adult mouse brain. A: Immunohistochemistry using anti-Egr-1 antibody marks cells within and around the brain ventricles. B–D: Immunofluorescence analysis of brain tissue sections stained with anti-Egr-1 and GFAP antibodies. Egr-1-expressing cells next to the ventricles (B, green color) stain positive for the astrocytic marker GFAP (C, red color). Double-labeled cells in D appear in yellow/orange color. E–G: Egr-1-positive cells found in the corpus callosum (E, green color) also stain positive for GFAP (F, red color). Double-labeled astrocytes (G) appear in yellow/orange. Images in F and G represent a higher magnification of the boxed area in E. H–J: Neurons in corpus callosum (H) and hippocampus (I, J) marked by anti-NeuN antibody (in green) express low, or no Egr-1 (in red). K–Q: Immunohistochemistry analysis shows that a subset of vascular structures in the brain stain positive with anti-Egr-1 antibody (K). Egr-1-expressing cells in the vascular wall (L and O, red) stain positive with anti-smooth muscle actin antibody (M, green color). Double-labeled cells (N) appear in yellow/orange. Endothelial cells stained with anti-CD31 antibody (P, green) show less Egr-1 expression in the superimposed images (Q). v, ventricle; CC, corpus callosum; Hip., hippocampus. Scale bars = 25 μm.
Egr-1 antibody staining also marks a subset of vascular structures showing a coin-stack appearance characteristic of smooth muscle cells (Figure 1K). Double labeling using anti-Egr-1 antibody and antibodies against endothelial (PECAM-1 or CD31) and smooth muscle (SM) cell markers (SM-specific actin) detect Egr-1 expression in a subpopulation of vascular smooth muscle cells (Figure 1, L–Q). Higher magnification images also show low levels of Egr-1 expression within endothelial cells (Supplemental Figure S1 at http://ajp.amjpathol.org).
Egr-1 Expression in Astrocytes after Stroke
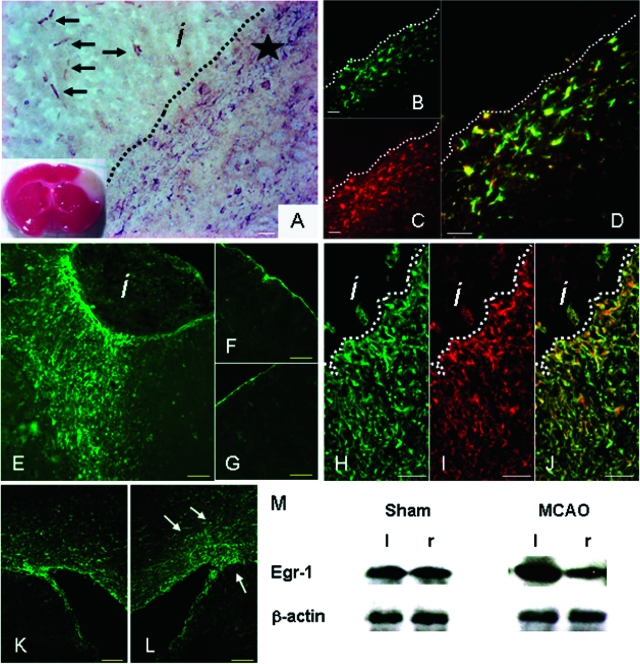
After experimental cerebral ischemia caused by permanent occlusion of the middle cerebral artery (MCAO), changes in Egr-1 expression appear after 12 hours within infarcted tissue and in the infarct border zone (not shown). The first peak of Egr-1 expression occurs 4 days after occlusion (Figure 2A). Cells with high Egr-1 levels were identified as astrocytes in the infarct border zone (GFAP-positive; Figure 2, B–D) and as endothelial cells within the infarcted tissue (CD31-positive, not shown). Ten days after MCAO, high expression of Egr-1 protein is maintained in cells at the border zone surrounding the infarct site (see below), but gradually lost in cells within the infarct area. Six weeks later, large numbers of Egr-1-expressing cells persist around the infarct region as compared with the corresponding area of the contralateral hemisphere or sham-operated controls (Figure 2, E–G). Consistent with earlier time points, Egr-1-positive cells associated with the glial scar are mostly astrocytes as revealed by double Egr-1/GFAP antibody staining (Figure 2, H–J). In addition to astrocytes, we detected Egr-1 expression in a small number of microglial cells in the glial scar area (Supplemental Figure S2 at http://ajp.amjpathol.org). In contrast, we did not observe double-stained nestin/Egr-1-positive cells in the peri-infarct area, although they are detectable in the vicinity of the ventricular wall (Supplemental Figure S2 at http://ajp.amjpathol.org). Interestingly, the number of Egr-1-positive cells around the ventricles of the infarcted hemispheres increases after MCAO compared with corresponding contralateral tissue (Figure 2, K and L).
Figure 2.
Egr-1 expression in adult mouse brain after cerebral ischemia. A: Cerebral ischemia was induced by permanent occlusion of the MCA. The infarcted tissue appears white in the TTC-stained brain slice (inset). Immunohistochemistry using anti-Egr-1 antibody 4 days after MCAO detects Egr-1 expression in cells within the infarct area (arrows) and around the infarct border zone (star). B–D: Immunofluorescence analysis of brain tissue sections stained with anti-Egr-1 and GFAP antibodies 4 days after MCAO. Egr-1-expressing cells in the border zone (B, green color) stain positive for GFAP (C, red color). D: Double-labeled astrocytic cells appear yellow/orange in the superimposed images. E–G: High Egr-1 expression persists in cells around the injury site 6 weeks after infarction. Low-magnification images reveal strong Egr-1 expression (green) in cells accumulating around the infarct region (E) compared with the corresponding area of the contralateral hemisphere (F), or the ipsilateral area of sham-operated animals (G). H–J: Confocal microscopy images of brain tissue sections stained with anti-Egr-1 and GFAP antibodies 6 weeks after MCAO. Egr-1-positive cells (H, green) around and within the glial scar stain positive for GFAP (I, red). Double-labeled astrocytes appear yellow/orange (J). K and L: Confocal microscopy images of brain tissue sections stained with anti-Egr-1 antibody. K: The number of Egr-1-positive cells (in green) around the ventricles of infarcted hemispheres increases after MCAO (L, arrows) compared with contralateral controls. M: Western blotting detects higher levels of Egr-1 protein in scar tissue isolated from infarcted (left, l) hemisphere as compared with corresponding noninfarcted area of the contralateral side (right, r). β-Actin protein levels serve as control. Sham-operated animals have comparable Egr-1 amounts on both brain sides. Quantitative image analysis shows that Egr-1 protein levels are 2.3-fold higher in the peri-infarct region compared with control tissue (SD, 0.3), whereas sham-operated animals show equivalent amounts of Egr-1 protein in the corresponding areas of both hemispheres (0.98-fold difference; SD, 0.12). i: infarct; dotted lines demarcate infarct and peri-infarct areas. Scale bars: 100 μm (A, E–G, K, L); 25 μm (B–D, H–J).
To further evaluate the high levels of Egr-1 in the peri-infarct area, scar tissue was isolated 10 days after MCAO and analyzed by Western blotting. Similar sized tissues were isolated from both sham-operated animals and corresponding areas in the control noninfarcted hemispheres to serve as controls. Blotting results show an accumulation of Egr-1 protein in the scar area (Figure 2M), consistent with our immunofluorescence data (Figure 2M).
Egr-1 Regulates Genes Involved in Gliosis
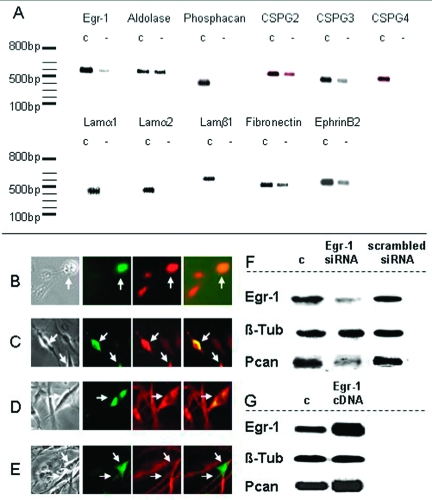
The robust expression of Egr-1 in reactive astrocytes within the glial scar raised the possibility that Egr-1 might regulate expression of scar components. To test this hypothesis, we investigated the effects of Egr-1 loss-of-function in primary cultures of human astrocytes using a mixture of siRNAs directed against Egr-1. Results show that the siRNA-mediated knockdown leads to a decrease of Egr-1 mRNA levels (Figure 3A). This reduction is accompanied by decreased mRNA levels of a number of genes involved in glial scar construction including several laminins, fibronectin, ephrinB2, and ECM proteoglycans such as phosphacan and chondroitin sulfate proteoglycan 2, 3, and 4 (CSPG2 or versican, CSPG3 or neurocan, and CSPG4 or NG2). An alternative, commercially available siRNA cocktail was also used to independently knockdown Egr-1 with similar results, indicating that this effect is not attributable to nonspecific RNA interference.
Figure 3.
Egr-1 regulates genes involved in gliosis. A: Egr-1 loss-of-function effects on human astrocytes transfected with siRNAs against Egr-1. RT-PCR analysis shows that knockdown of Egr-1 leads to reduction in the expression levels of genes encoding ECM components of the glial scar. c, control mock-transfected cells; −, Egr-1 knockdown. DNA size markers are shown on the left. Gene name abbreviations are as follows: CSPG2, 3, 4, chondroitin sulfate proteoglycan 2, 3, 4, respectively; Lamα1, laminin α1; Lamα2: laminin α2; Lamβ1: laminin β1. Real-time PCR quantification of the effects of the Egr-1 knockdown on the expression of putative gene targets shows a 60% drop in phosphacan RNA levels in Egr-1 siRNA-treated astrocytes. Adjusted for aldolase, relative phosphacan expression was 12.96 ± 1.21 U in controls versus 5.05 ± 0.89 U after Egr-1 knockdown (n = 5, P < 0.001). B–E: Immunofluorescence analysis after Egr-1 overexpression in astrocytes transfected with the CMV-Egr-1-IRES-EGFP construct. Transfected, EGFP-positive cells (green, marked by arrows), stain more intensely with antibodies recognizing laminin α1 (B, red) and phosphacan (C, red, anti-RPTPβ) than nontransfected neighboring cells. No difference in expression levels of GFAP (D, red) or β-tubulin (E, red) between Egr-1-overexpressing cells (green, marked by arrows) and nontransfected cells. Superimposed images (far right) confirm that transfected cells express higher levels of putative Egr-1 targets, but similar levels of other proteins. F: Western blotting of proteins isolated from astrocytes transfected with siRNAs against Egr-1 (Egr-1 siRNA), control siRNA (scrambled siRNA), or mock-transfected cells (c). siRNAs against Egr-1 diminish effectively Egr-1 protein levels and lead to down-regulation of phosphacan (Pcan, detected with the KAF13 antibody). β-Tubulin (β-Tub) levels remain unaffected serving as control. Quantification of blot images shows that Egr-1 protein levels are reduced 3.0-fold versus control (mock-transfected cells; SD, 0.4) and 2.8-fold (SD, 0.2) versus scrambled siRNA-transfected cells; phosphacan protein levels are down-regulated 2.18-fold versus control (mock; SD 0.1) and 2.23 times (SD 0.1) versus scrambled siRNA. G: Western blotting of proteins isolated from astrocytes transfected with the CMV-Egr-1-IRES-EGFP (Egr-1 cDNA) construct or the empty vector (c). Egr-1 protein levels increase 2.08-fold (SD, 0.4) leading to 1.42-fold up-regulation of phosphacan (Pcan; SD, 0.14). β-Tubulin (β-Tub) serves as control.
To further assess the influence of Egr-1 on these putative gene targets, we transfected astrocytes with an expression vector carrying an Egr-1/EGFP bicistronic insert under the CMV enhancer (CMV-Egr-1-IRES-EGFP) and stained the cells with antibodies against laminin α1, phosphacan, GFAP, and β-tubulin. Immunofluorescence analysis shows that transfected, EGFP-positive astrocytes express higher levels of laminin and phosphacan than neighboring untransfected cells indicating that gain of Egr-1 function leads to up-regulation of candidate gene targets (Figure 3, B and C). In contrast, we observed no increase in the levels of GFAP and β-tubulin (Figure 3, D and E).
To test whether mRNA changes result in altered levels of corresponding peptides, we isolated proteins from astrocytes transfected with anti-Egr-1 siRNAs. Mock-transfected astrocytes and astrocytes transfected with scrambled siRNAs served as controls. Western blots show that knockdown of Egr-1 leads to a significant decrease in the amount of endogenous Egr-1 protein and its putative target phosphacan, whereas protein levels for β-tubulin are unaffected (Figure 3F). Conversely, overexpression of Egr-1 results in increased levels of phosphacan protein (Figure 3G). Taken together, the gain and loss of Egr-1 function approaches in cultured astrocytes indicate that Egr-1 is involved in the regulation of genes encoding structural components of the glial scar.
Egr-1-Positive Astrocytes Express Putative Egr-1 Target Genes in Vivo
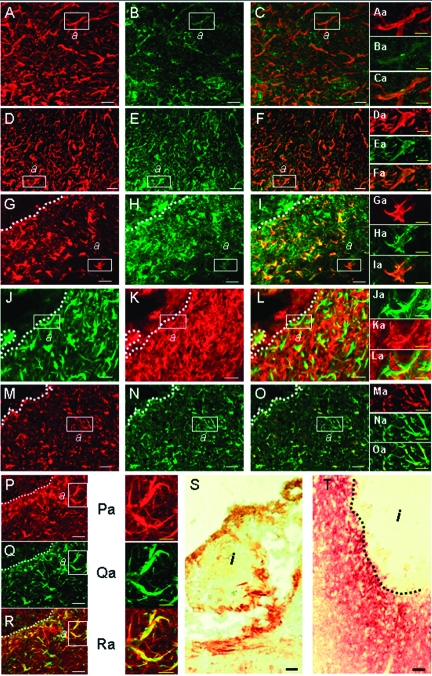
To test if the spatial and temporal patterns of Egr-1 expression in vivo reflect the molecular events associated with glial scar formation, we stained sections of control tissue and tissue 10 days after MCAO with antibodies recognizing Egr-1, GFAP, phosphacan, NG2, laminin α1, ephrinB1/B2, and neurocan (Figure 4). Similar staining of tissues isolated from sham-operated animals served as controls (Supplemental Figure S3 at http://ajp.amjpathol.org).
Figure 4.
Egr-1 and its putative target genes show similar spatial and temporal expression patterns in vivo after focal cerebral ischemia. A–C: Confocal microscopy images of brain tissue show Egr-1 protein expression (A, red color) in the gliotic tissue 10 days after MCAO. Egr-1-positive cells are also expressing phosphacan (B, in green, antibody KAF13 recognizing both the secreted and membrane-bound splice variants of phosphacan); the double-labeling image reveals expression overlap of Egr-1 and phosphacan (C, yellow). Aa–Ca: Higher magnification of boxed areas marked (a) in A–C shows co-expression of Egr-1 (Aa) and phosphacan (Ba) inside the gliotic tissue; the superimposed images are shown in Ca. D–F: Immunofluorescence analysis indicates that Egr-1-expressing cells in the glial scar (D, red color) also express phosphacan (E, green color, antibody PTP1 recognizing only membrane-bound phosphacan); the double-labeling image is shown in F. Da–Fa: Higher magnification of boxed areas (a) in D–F shows co-expression of Egr-1 (Da) and phosphacan (Ea) inside the gliotic tissue; the superimposed images are shown in Fa. G–I: Immunofluorescence analysis shows expression of Egr-1 (G, red color) and NG2 (H, green color). The corresponding superimposed images (I) show expression overlap (yellow). Ga–Ia: Higher magnification of boxed areas (a) in G–I shows co-expression of Egr-1 (Ga) and NG2 (Ha) inside the gliotic tissue; the superimposed images are shown in Ia. J–L: Expression of GFAP (J, green color) and NG2 (K, red color) and the corresponding superimposed images (L). Ja–La: Higher magnification of boxed areas (a) in J–L shows co-expression of GFAP (Ja) and NG2 (Ka) inside the gliotic tissue; the superimposed images are shown in La. M–O: Immunofluorescence analysis showing expression of GFAP (M, red color) and laminin α1 (N, green color); the corresponding superimposed images are shown in O. Ma–Oa: Higher magnification of boxed areas (a) in M–O shows co-expression of GFAP (Ma) and laminin α1 (Na) inside the gliotic tissue; the superimposed images are shown in Oa. P–R: Confocal microscopy images show that Egr-1-positive cells (P, red color) in the infarct border zone also express laminin α1 (Q, green color); the superimposed images are shown in R. Pa–Ra: Higher magnification of boxed areas (a) in P–R shows co-expression of Egr-1 (Pa) and laminin α1 (Qa); the superimposed images are shown in Ra. S: Immunohistochemistry staining of ephrin B1/B2 (brown color) in mouse brain 6 weeks after MCAO. Ephrin B1/B2 expression predominates in the zone that directly surrounds the infarcted area. T: Immunohistochemistry staining of neurocan (brown/red color) in gliotic rat brain tissue 10 days after MCAO. Dotted lines demarcate the infarct areas (i). Scale bars: 25 μm (A–T); 10 μm (Aa–Ra).
The results show that Egr-1-positive cells co-localize with cells expressing phosphacan and NG2 (Figure 4, A–I and Aa–Ia). The Egr-1/phosphacan co-localization is evident with two different phosphacan-specific antibodies, one recognizing all phosphacan splice variants (KAF13) and one recognizing only the transmembrane form of phosphacan (PTP1). As expected, KAF13 stains both cells and extracellular space, whereas PTP1 marks primarily cellular structures (Figure 4, A–F and Aa–Fa).
A number of NG2-expressing cells stain for GFAP indicating that they are astrocytes (Figure 4, J–L and Ja–La). GFAP- and Egr-1-positive cells are also marked with antibodies against laminin α1 (Figure 4, M–R and Ma–Ra). Further staining showed that ephrin B1/B2 and neurocan are expressed at high levels by cells within the glial scar (Figure 4, S and T). Neurocan expression was evaluated in tissue sections from a rat stroke model because the available anti-neurocan antibody is specific for the rat antigen. Immunohistological analysis showed that Egr-1 induction in rat brain after stroke follows a similar pattern as in mouse, ie, Egr-1 accumulates in astrocytes at day 10 (data not shown). In summary, our results show that there is spatial and temporal overlap in the glial scar area in vivo between strong levels of Egr-1 protein in astrocytes and expression of putative Egr-1 targets identified in the loss- and gain-of-function experiments in cultured astrocytic cells. This further supports the role of Egr-1 in the transcriptional regulation of genes involved in glial scar formation.
Egr-1 Binds to a Site within the Human Phosphacan Gene Promoter
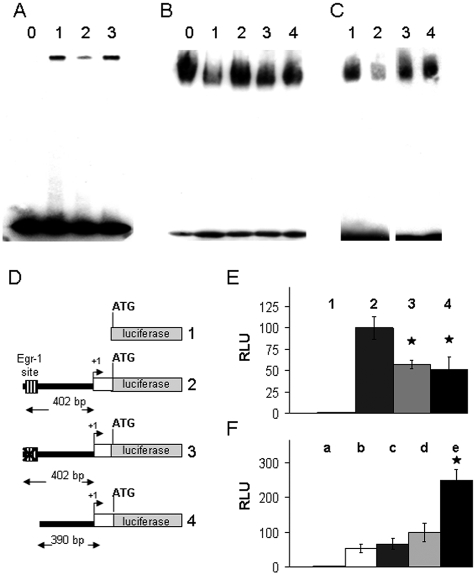
To investigate if Egr-1 regulates expression of target genes directly, we examined the promoter region of the human phosphacan gene for candidate Egr-1 binding sites. We identified an Egr-1 consensus binding sequence (GCGGGGGCG)17 ∼280 bases upstream from the transcription initiation site. To test if Egr-1 binds to the candidate site, we synthesized and biotinylated double-stranded OGNs spanning this sequence. The biotinylated OGN (called Phos-1) was incubated with nuclear extracts prepared from cultured human astrocytes and DNA/protein complexes were analyzed in electrophoresis mobility shift assays. Figure 5A shows that a specific DNA-protein complex assembles when nuclear extracts are combined with the biotinylated DNA probe. The protein binding intensity is reduced when a nonbiotinylated, specific competitor OGN carrying a bona fide Egr-1 binding site from the fibronectin promoter17 is added to the mixture. In contrast, the complex is not affected when the same OGN containing a mutated Egr-1 consensus site is used, indicating that the observed shift is attributable to binding of the Egr-1 protein (Figure 5A).
Figure 5.
Egr-1 binds and transactivates the human phosphacan promoter. A: EMSAs with biotin-labeled OGNs carrying the putative phosphacan promoter Egr-1 site (Phos-1) were incubated with: 0, no nuclear extract; 1, nuclear extract from astrocytes; 2, nuclear extract from astrocytes plus unlabeled OGN carrying the fibronectin promoter Egr-1 site; and 3, nuclear extract from astrocytes plus the same fibronectin promoter OGN, but with a mutated Egr-1 site. The OGN with the fibronectin promoter Egr-1 site (lane 2) competes effectively with binding to Phos-1. B: EMSA using the Phos-1 OGN and nuclear extracts from astrocytes in the presence of antibodies against different Egr family members. The numbered lanes represent the following conditions: 0, no antibody; 1, anti-Egr-1; 2, anti-Egr-2; 3, anti-Egr-3; and 4, anti-Egr-4. Only the anti-Egr-1 antibody in lane 2 hinders the nuclear protein/Phos-1 DNA binding complex. C: EMSA using Phos-1 and OCT-1 OGN and nuclear extracts isolated from wild-type (WT) or Egr-1-deficient mouse brain tissue. Numbered lanes represent: 1, Phos-1 OGN incubated with nuclear extract from WT brain tissue; 2, Phos-1 OGN incubated with nuclear extract from Egr-1−/− brain tissue; 3,: OCT-1 OGN incubated with nuclear extract from WT brain; and 4, OCT-1 OGN incubated with nuclear extract from Egr-1−/− brain tissue. Nuclear extracts from Egr-1−/− brain tissue (lane 2) produce less Phos-1 shift. D: Schematic drawing of the four constructs used to study the activity of the phosphacan promoter Egr-1 binding site. Construct 1: pGL3 basic, empty vector; construct 2: contains the 402-bp promoter fragment of the phosphacan gene cloned in pGL3; construct 3: same as 2 except point mutations were introduced to destroy the Egr-1 site; and construct 4: same as 2 except for a short nucleotide deletion that removes the Egr-1 site. E: The constructs outlined in D were transfected in primary human astrocytes and cell extracts were assayed for luciferase activity 24 hours later. Bars labeled 1 to 4 represent the activities of constructs 1 to 4, respectively. Mann-Whitney rank sum test shows a statistically significant difference between construct 2 and constructs 3 and 4, P < 0.001. F: Luciferase assay in HeLa cells after transfection of: a, construct 1, empty vector; b, construct 4 lacking the Egr-1 site; c, construct 4 together with the Egr-1 expression vector; d, construct 2 including the Egr-1 site; e, construct 2 together with the Egr-1 expression vector. Mann-Whitney rank sum test shows a statistical significant difference between d and e, P < 0.001. RLU, relative luciferase units. The activity of construct 2 (bars 2 in E and d in F) in one assay was set arbitrarily as 100.
To further probe the identity of the protein binding to the putative Egr-1 site, we incubated the Phos-1 OGN and astrocytic nuclear extracts in the presence of antibodies specific for Egr-1 that interfere with the formation of the corresponding DNA/protein complex.17 As a control, we used antibodies against other distinct members of the Egr family (Egr-2, Egr-3, and Egr-4). Among them, only the anti-Egr-1 antibody disrupts DNA/protein complex formation suggesting that Egr-1 is the primary nuclear protein in astrocytes binding to this site (Figure 5B). Moreover, we prepared whole brain nuclear extracts from wild-type and Egr-1-deficient mice. We observed less protein binding to the Phos-1 OGN using nuclear extracts from Egr-1−/− mice compared with wild-type controls; in contrast, binding of the ubiquitous Oct-1 transcription factor to an OGN containing the octamer motif (ATTTGCAT) of the AHSP gene promoter,30 shows no difference between Egr-1−/− and wild-type samples (Figure 5C). The in vitro binding assays support the idea that the identified site within the phosphacan gene promoter binds to Egr-1.
Egr-1 Regulates the Activity of the Phosphacan Promoter
To investigate if the putative Egr-1 binding site in the phosphacan promoter contributes to Egr-1-mediated transcriptional activity, we cloned the corresponding promoter fragments upstream from the luciferase reporter gene and measured promoter activity using the following constructs: pGL3Basic, the empty luciferase reporter vector as control; a construct of the human phosphacan gene promoter containing the putative Egr-1 binding site in front of the luciferase gene (402 bp long, containing the promoter and most of the 5′ UTR sequences); a 402-bp derivative with an engineered mutation in the Egr-1 binding site; and, a 390-bp long fragment with the Egr-1 binding site deleted (Figure 5D). As shown in Figure 5E, luciferase activity in extracts of transfected primary astrocytes is reduced when the Egr-1 site is either mutated or deleted compared with the activity of the intact construct.
In a second set of experiments, the plasmids containing the intact 402-bp fragment or the 390-bp fragment with the Egr-1 site deletion were transfected together with the Egr-1 expression vector in HeLa cells. Luciferase assays show that Egr-1 induces expression of the phosphacan promoter-driven reporter gene and that induction depends on the presence of the Egr-1 site (Figure 5F). Taken together, the binding assays and the promoter activity analysis indicate that the area 280 nucleotides upstream from the transcription initiation site of the phosphacan gene contributes to transcriptional activity, binds to Egr-1, and takes part in the transactivation of the phosphacan promoter by Egr-1.
Egr-1 Binds to the Phosphacan Promoter in Vivo
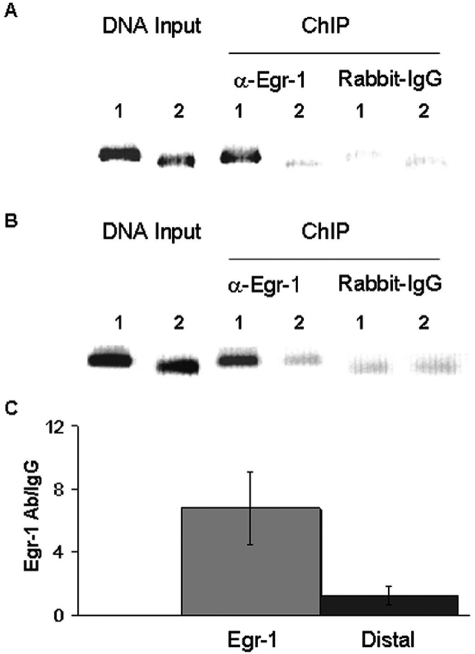
To examine if Egr-1 binds to the phosphacan gene promoter in vivo, HeLa cells were transiently transfected with the Egr-1 full-length human cDNA clone and subjected to ChIP with anti-Egr-1 antibody and rabbit IgG isotype as control. The precipitated Egr-1-DNA complexes were analyzed by PCR with primers flanking the Egr1 binding site in the phosphacan promoter (Table 1). Using chromatin precipitated with the anti-Egr1 antibody as template, we obtained a strong band after PCR amplification, but not with IgG control (Figure 6A). Primers amplifying a randomly selected distal area 4.5 kb upstream from the putative Egr-1 site of the phosphacan promoter show no preferential amplification after immunoprecipitation. We also detected an approximately sevenfold stronger band with the Egr-1 site flanking primers after stimulating HeLa cells with phorbol ester and ionomycin to activate endogenous Egr-1 expression (Figure 6, B and C).31 These data indicate that Egr-1 binds to the identified site in vivo, further supporting a role for Egr-1 in the regulation of phosphacan gene expression.
Figure 6.
Egr-1 binds to the phosphacan promoter in vivo. A: HeLa cells were transiently transfected with Egr-1 full-length human cDNA and subjected to ChIP with anti-Egr-1 antibody or rabbit IgG isotype control. Specific primers were used to amplify the area containing the putative Egr-1 site (lanes 1), or a distal, 4.5-kb upstream fragment (lanes 2). Genomic DNA input shows that the primers amplify bands of the expected size. Immunoprecipitation of sheared chromatin (ChIP) with anti-Egr-1 (α-Egr-1) produces a band of stronger intensity as compared with the distal site product and to control isotype IgG. B: HeLa cells were stimulated for 2 hours with PMA and ionomycin to induce endogenous Egr-1 and subjected to ChIP with anti-Egr-1 antibody or rabbit IgG isotype control. Precipitated DNA fragments were amplified with the same primers as in A surrounding the Egr-1 site (lanes 1) or the distal area (lanes 2). Genomic DNA input shows that primers amplify bands of the expected size. Immunoprecipitation of sheared chromatin (ChIP) with anti-Egr-1 (α-Egr-1) produces a band of stronger intensity as compared with the distal site product and to control isotype IgG. C: Immunoprecipitated DNA from the analysis in B was amplified and quantified by real-time PCR using primers surrounding the Egr-1 binding site or the distal area. Fold differences in the amounts of PCR products obtained with anti-Egr-1 antibody relative to isotype controls were calculated as described in Materials and Methods. Average values with SEM from five independent experiments are depicted. There is ∼6- to 10-fold higher amplification of the Egr-1 site fragment compared with the control fragment from the distal area.
Egr-1-Deficient Mice Have Reduced Phosphacan in Scar Tissue after Experimental Stroke
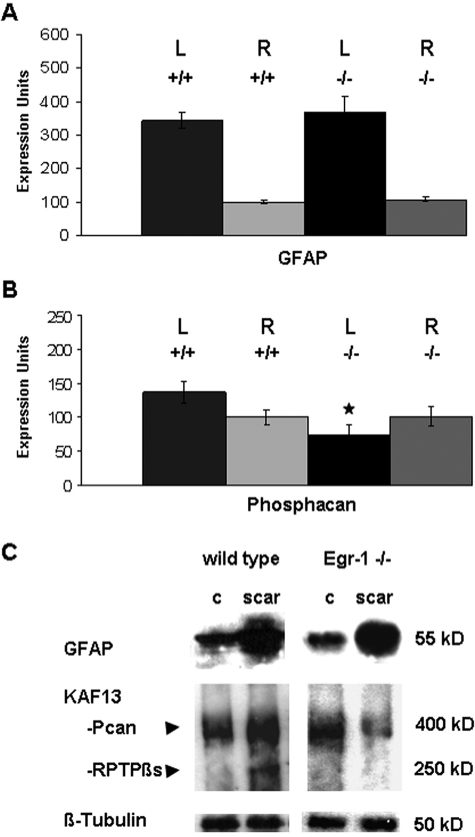
To investigate if Egr-1 activity contributes to phosphacan expression after stroke, we compared phosphacan RNA and protein levels in infarcted brain hemispheres of Egr-1-deficient mice to wild-type littermates. Ten days after MCAO, we identified higher levels of GFAP mRNA in the infarcted hemisphere as previously described.32,33 There are no measurable differences in the amount and GFAP induction levels between wild-type and Egr-1-deficient littermates (Figure 7A). Conversely, we detected a small increase in phosphacan at whole tissue level in wild-type animals after MCAO, but phosphacan RNA levels were reduced in the infarcted hemisphere of Egr-1-deficient mice compared with wild types, as well as compared with either infracted tissue from wild-type mice or noninfarcted tissue from Egr-1−/− and wild-type animals (Figure 7B).
Figure 7.
Phosphacan accumulation is impaired in the glial scar of Egr-1-deficient mice. A and B: Measurement of GFAP (A) and phosphacan (B) RNA expression levels 10 days after permanent occlusion of the MCA. RNA samples were isolated from wild-type (+/+) and Egr-1-deficient (−/−) mice and from the infarcted, left brain (L) and contralateral, right brain (R) hemispheres. Results were normalized to β-actin RNA expression levels setting arbitrarily the value of one wild-type sample from the right hemisphere as 100. Statistical analysis was performed with the Mann-Whitney rank sum test. Wild-type and Egr-1-deficient mice show comparable GFAP induction. Egr-1−/− mice show a statistically significant reduction of phosphacan levels in the infarcted hemisphere compared with the contralateral site (P = 0.001). The expression levels of phosphacan are also statistically significant different between the infarcted hemispheres of Egr-1−/− mice and their wild-type littermates (P ≤ 0.001). In contrast, there is no statistically significant change of phosphacan expression between both hemispheres (P = 0.163) in wild-type mice and between noninfarcted hemispheres of WT and KO mice (P = 0.380). C: Western blot analysis of brain tissue isolated from the glial scar 10 days after occlusion of the MCA and control (c) tissue from the contralateral brain hemisphere. Quantitative image analysis demonstrates that GFAP up-regulation after stroke is comparable in Egr-1−/− and wild-type brain tissue, ie, 4.4 times in wild-type samples (SD, 0.6) and 4.7 times in Egr-1−/− mice (SD, 1.8). The levels of the two phosphacan isoforms detected by the KAF13 antibody, namely the secreted form of phosphacan (Pcan) as well as the transmembrane isoform receptor protein tyrosine phosphatase β (RPTPβs) are up-regulated in the glial scar of wild types by 1.49-fold (SD, 0.14), but not in the glial scar of Egr-1-deficient animals (0.89-fold; SD, 0.26). β-Tubulin serves as loading control. Protein sizes are indicated in kDa.
The quantitative RNA analysis suggested that wild-type and Egr-1−/− mice show similar activation of GFAP, but Egr-1−/− mice may accumulate less phosphacan after experimental stroke. To further test this notion in the local injury area, we isolated scar tissue from control and Egr-1−/− mice and analyzed protein content with antibodies against GFAP and phosphacan. As shown in Figure 7C, wild-type and Egr-1−/− samples have comparable increases in GFAP. In contrast, there is less measurable phosphacan protein in scar tissue of Egr-1−/− mice than wild-type controls. Taken together, both phosphacan RNA and protein levels are lower in the postischemic brain hemisphere of Egr-1-deficient mice compared with wild-type controls indicating that Egr-1 activity contributes to phosphacan gene expression after MCAO.
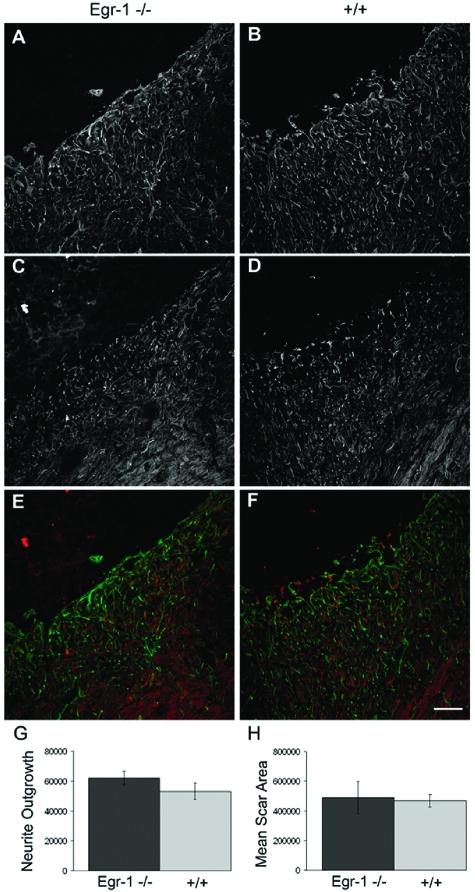
To test if the lower phosphacan protein levels in scar tissue affect the injury process, we compared the infarct areas of Egr-1−/− and wild-type mice. We found that the overall glial scar size and the extent of axon outgrowth are not significantly different between wild-type and Egr-1−/− siblings suggesting that the reduction of phosphacan levels had no measurable effect (Figure 8). Of note, we did not detect major changes in the mRNA expression levels of Egr-2, -3, and -4 either before or after MCAO in Egr-1−/− mice (compared with wild-type animals), making it unlikely that they compensate for the Egr-1 deficiency (Supplemental Figure S4 at http://ajp.amjpathol.org).
Figure 8.
Neurite outgrowth and scar size after MCAO appear normal in Egr-1-deficient mice. Analysis of neurite outgrowth and scar size area 10 days after MCAO in Egr-1-deficient (Egr-1−/−) and wild-type (+/+) mice. A–F: Confocal microscopy images of serial sections from infarcted brain tissue that were stained with antibody against the glial marker GFAP to define the infarction border area and the glial scar (A, B), as well as with anti-neurofilament antibody to mark neurites (C, D). Superimposed images are shown in E and F (GFAP in green, neurofilament protein in red). G and H: Quantitative analysis of the staining intensity of the 160-kDa neurofilament marker inside the infarcted tissue to measure neurite outgrowth (G) and GFAP staining to assess scar size area (H). In both cases, there are no statistically significant differences between Egr-1-deficient mice and wild-type littermates (Mann-Whitney rank sum test, P ≥ 0.05).
Discussion
Stroke is the foremost reason of long-term disability and the third leading cause of death in the United States. Approximately 80% of all stroke cases are caused by focal ischemia and there are limited therapies approved for the treatment of acute ischemic stroke.34 Advances in stroke therapy require a better understanding of the molecular mechanisms that control and coordinate the repair mechanisms of ischemic injury in the brain.
The transcription factor Egr-1 regulates the expression of a large number of genes encoding chemokines, angiogenic factors, ECM proteins, and cell adhesion molecules.35 Moreover, Egr-1 has been implicated as a key control gene after ischemic injury in the lung, and it is up-regulated in the brain after experimental stroke suggesting that Egr-1 plays an important regulatory role in poststroke events.14,24
To test this hypothesis, we have investigated the expression of Egr-1 in a mouse model of brain ischemia caused by permanent occlusion of the MCA. Permanent occlusion was better tolerated than reopening of the closed vessel and was used in this study to allow long-term observations of the molecular and cellular events associated with injury. Our results demonstrate that under normal conditions in the adult brain, Egr-1 protein is expressed in smooth muscle cells in a subset of large blood vessels, and in astrocytes located in and around the ventricular walls, in the corpus callosum, the striatum, and the external capsule. Our immunofluorescence results show lower levels of Egr-1 in neuronal and endothelial cells.
The astrocytic expression pattern changes rapidly after ischemic injury and we observed strong Egr-1 expression in most of the astrocytes accumulating around the affected tissue. Egr-1 protein is first detected 12 to 24 hours after ischemia and becomes stronger approximately at day 4 in endothelial cells within the infarct site and in astrocytes in the peri-infarct area. Egr-1 protein levels subside in the endothelium, but remain high in astrocytes within the forming gliotic scar tissue for several weeks after MCAO.
We found that siRNA-mediated knockdown of Egr-1 in human astrocytes in culture lowers the expression of many genes that encode ECM components of the glial scar including laminin genes and fibronectin. In parallel, there is a considerable reduction in the expression levels of chondroitin sulfate proteoglycan-encoding genes, which are the main components of the glial scar. CSPGs block axon outgrowth and play a crucial role in the regeneration failure on the surface of the glial scar.2,36 EphrinB2, which may also inhibit axonal growth,37 is down-regulated as well. The data presented here indicate that Egr-1 might be a critical control gene of gliosis after MCAO.
Using sequence analysis, DNA binding assays, and promoter deletion studies, we located a putative binding site in the human phosphacan gene promoter that interacts physically with the Egr-1 protein in vitro and in vivo. The identified motif is required for transactivation by Egr-1 in cultured astrocytes suggesting that phosphacan is a direct Egr-1 target. This interaction appears to be important for up-regulation of phosphacan gene expression in astrocytes after ischemic injury in the brain because we detected less phosphacan in the infarcted tissue of Egr-1-deficient mice after experimental stroke relative to wild-type mice.
The identification of Egr-1 as a control factor may lead to a better understanding of the regulatory mechanisms of glial scar formation and neuronal plasticity. For example, transforming growth factor-β is known to induce CSPG molecules in the central nervous system, and transforming growth factor-β has been shown to up-regulate Egr-1 in several cell types.38,39 Thus, it is possible that the transforming growth factor-β-mediated stimulation of glial scar proteins occurs through induction or maintenance of Egr-1 expression in astrocytes. Moreover, because both phosphacan and Egr-1 have been implicated in neuronal plasticity, it is possible that controlling expression of phosphacan or other ECM components might be one way that Egr-1 regulates plasticity.23,40
It is noteworthy that knife injury in brain tissue causes an accumulation of astrocytes around the wound area, but a transient decrease in phosphacan deposition around the injury site at early time points, which recovers 14 days later.41 We also observed a decrease in phosphacan levels in Egr-1 knockout mice after MCAO that does not occur in wild-type controls. It is possible that phosphacan expression is differentially regulated by Egr-1 in certain types and stages of injury or in specific areas of the central nervous system. To this end, it will be interesting to examine the expression pattern of Egr-1 in different model systems of brain injury.
Histological analysis of infarct sites shows that glial scar size is not significantly different between wild-type and Egr-1 siblings. It is possible that the Egr-1 deficiency does not fully abrogate production of phosphacan and other ECM proteins because additional factors contribute to the transcriptional control of these genes after stroke. For example, the transcription factor Sox9 has been recently shown to regulate expression of laminin γ1 and genes involved in the CSPG biosynthetic pathway in primary astrocytes.42 Further detailed analysis of the phosphacan promoter/enhancer sequences could identify additional regulators of phosphacan gene expression after tissue injury.
It is also possible that the lack of apparent phenotype in scar size or neurite outgrowth in Egr-1−/− mice is attributable to the considerable redundancy among ECM proteins and/or the existence of compensatory mechanisms. A characteristic example is that mice completely lacking four distinct ECM components, namely neurocan, brevican, tenascin-R, and tenascin-C are viable and, although they have aberrant ECM structure and deposition, the basic ECM organization is maintained probably through up-regulation of other matrix components.43 Furthermore, there is evidence that the CSPG milieu contains both inhibitory and promotional cues for axonal growth, so a proportional reduction in scar size and deposition of scar components might not alter the balance between opposing signals.39,41,44
Excessive gliosis after brain ischemia or after mechanical spinal cord injury creates a formidable barrier to axonal growth and recovery in the nervous system. Besides a negative role in neuronal tissue regeneration,45 the astrocytic wall is important to isolate necrotic areas and prevent spreading of cytotoxic products released from dead cells to healthy neighboring tissue.4,46 The importance of this function was most evident in double knockout mice for the intermediate filament proteins vimentin and GFAP, which have defects in reactive astrocytes leading to abnormal gliosis.47 Although these mice show improved integration of neural grafts and posttraumatic regeneration, they have considerably larger infarct sizes than wild-type or single knockout counterparts underscoring the dual protective and harmful effects of the glial scar.
The identification of Egr-1 as a likely control gene in the gliotic process might open the way to understand the transcriptional mechanisms regulating glial scar production. Further dissection of the regulatory elements in this genetic pathway might present an opportunity to separate the neuroprotective from the neurosuppressive functions of reactive astrocytes. This information could lead to new approaches to modulate the time span and severity of gliosis, thus fine-tuning neuronal tissue repair and regeneration.
Acknowledgments
We thank A. Faissner for providing the anti-phosphacan antibodies, S. Dietzel for help with confocal microscopy, and K. Kain for critical reading of the manuscript.
Footnotes
Address reprint requests to Antonis K. Hatzopoulos, Ph.D., Department of Medicine and Cell and Developmental Biology, Vanderbilt University, Division of Cardiovascular Medicine, PRB 383, 2220 Pierce Ave., Nashville, TN 37232-6300. E-mail: antonis.hatzopoulos@vanderbilt.edu.
Supported by the Deutsche Forschungsgemeinschaft (Priority Program Angiogenesis SPP1069, grant HA2983/1 to A.K.H.), the National Institutes of Health (grant HL083958 to A.K.H.), the Vanderbilt University (development funds to A.K.H.), and the Friedrich-Baur Stiftung (to H.B.).
Supplemental material for this article can be found on http://ajp. amjpathol.org.
References
- Anderson MF, Blomstrand F, Blomstrand C, Eriksson PS, Nilsson M. Astrocytes and stroke: networking for survival? Neurochem Res. 2003;28:293–305. doi: 10.1023/a:1022385402197. [DOI] [PubMed] [Google Scholar]
- McKeon RJ, Schreiber RC, Rudge JS, Silver J. Reduction of neurite outgrowth in a model of glial scarring following CNS injury is correlated with the expression of inhibitory molecules on reactive astrocytes. J Neurosci. 1991;11:3398–3411. doi: 10.1523/JNEUROSCI.11-11-03398.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JW, Asher RA. The glial scar and central nervous system repair. Brain Res Bull. 1999;49:377–391. doi: 10.1016/s0361-9230(99)00072-6. [DOI] [PubMed] [Google Scholar]
- Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci. 2004;5:146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- McKeon RJ, Jurynec MJ, Buck CR. The chondroitin sulfate proteoglycans neurocan and phosphacan are expressed by reactive astrocytes in the chronic CNS glial scar. J Neurosci. 1999;19:10778–10788. doi: 10.1523/JNEUROSCI.19-24-10778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LL, Margolis RU, Tuszynski MH. The chondroitin sulfate proteoglycans neurocan, brevican, phosphacan, and versican are differentially regulated following spinal cord injury. Exp Neurol. 2003;182:399–411. doi: 10.1016/s0014-4886(03)00087-6. [DOI] [PubMed] [Google Scholar]
- Tang X, Davies JE, Davies SJ. Changes in distribution, cell associations, and protein expression levels of NG2, neurocan, phosphacan, brevican, versican V2, and tenascin-C during acute to chronic maturation of spinal cord scar tissue. J Neurosci Res. 2003;71:427–444. doi: 10.1002/jnr.10523. [DOI] [PubMed] [Google Scholar]
- Pindzola RR, Doller C, Silver J. Putative inhibitory extracellular matrix molecules at the dorsal root entry zone of the spinal cord during development and after root and sciatic nerve lesions. Dev Biol. 1993;156:34–48. doi: 10.1006/dbio.1993.1057. [DOI] [PubMed] [Google Scholar]
- Sukhatme VP, Cao XM, Chang LC, Tsai-Morris CH, Stamenkovich D, Ferreira PC, Cohen DR, Edwards SA, Shows TB, Curran T, Le Beau M, Adamson ED. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- Khachigian LM, Lindner V, Williams AJ, Collins T. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- Gashler A, Sukhatme VP. Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog Nucleic Acid Res Mol Biol. 1995;50:191–224. doi: 10.1016/s0079-6603(08)60815-6. [DOI] [PubMed] [Google Scholar]
- Cui MZ, Parry GC, Oeth P, Larson H, Smith M, Huang RP, Adamson ED, Mackman N. Transcriptional regulation of the tissue factor gene in human epithelial cells is mediated by Sp1 and EGR-1. J Biol Chem. 1996;271:2731–2739. doi: 10.1074/jbc.271.5.2731. [DOI] [PubMed] [Google Scholar]
- Khachigian LM, Anderson KR, Halnon NJ, Gimbrone MA, Jr, Resnick N, Collins T. Egr-1 is activated in endothelial cells exposed to fluid shear stress and interacts with a novel shear-stress-response element in the PDGF A-chain promoter. Arterioscler Thromb Vasc Biol. 1997;17:2280–2286. doi: 10.1161/01.atv.17.10.2280. [DOI] [PubMed] [Google Scholar]
- Yan SF, Fujita T, Lu J, Okada K, Shan Zou Y, Mackman N, Pinsky DJ, Stern DM. Egr-1, a master switch coordinating upregulation of divergent gene families underlying ischemic stress. Nat Med. 2000;6:1355–1361. doi: 10.1038/82168. [DOI] [PubMed] [Google Scholar]
- Haas TL, Stitelman D, Davis SJ, Apte SS, Madri JA. Egr-1 mediates extracellular matrix-driven transcription of membrane type 1 matrix metalloproteinase in endothelium. J Biol Chem. 1999;27:22679–22685. doi: 10.1074/jbc.274.32.22679. [DOI] [PubMed] [Google Scholar]
- Yan SF, Zou YS, Gao Y, Zhai C, Mackman N, Lee SL, Milbrandt J, Pinsky D, Kisiel W, Stern D. Tissue factor transcription driven by Egr-1 is a critical mechanism of murine pulmonary fibrin deposition in hypoxia. Proc Natl Acad Sci USA. 1998;95:8298–8303. doi: 10.1073/pnas.95.14.8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yao J, Mercola D, Adamson E. The transcription factor EGR-1 directly transactivates the fibronectin gene and enhances attachment of human glioblastoma cell line U251. J Biol Chem. 2000;275:20315–20323. doi: 10.1074/jbc.M909046199. [DOI] [PubMed] [Google Scholar]
- Sharp FR, Ran R, Lu A, Tang Y, Strauss KI, Glass T, Ardizzone T, Bernaudin M. Hypoxic preconditioning protects against ischemic brain injury. NeuroRx. 2004;1:26–35. doi: 10.1602/neurorx.1.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Hyman MC, Lawrence DA, Pinsky DJ. Molecular regulation of the PAI-1 gene by hypoxia: contributions of Egr-1, HIF-1alpha, and C/EBPalpha. FASEB J. 2007;21:935–949. doi: 10.1096/fj.06-6285com. [DOI] [PubMed] [Google Scholar]
- Lee SL, Sadovsky Y, Swirnoff AH, Polish JA, Goda P, Gavrilina G, Milbrandt J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1). Science. 1996;273:1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- Abdulkadir SA, Qu Z, Garabedian E, Song SK, Peters TJ, Svaren J, Carbone JM, Naughton CK, Catalona WJ, Ackerman JJ, Gordon JI, Humphrey PA, Milbrandt J. Impaired prostate tumorigenesis in Egr1-deficient mice. Nat Med. 2001;7:101–107. doi: 10.1038/83231. [DOI] [PubMed] [Google Scholar]
- Harja E, Bucciarelli LG, Lu Y, Stern DM, Zou YS, Schmidt AM, Yan SF. Early growth response-1 promotes atherogenesis: mice deficient in early growth response-1 and apolipoprotein E display decreased atherosclerosis and vascular inflammation. Circ Res. 2004;94:333–339. doi: 10.1161/01.RES.0000112405.61577.95. [DOI] [PubMed] [Google Scholar]
- James AB, Conway AM, Morris BJ. Genomic profiling of the neuronal target genes of the plasticity-related transcription factor—Zif268. J Neurochem. 2005;95:796–810. doi: 10.1111/j.1471-4159.2005.03400.x. [DOI] [PubMed] [Google Scholar]
- Lu A, Tang Y, Ran R, Clark JF, Aronow BJ, Sharp FR. Genomics of the periinfarction cortex after focal cerebral ischemia. J Cereb Blood Flow Metab. 2003;23:786–810. doi: 10.1097/01.WCB.0000062340.80057.06. [DOI] [PubMed] [Google Scholar]
- Welsh FA, Sakamoto T, McKee AE, Sims RE. Effect of lactacidosis on pyridine nucleotide stability during ischemia in mouse brain. J Neurochem. 1987;49:846–851. doi: 10.1111/j.1471-4159.1987.tb00971.x. [DOI] [PubMed] [Google Scholar]
- Beck H, Voswinckel R, Wagner S, Ziegelhoeffer T, Heil M, Helisch A, Schaper W, Acker T, Hatzopoulos AK, Plate KH. Participation of bone marrow-derived cells in long-term repair processes after experimental stroke. J Cereb Blood Flow Metab. 2003;23:709–717. doi: 10.1097/01.WCB.0000065940.18332.8D. [DOI] [PubMed] [Google Scholar]
- Culmsee C, Stumm RK, Schafer MK, Weihe E, Krieglstein J. Clenbuterol induces growth factor mRNA, activates astrocytes, and protects rat brain tissue against ischemic damage. Eur J Pharmacol. 1999;379:33–45. doi: 10.1016/s0014-2999(99)00452-5. [DOI] [PubMed] [Google Scholar]
- Klausmeyer A, Garwood J, Faissner A. Differential expression of phosphacan/RPTPbeta isoforms in the developing mouse visual system. J Comp Neurol. 2007;504:659–679. doi: 10.1002/cne.21479. [DOI] [PubMed] [Google Scholar]
- Wei J, Blum S, Unger M, Jarmy G, Lamparter M, Geishauser A, Vlastos GA, Chan G, Fischer KD, Rattat D, Debatin KM, Hatzopoulos AK, Beltinger C. Embryonic endothelial progenitor cells armed with a suicide gene target hypoxic lung metastases after intravenous delivery. Cancer Cell. 2004;5:477–488. doi: 10.1016/s1535-6108(04)00116-3. [DOI] [PubMed] [Google Scholar]
- Gallagher PG, Liem RI, Wong E, Weiss MJ, Bodine DM. GATA-1 and Oct-1 are required for expression of the human alpha-hemoglobin-stabilizing protein gene. J Biol Chem. 2005;280:39016–39023. doi: 10.1074/jbc.M506062200. [DOI] [PubMed] [Google Scholar]
- Cron RQ, Bandyopadhyay R, Genin A, Brunner M, Kersh GJ, Yin J, Finkel TH, Crow MK. Early growth response-1 is required for CD154 transcription. J Immunol. 2006;176:811–818. doi: 10.4049/jimmunol.176.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung WM, Wang CK, Kuo JS, Lin TN. Changes in the level of glial fibrillary acidic protein (GFAP) after mild and severe focal cerebral ischemia. Chin J Physiol. 1999;42:227–235. [PubMed] [Google Scholar]
- Carmichael ST, Archibeque I, Luke L, Nolan T, Momiy J, Li S. Growth-associated gene expression after stroke: evidence for a growth-promoting region in peri-infarct cortex. Exp Neurol. 2005;193:291–311. doi: 10.1016/j.expneurol.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Janardhan V, Qureshi AI. Mechanisms of ischemic brain injury. Curr Cardiol Rep. 2004;6:117–123. doi: 10.1007/s11886-004-0009-8. [DOI] [PubMed] [Google Scholar]
- Fu M, Zhu X, Zhang J, Liang J, Lin Y, Zhao L, Ehrengruber MU, Chen YE. Egr-1 target genes in human endothelial cells identified by microarray analysis. Gene. 2003;315:33–41. doi: 10.1016/s0378-1119(03)00730-3. [DOI] [PubMed] [Google Scholar]
- Galtrey CM, Fawcett JW. The role of chondroitin sulfate proteoglycans in regeneration and plasticity in the central nervous system. Brain Res Rev. 2007;54:1–18. doi: 10.1016/j.brainresrev.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Bundesen LQ, Scheel TA, Bregman BS, Kromer LF. Ephrin-B2 and EphB2 regulation of astrocyte-meningeal fibroblast interactions in response to spinal cord lesions in adult rats. J Neurosci. 2003;23:7789–7800. doi: 10.1523/JNEUROSCI.23-21-07789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SJ, Ning H, Ishida W, Sodin-Semrl S, Takagawa S, Mori Y, Varga J. The early-immediate gene EGR-1 is induced by transforming growth factor-beta and mediates stimulation of collagen gene expression. J Biol Chem. 2006;281:21183–21197. doi: 10.1074/jbc.M603270200. [DOI] [PubMed] [Google Scholar]
- Moon LD, Fawcett JW. Reduction in CNS scar formation without concomitant increase in axon regeneration following treatment of adult rat brain with a combination of antibodies to TGFbeta1 and beta2. Eur J Neurosci. 2001;14:1667–1677. doi: 10.1046/j.0953-816x.2001.01795.x. [DOI] [PubMed] [Google Scholar]
- Miyata S, Akagi A, Hayashi N, Watanabe K, Oohira A. Activity-dependent regulation of a chondroitin sulfate proteoglycan 6B4 phosphacan/RPTPbeta in the hypothalamic supraoptic nucleus. Brain Res. 2004;1017:163–171. doi: 10.1016/j.brainres.2004.05.034. [DOI] [PubMed] [Google Scholar]
- Dobbertin A, Rhodes KE, Garwood J, Properzi F, Heck N, Rogers JH, Fawcett JW, Faissner A. Regulation of RPTPbeta/phosphacan expression and glycosaminoglycan epitopes in injured brain and cytokine-treated glia. Mol Cell Neurosci. 2003;24:951–971. doi: 10.1016/s1044-7431(03)00257-4. [DOI] [PubMed] [Google Scholar]
- Gris P, Tighe A, Levin D, Sharma R, Brown A. Transcriptional regulation of scar gene expression in primary astrocytes. Glia. 2007;55:1145–1155. doi: 10.1002/glia.20537. [DOI] [PubMed] [Google Scholar]
- Rauch U, Zhou XH, Roos G. Extracellular matrix alterations in brains lacking four of its components. Biochem Biophys Res Commun. 2005;328:608–617. doi: 10.1016/j.bbrc.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Jones LL, Sajed D, Tuszynski MH. Axonal regeneration through regions of chondroitin sulfate proteoglycan deposition after spinal cord injury: a balance of permissiveness and inhibition. J Neurosci. 2003;23:9276–9288. doi: 10.1523/JNEUROSCI.23-28-09276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge JS, Silver J. Inhibition of neurite outgrowth on astroglial scars in vitro. J Neurosci. 1990;10:3594–3603. doi: 10.1523/JNEUROSCI.10-11-03594.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner JR, Herrmann JE, Woo MJ, Tansey KE, Doan NB, Sofroniew MV. Reactive astrocytes protect tissue and preserve function after spinal cord injury. J Neurosci. 2004;24:2143–2155. doi: 10.1523/JNEUROSCI.3547-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lundkvist A, Andersson D, Wilhelmsson U, Nagai N, Pardo AC, Nodin C, Ståhlberg A, Aprico K, Larsson K, Yabe T, Moons L, Fotheringham A, Davies I, Carmeliet P, Schwartz JP, Pekna M, Kubista M, Blomstrand F, Maragakis N, Nilsson M, Pekny M. Protective role of reactive astrocytes in brain ischemia. J Cereb Blood Flow Metab. 2008;28:468–481. doi: 10.1038/sj.jcbfm.9600546. [DOI] [PubMed] [Google Scholar]